Abstract
As well-known antibiotic-producing and filamentous bacteria, streptomycetes can be an ideal model to study the effects of microgravity on microbial development and antibiotic production. In this study, the model organism Streptomyces coelicolor A3(2) was exposed to simulated microgravity (SMG) on a rotating clinostat and microgravity (μg) on the Shenzhou-8 spacecraft. The strain exhibited some similar responses under both conditions. Compared with the controls, its life cycle in agar medium was shortened relatively, and the sporulation process was accelerated with higher accumulation of the gray spore pigment; the liquid cultures yielded more cell biomass, coupled with thicker, more fragmented, and well-dispersed hyphae of the μg spaceflight samples. Global transcriptional analysis verified that most of the differentially expressed genes involved in morphological differentiation of S. coelicolor were upregulated during days 4–6 under SMG conditions, notably the whi genes (whiD, sigF, and whiE). Production of actinorhodin (ACT) in agar cultures decreased under both conditions while undecylprodigiosin (RED) was produced earlier, which were consistent with the transcriptional levels of act and red gene clusters. Meanwhile, expression of the gene clusters for calcium-dependent antibiotic (CDA), methylenomycin (MMY), and a cryptic polyketide (CPK) was unchanged, downregulated, and upregulated, respectively, the latter of which might contribute to the enhanced activity of S. coelicolor against Bacillus subtilis under microgravity. Our study provides new insights into the morphological and secondary metabolic responses of streptomycetes to microgravity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Microbes have evolved for some 3.8 billion years and can survive in many extreme environments including space, but how they respond and adapt themselves to these environments, especially microgravity in space, remains unclear. Spaceflight and ground-simulated microgravity (SMG) experiments have suggested that microgravity can affect cellular processes and functions in microorganisms, such as cell growth (Kacena et al. 1999; Lawal et al. 2013) and differentiation (Mennigmann and Lange 1986; Van Mulders et al. 2011), biofilm formation (Crabbé et al. 2008; Lynch et al. 2006), gene expression (Crabbé et al. 2011; Wilson et al. 2007; Wilson et al. 2002b), secondary metabolism (Demain and Fang 2001; Lam et al. 1998), cell virulence (Crabbé et al. 2010; Lawal et al. 2010), and cell resistance (Lynch et al. 2004; Wilson et al. 2002a). Although the effects of microgravity on microbial growth and metabolism have been studied for more than 50 years, systematic and comprehensive studies of the genetic and phenotypic responses of microorganisms to microgravity environment in space are still insufficient due to technological and logistical hurdles. Data of microbial growth and metabolism in space were often based on small samples and lack of proper control groups (Herranz et al. 2013), thus leading to compromised experimental results. Meanwhile, the selected strains, cultivation patterns, and methods of microgravity varied according to purposes of research, and conflicting results were often reported in different experiments (Benoit and Klaus 2007). Previous spaceflight experiments have shown that suspension cultures of Escherichia coli and Bacillus subtilis exhibited increased cell growth in the spaceflight environment (Kacena et al. 1999; Klaus et al. 1997; Mennigmann and Lange 1986). However, some other studies showed that the E. coli and B. subtilis grown on solid agar on Space Shuttle Mission STS-63 did not experience an increased final cell mass, and changes in other growth characteristics might occur when grown under various gravitational conditions (Kacena and Todd 1997; Kacena et al. 1997).
At present, long-term real microgravity can be achieved only in spaceflight. Recently, taking advantage of the cooperation between China and Germany in the scientific utilization of China Manned Space Program, 17 biological experiments were accomplished in the German SIMBOX (Science in Microgravity Box) onboard the Chinese spacecraft Shenzhou-8 (Preu and Braun 2014), and a number of new findings have been obtained (Nasir et al. 2014; Paulsen et al. 2014; Pietsch et al. 2013). Shenzhou-8 was launched atop a Long March-2 F carrier rocket at the Jiuquan Satellite Launch Center (Jiuquan, China) on November 1, 2011 and landed after a 16.5-day mission on the main landing site at the Gebi desert in Inner Mongolia. Because of the rarity and costliness of microgravity experiments in space, several ground-based facilities (GBFs) with different physical concepts have been developed to simulate microgravity effects on the ground (Herranz et al. 2013). Clinostats, for instance, can generate along with rotation a continuous change of the direction of the gravity vector on the sample, resulting in the gravity vector being time-averaged to near zero (Klaus 2001). The two-dimensional (2D) clinostat, a classical and well-established paradigm, has been widely used till today to study the effects of microgravity on biological samples. Moreover, several studies have shown that results from various model systems using 2D clinorotation are similar to those found under real microgravity conditions (Brungs et al. 2011; Eiermann et al. 2013; Hemmersbach et al. 2006; Thiel et al. 2012).
Streptomycetes are high genome G + C, gram-positive, filamentous bacteria that produce a variety of bioactive natural products and have a complex life cycle, undergoing development and differentiation from spores to substrate hyphae, aerial hyphae, spore chains, and mature spores (Fig. 1) (Bentley et al. 2002; Flardh and Buttner 2009). Meanwhile, morphological differentiation and secondary metabolism of streptomycetes are generally sensitive to extracellular environmental signals and stresses, including nutrients, heat, osmotic pressure, and so on (Bibb 2005; Viollier et al. 2003). Therefore, streptomycetes can be an ideal model to study the microbial responses in morphology and secondary metabolism to microgravity environment. However, effects of microgravity on morphological development and differentiation of streptomycetes have been little studied and described. Regarding the secondary metabolism, previous studies demonstrated that the production of β-lactam antibiotics by Streptomyces clavuligerus and rapamycin by Streptomyces hygroscopicus was inhibited under SMG using NASA rotating-wall bioreactors (RWBs) (Fang et al. 1997; Fang et al. 2000), and the productivity of actinomycin D by Streptomyces plicatus was increased during the US Space Shuttle Mission STS-80 (Lam et al. 2002), corroborated by later findings for the early sample points (days 8 and 12) aboard the International Space Station (Benoit et al. 2006). Collectively, these studies suggest that microgravity environment could alter secondary metabolism in streptomycetes. Nevertheless, molecular evidence for the metabolic phenotypes has never been reported.
The model strain Streptomyces coelicolor A3(2) is a representative soil-dwelling, antibiotic-producing streptomycete, with its genome containing more than 20 secondary metabolite clusters and 965 genes encoding proteins predicted to have a regulatory role (Bentley et al. 2002). In this study, we investigated the growth, morphological and secondary metabolic responses of S. coelicolor A3(2) exposed to ground SMG by clinostat rotation and real microgravity environment by Shenzhou-8 space mission, respectively. We observed interesting similarities in acceleration of morphological differentiation, increase of bacteriostatic activity, and variations of secondary metabolite production in both SMG and space microgravity environments. The phenotypic changes were further correlated to the genetic responses at transcriptional level by microarray gene expression profiling and qRT-PCR analysis.
Materials and methods
Strains and growth conditions
S. coelicolor A3(2) (=DSM 40783 = JCM 4979), S. coelicolor M145 (=ATCC BAA-471), and B. subtilis CGMCC 1.2428 were revived on yeast-starch agar (yeast extract, 2.0 g/L; soluble starch, 10.0 g/L; agar, 15.0 g/L) at 28 °C and maintained as suspensions of spores (streptomycetes, 108 spores/mL) or cells (bacillus, 106 cells/mL) in 20 % (v/v) glycerol at −20 °C. The strains were cultured in experiment unique equipment (EUE, Astrium EADS, Germany; http://www.astrium-na.com/2012AstriumSpaceBiologyProductCatalog.pdf) each with eight cell culture chambers, four on one side for solid culture and four on the other side for liquid culture, sealed with gas permeable biofoil (Astrium EADS, Germany) (Fig. S1; Electronic supplementary material). The streptomycetes were grown as pure cultures and cocultures with B. subtilis, and colonies were grown by spotting 5-μL spore suspensions each spot onto yeast-starch agar (3 mL per chamber of the EUE). For solid coculture, 10-μL cell suspensions of B. subtilis were well mixed into 50-mL yeast-starch agar medium before pouring into the chamber. For liquid culture, 100-μL streptomycete spore suspensions were inoculated into 3-mL yeast-starch liquid medium in the chamber, followed by inoculating 5-μL B. subtilis cell suspensions for coculture. The strains were also cultured on agar plates (3.0 cm of radius and 1 cm of thickness, containing 15-mL yeast-starch agar medium, sealed with parafilm) and in centrifuge tubes (50 mL, containing 30-mL yeast-starch liquid medium) for clinostat experiments.
Clinostat-SMG experiment
A 2D clinostat with a horizontal axis and a radius of 15.0 cm, manufactured by the National Space Science Center, Chinese Academy of Sciences, was used in this study to simulate microgravity effects on the ground (Fig. S2). The EUEs, plates, and centrifuge tubes containing test group samples were fixed to the clinostat and rotated at 10.0 rpm and an average rotation radius of 10.0 cm. This created a residual gravity level of (7.8–14.5) × 10−3 g according to the reference method (van Loon 2007). The control groups were grown under the static 1-g condition, and all other conditions were the same. The temperature for the SMG experiment was 28 °C. The samples were collected every 24 h by scraping the surface mycelium of agar cultures or centrifuging the cells of liquid cultures for further analyses.
Shenzhou-8 spaceflight experiment
The strains were incubated in SIMBOX (Astrium EADS, Germany) during the Shenzhou-8 space mission (μg = 10−3–10−4 g). SIMBOX is an advanced space incubator with 42 separate slots for experimental containers and a 1-g centrifuge to simulate gravity in space (Prasad et al. 2004; Preu and Braun 2014). The microbial samples were inoculated into the EUE about 12 h before the launch of Shenzhou-8, and two EUEs were loaded into the SIMBOX about 10 h before launch, one in a static slot (μg position, μg) and one in a centrifuge slot (simulated 1-g position, S-1 g) (Fig. S3). The centrifuge was started after launch, and the temperature of SIMBOX throughout the mission was maintained at 23 ± 0.5 °C. The Shenzhou-8 spacecraft reentered and landed after 16.5 days of spaceflight, and the microbial samples in SIMBOX were collected 6 h after landing at Payload Integration Test Center (PITC) of General Establishment of Space Science and Application (GESSA), Chinese Academy of Sciences (Beijing, China).
The corresponding ground controls of the experiment were conducted 1 day later at the PITC according to the spaceflight scenario. And, the parallel clinostat-SMG experiment was executed at our laboratory in Beijing following the same procedure except for temperature.
Cultural and morphological characterization
The cultural and morphological properties of streptomycetes were examined both by the eye and microscopically following 2-, 4-, 6-, and 16.5-day clinostat-SMG experiment and recovery of the spaceflight samples. For scanning electron microscopy (SEM), the samples were fixed in 2.5 % glutaraldehyde (in 0.1 M phosphate buffer, pH 7.0), dehydrated in 50, 70, 85, 95, and 100 % methanol successively, subjected to CO2 critical point drying and sputter-coated with gold, and then observed using a QUANTA 200 (30 kV) SEM (FEI). To measure the growth rate of S. coelicolor A3(2), triplicate samples of mycelium were washed twice with distilled water and collected on a preweighed filter by vacuum filtration before weighing (fresh cell weight). The filters with the mycelium were reweighed after freeze-drying at −80 °C for 24 h (dry cell weight).
Bacteriostatic activity assay
Bacteriostatic activity of S. coelicolor A3(2) under simulated and space microgravity conditions was assayed by calculating colony-forming units (CFUs/mL) of the cocultured indicator strain B. subtilis CGMCC 1.2428. For the agar culture, half of the 3-mL agar block was cut uniformly into 32 small pieces, and B. subtilis CFUs in each piece were counted directly under a microscope; for the liquid culture, the surviving B. subtilis CFUs were quantified by serial dilution plating. CFUs of pure cultures of B. subtilis were also calculated to determine the effect of microgravity on the growth of the indicator.
Chemical analysis
The spore pigment (TW95a, a product of the type II PKS gene cluster named whiE) of S. coelicolor A3(2) grown on yeast-starch agar medium was extracted from the aerial spore mass and measured by OD404 according to its characteristic UV absorption spectrum (Yu et al. 1998). A strong correlation (r 2 = 0.9991) was found between the concentration of the spore pigment and OD404 (Fig. S4). Undecylprodigiosin (RED) and actinorhodin (ACT) were extracted from agar cultures and quantified spectrophotometrically as previously described (Gao et al. 2012; Kang et al. 1998).
RNA extraction, labeling, and microarray analysis
The S. coelicolor A3(2) DNA microarrays were custom designed using the Agilent eArray 5.0 program according to the manufacturer’s recommendations (http://earray.chem.agilent.com/earray/). The genechip specification was 8*15 K (Agilent). The genome sequence was downloaded from http://www.ncbi.nlm.nih.gov/genome?Db =genome&Cmd = Search&Term = NC_003888, NC_003903.1, NC_003904.1. Each gene in the chromosome and two plasmids was represented by one 60-nt oligonucleotide probe. Out of the total 8116 genes, 680 genes associated with morphological differentiation and secondary metabolism were replicated nine times each. Total RNA of S. coelicolor A3(2) was extracted from the 4- and 6-day samples of SMG experiment and the 16.5-day samples of spaceflight experiment by using TRIZOL (Invitrogen) and purified using the RNeasy Mini Kit (Qiagen). The quality and quantity of RNA were examined by 1 % agarose gel electrophoresis (Agilent Bioanalyzer 2100) and with a UV spectrophotometer (NanoDrop ND-1000, USA). Two micrograms of RNA was reverse-transcribed into cDNA and then transcribed into cRNA using the Low RNA Input Linear Amplication Kit (Agilent). After purification, 4 μg of cRNA was labeled with Cy3 NHS ester (GE Healthcare) and further purified. The labeled cRNAs were hybridized to the Agilent GeneChip arrays using the Gene Expression Hybridization Kit (Agilent) at 65 °C for 17 h with a rotation at 10 rpm. Arrays were washed twice using the Gene Expression Wash Buffer Kit (Agilent), and then scanned with an Agilent Microarray Scanner System (G2565BA, Agilent) at a resolution of 5 μm, dual pass at 100 and 10 % PMT settings. The two data sets were combined automatically, and signal intensities were normalized using the Feature Extraction Software (Agilent). Data were analyzed using GeneSpring Software 5.0 (Agilent). Microarray analyses were performed on three (SMG experiment) or one (spaceflight experiment) biological replicates. Differentially expressed genes were selected with P < 0.05 and fold change (FC) ≥2.0 in a t test. Pathways were analyzed by the SAS pathway enrichment suite (Shanghai Biotechnology Corporation, Shanghai, China) using the genes with an FC of ≥2.0. Genes with an FC of ≥1.5 but less than 2.0 were analyzed when needed. Microarray data obtained in this study have been deposited in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession numbers GSE53748, GSE53749, and GSE53750.
Real-time qRT-PCR analysis
The qRT-PCR primers used in this study are listed in Table S1 (Electronic supplementary material). First-strand cDNA synthesis was carried out with the Superscript III first-strand Synthesis System (Invitrogen) using 500 ng total RNA following the manufacturer’s instructions. All cDNA synthesis reactions included a replicate reaction without reverse transcriptase to ensure the complete removal of contaminating DNA from the RNA samples. The PCR was done with SYBR®Premix Ex Taq™ (Takara, Dalian, China) in a reaction mixture containing 8.8 μL cDNA, 10 μL SYBR®Premix Ex Taq™ (2×), 0.4 μL ROX Reference Dye (50×), and 0.2 μM of each pair of primers. Thermal cycling conditions were 50 °C for 2 min and 95 °C for 10 min, followed by 40 three-step amplification cycles consisting of denaturation at 95 °C for 10 s, annealing at 60 °C for 30 s, and extension at 72 °C for 30 s. A final dissociation stage was run to generate a melting curve and consequently verify the specificity of the amplification products. PCRs were carried out in triplicate, and each experiment was repeated three times, using an ABI Prism 7000 sequence detection system (Applied Biosystems, CA, USA). The transcriptional levels of target genes were normalized internally to the level of hrdB gene.
Statistical analysis
Data collected in this study were evaluated by the Student’s t test method. Unless otherwise stated, the statistical analysis of Microsoft Excel 2010 for Windows was used. Probability values of P < 0.05 were considered to be significant. A gene’s expression was considered to be significantly altered if the transcript number changed ≥2-fold with P < 0.05 in a Student’s t test.
Results
Effects of SMG and spaceflight on morphological differentiation and growth of S. coelicolor
It was obvious from the cultural characteristics of S. coelicolor A3(2) that the colony color of SMG samples turned to red and then gray earlier than that of the 1-g controls, with higher accumulation of the gray spore pigment on colonies from day 4 (Fig. 2a). The spaceflight samples were examined after landing and recovery. Again, the 16.5-day μg agar cultures of S. coelicolor A3(2) also accumulated relatively more gray spore pigment on colony surfaces than the S-1 g and 1-g controls (Fig. 2b). Similar results were found for the μg spaceflight sample of S. coelicolor M145 as well, which developed even more abundant aerial hyphae than the two controls (Fig. 2b).
Colony features of S. coelicolor cultured on yeast-starch agar under SMG and spaceflight conditions compared to the controls. a Colony features under SMG after 1, 2, 3 (reverse side), 4, 6 and 16.5 days (front side). b Colony features after the 16.5-day spaceflight experiment. SCOA3(2) framed in red indicates S. coelicolor A3(2) cultured purely; SCOA3(2) * framed in blue indicates S. coelicolor A3(2) cocultured with the indicator strain B. subtilis; SCOM145 * framed in yellow indicates S. coelicolor M145 cocultured with B. subtilis. 1g static 1-g conditions on the ground; μg the μg position in a static slot of the SIMBOX on Shenzhou-8; S-1 g the simulated 1-g position in a centrifuge slot of the SIMBOX on Shenzhou-8
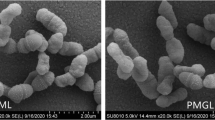
Micrographs revealed that, under SMG conditions, S. coelicolor A3(2) on agar medium produced aerial hyphae earlier, which spiraled and fragmented into spores ahead of the 1-g controls, and the amount of mature spores characteristic of short- and blunt-rod shape was obviously higher than that of the controls after 6-day growth (Fig. 3a). However, the morphological feature of mycelium in SMG liquid cultures was similar to the controls, with mycelium settled to the base of the growth chamber and aggregated into clusters (data not shown). Micrographs of the 16.5-day spaceflight samples also showed more mature spores of agar cultures in the μg group than in the S-1 g and 1-g control groups that still contained some long rod-shaped spores (Fig. 3b). Moreover, mycelium in the μg liquid cultures showed high uniformity and dispersity, while mycelium in the two controls aggregated into large and dense clusters (Fig. 4a); and the hyphae in the μg liquid cultures were thick and fragmented obviously into segments compared to the two controls (Fig. 4b).
Microscopic morphological features of S.coelicolor A3(2) grown on yeast-starch agar under SMG and spaceflight conditions compared to the controls. a SEM micrographs after growth under SMG and 1-g conditions for 2, 4, 6, and 16.5 days. b SEM micrographs after the 16.5-day spaceflight experiment. 1g static 1-g conditions on the ground; μg the μg position in a static slot of the SIMBOX on Shenzhou-8; S-1 g the simulated 1-g position in a centrifuge slot of the SIMBOX on Shenzhou-8
Cultural and morphological features of S. coelicolor A3(2) grown in yeast-starch liquid medium after the 16.5-day spaceflight experiment. a Cultural features in the EUEs. SCOA3(2) framed in red indicates S. coelicolor A3(2) cultured purely; SCOA3(2) * framed in blue indicates S. coelicolor A3(2) cocultured with B. subtilis. FM6-001, FM6-002, and FM6-003 are the serial numbers of EUEs in the mission. b SEM micrographs of S. coelicolor A3(2) mycelium. Arrows indicate fragmentations of the mycelium. 1g static 1-g conditions on the ground; μg the μg position in a static slot of the SIMBOX on Shenzhou-8; S-1 g the simulated 1-g position in a centrifuge slot of the SIMBOX on Shenzhou-8
The effect of microgravity on growth of S. coelicolor was also investigated by measuring the cell weight. As shown in Fig. 5, similar growth curves were observed for the agar cultures of SMG and 1-g control groups, but in liquid cultures, cells under SMG conditions grew more rapidly than the 1-g control cells. The recovered 16.5-day μg spaceflight samples showed in colony diameter no obvious difference from the S-1 g spaceflight and 1-g ground control groups, while the liquid cultures of μg samples accumulated more cell mass (158.73 ± 0.55 mg/chamber, mean ± root mean square error [RMSE], measured by fresh cell weight) than the S-1 g (133.50 ± 0.51) and 1-g (124.80 ± 0.74) controls.
Effects of SMG and spaceflight on bacteriostatic activity and secondary metabolite production
S. coelicolor A3(2) has been reported to produce at least five antibiotics: actinorhodin (ACT), undecylprodigiosins (RED), calcium-dependent ionophore antibiotic (CDA), methylenomycin (MMY), and cryptic polyketide (CPK) (Liu et al. 2013), of which ACT (Wright and Hopwood 1976), CDA (Lakey et al. 1983), CPK (Gottelt et al. 2010), and MMY (Haneishi et al. 1974) exhibited bacteriostatic activity. To determine the overall bacteriostatic activity under SMG and spaceflight, B. subtilis was used as an indicator and cocultured with S. coelicolor A3(2). According to statistical analysis of B. subtilis CFUs, the final number (CFUs/mL, mean ± standard deviation [SD]) of B. subtilis viable cells in agar and liquid samples under SMG conditions were (1.637 ± 0.022) × 103 and (2.907 ± 0.018) × 104, respectively, while those in the 1-g control groups were (3.044 ± 0.028) × 103 and (9.367 ± 0.088) × 104, respectively, which were significantly higher (P < 0.05). On the contrary, the pure culture of B. subtilis exhibited increased cell growth under SMG compared to the 1-g control, particularly in the liquid medium (Fig. S5). The 16.5-day μg spaceflight samples contained B. subtilis CFUs/mL of (8.070 ± 0.230) × 102 and (8.978 ± 0.222) × 103 in agar and liquid chambers, respectively, while the S-1 g controls yielded (1.757 ± 0.018) × 103 and (2.486 ± 0.034) × 104, and the 1-g controls yielded (2.539 ± 0.027) × 103 and (7.022 ± 0.051) × 104, respectively, which were also significantly higher (P < 0.05). These results indicated that S. coelicolor exhibited stronger bacteriostatic activity against B. subtilis under microgravity conditions.
In accordance with the observed morphological characteristics, measuring the gray spore pigment TW95a (a product of the whiE gene cluster) of colonies at OD404 quantitatively revealed that the spore pigment was accumulated faster and more under SMG conditions than under 1 g (Fig. 6a). The blue aromatic polyketide antibiotic ACT and the red oligopyrrole prodiginine antibiotic RED, two well-known secondary metabolites of S. coelicolor, were also measured for the agar cultures. The production of ACT (OD633/mg biomass) was reduced with less final accumulation under SMG conditions (Fig. 6b), while RED (OD530/mg biomass) was produced earlier with nearly the same final accumulation (Fig. 6c). In the spaceflight experiment, both the ACT and RED productions (OD/chamber, mean ± RMSE) in the recovered μg samples (0.316 ± 0.002; 0.505 ± 0.001) were relatively less than those in the S-1 g spaceflight (0.372 ± 0.002; 0.534 ± 0.003) and 1-g ground control (0.453 ± 0.003; 0.591 ± 0.002) groups.
Secondary metabolite analysis of S. coelicolor A3(2) cultured on yeast-starch agar under SMG and 1-g conditions. The productions of secondary metabolites were quantified spectrophotometrically after growing for 1–8 and 16.5 days. a The gray spore pigment TW95a measured by OD404. b ACT measured by OD633. c RED measured by OD530
Transcriptome variation under SMG and spaceflight conditions
Microarray analysis revealed a significant fraction of gene expression variation (2.4–7.0 %, 2.0-fold threshold, P < 0.05) across the transcriptome under SMG and spaceflight conditions, as compared to the 1-g and S-1 g controls. At day 4 under SMG conditions, 196 out of the total 8116 genes were significantly differentially expressed (≥2.0-fold, P < 0.05), with 159 genes upregulated and 37 genes downregulated, which were enriched in 26 out of the total 122 pathways. The differentially expressed genes increased to 552 at day 6 under SMG conditions, with 362 genes upregulated and 190 genes downregulated, enriched in 52 pathways. The16.5-day spaceflight samples showed 271 and 570 genes significantly differentially expressed compared to the control samples of S-1 g and 1 g, respectively, enriched in 48 and 64 pathways, respectively. Nineteen pathways were enriched in all four experimental cases, however, with no more than 50 % of genes in the category (Fig. 7). Besides, it is remarkable that the two pathways responsible for biosynthesis of type II polyketides (KEGG Pathway IDs: sco01056 and sco01057) were both significantly enriched under SMG conditions, with most of the genes (>85 %) differentially expressed at day 6 and 50 % genes for biosynthesis of the backbone altered 2 days earlier (Fig. 7). These results indicated that the biosynthesis of spore pigment TW95a and antibiotic ACT, the only two type II polyketides produced by S. coelicolor, were likely to be altered during cultivation under microgravity conditions, which is in line with the phenotypic results. Based on the whole genome transcriptional data and phenotypic data, we focused on the genes involved in morphological differentiation and secondary metabolism in S. coelicolor A3(2) for further analysis.
Comparative analyses of the enriched pathways based on microarray data. Pathways were analyzed by the SAS pathway enrichment suite (Shanghai Biotechnology Corporation, Shanghai, China) using the genes with an FC of ≥2.0. Nineteen pathways were enriched in all four experimental cases, and two pathways responsible for biosynthesis of type II polyketides were enriched in three cases, significantly under SMG conditions.“*” indicates enrichment test P value <0.05
Effects of SMG and spaceflight on genes involved in morphological differentiation
A series of the differentially expressed involved in morphological differentiation and development of streptomycetes, such as chp, dev, ku-lig, pbp, rdl, sig, ssg, sortase gene, wbl, whi, and so on (Table 1 and Table S2), mainly associated with aerial hyphae erection, sporulation, spore germination, cell wall structure, spore structure, and development-associated secondary sigma factors (Kirby et al. 2011). Among these genes, 31 were significantly upregulated and three downregulated at day 4 in SMG, and these numbers changed to 23 and eight at day 6 in SMG. It is noticeable from Table 1 that the transcription of most whi genes (16 in total) were upregulated at days 4 and 6 in SMG, notably the late-stage whiE cluster of genes (SCO5314-SCO5320, whiEI-whiEVII except for whiEVIII) coding for biosynthesis of the type II polyketide spore pigment, and another two late-stage whi genes (whiD and sigF) that regulate positively the whiE cluster. The six early-to-mid whi genes (whiG, whiA, whiB, whiJ, whiH, whiI) were not significantly altered. Most of the other differentially expressed genes involved in the development of S. coelicolor A3(2) were also upregulated during days 4–6 under SMG conditions, notably those encoding secondary RNA polymerase sigma factors (sigB, sigL, and sigM) (Table S2).
As a further test, we used qRT-PCR to examine the transcription profiles of select genes at days 4 and 6 in SMG, including whiD, sigF, whiE (whiEI, whiEVII, and whiEVIII), whiH, whiI, whiG, ku, and ligD. The qRT-PCR results matched the microarray profiles, effectively validating our microarray data (Fig. S6a and 6b).
In the 16.5-day spaceflight samples, most of the differentially expressed involved in morphological differentiation and development were down-expressed, and a higher number of the genes were differentially regulated as compared to the 1-g control than to the S-1 g control. Genes for the BldK ABC transporter complex (bldKB, bldKC, and bldKD) were differentially expressed (upregulated) only in the case of 16.5-day μg versus 1 g. And, none of the whi genes were differentially expressed in the case of 16.5-day μg versus S-1 g, while whiA and whiG were upregulated and whiEIII and sigF were downregulated in the case of 16.5-day μg versus 1 g. These gene expression data were also confirmed by qRT-PCR results (Fig. S6c and 6d).
Effects of SMG and spaceflight on genes involved in antibiotic biosynthesis
Gene expression FCs from microarray results of the act gene cluster (SCO5071-5092) are shown in Table 2. According to the microarray analysis in the case of SMG versus 1 g, transcription of the whole 22 genes of the ACT biosynthetic gene cluster exhibited a downregulation trend at day 4, with the genes actVI-ORF3 (SCO5074), actII-ORF2 (SCO5083), and actII-ORF3 (SCO5084) downregulated over 2.0-fold, and exhibited obvious downregulation at day 6, with 20 genes downregulated over 2.0-fold and the other two genes (actII-ORF4 and actVB) downregulated over 1.5-fold (Table 2). Interestingly, at day 6, most of the late genes of ACT biosynthesis (actVA and actVI, involving the biosynthesis of ACT end-product) were altered more substantially than the early-to-mid genes (actI, actIII, actIV, and actVII, involving the biosynthesis of ACT backbone). In the 16.5-day spaceflight samples, genes in the central regulatory region (actII-ORF1 to ORF4) of the ACT cluster, except actII-ORF4, still showed a downregulation trend, and a couple of tailoring enzyme genes were upregulated as compared to the controls (Table 2).
In contrast, transcription of the whole 22 genes of the RED biosynthetic gene cluster (SCO5877-5898) exhibited no significant difference at days 4 and 6 in SMG, and only polyketide synthase gene redX (SCO5878) and thioesterase gene redJ (SCO5894) were differentially expressed over 2.0-fold in the 16.5-day spaceflight samples as compared to the controls (Table S3). The acyl carrier protein gene redQ was slightly downregulated (1.5 < FC < 2.0) in the cases of 6-day SMG versus 1 g and 16.5-day μg versus 1 g. The transcription of pathway-specific regulatory genes redD (SCO5877) and redZ (SCO5881) was not significantly altered in every case (Table S3).
The transcription profiles of representative genes actII-ORF2, actII-ORF3, actII-ORF4, redD, redZ, and redQ in each case were further examined by qRT-PCR, and the results matched our microarray profiles (Fig. S6).
Besides ACT and RED, expression of the gene clusters for another three antibotics in S. coelicolor A3(2), CDA, MMY, and CPK, which have antimicrobial activity, were also analyzed, and the results are shown in Tables S4, S5, and S6, respectively. There were almost no significant differences at transcriptional level of the whole cda gene cluster (SCO3210-SCO3249) under either SMG or spaceflight microgravity, and ten out of the 40 genes were downregulated over 2.0-fold in the case of 16.5-day μg versus 1 g, including those involved in two-component system (TCS) (absA1/absA2) and ABC transporter (SCE8.16c and SCE8.17c) (Table S4). Transcription of the mmy gene cluster (SCO1.228c-SCP1.246) showed a decrease in the case of 6-day SMG versus 1 g, in that nine out of the 21 genes were downregulated over 2.0-fold (Table S5), which were mainly involved in biosynthetic process (mmyE, mmyG, mmyO, and mmyT) and regulation (mmyB and mmyR), but showed almost no significant differences in the other three cases. For CPK, seven out of the 20 genes in the cpk cluster (SCO6269-6288) were significantly upregulated at day 4 under SMG, including CPK biosynthetic enzyme genes (cpkD, cpkE, cpkG, cpkI, cpkJ, and cpkK) and CPK transporter gene (cpkF); three genes (cpkJ, cpkK, and cpkL) were upregulated in the case of 16.5-day μg versus 1 g; and no significant differences were found in the cases of 6-day SMG versus 1 g or 16.5-day μg versus S-1 g (Table S6).
Discussion
With the human exploration of space accelerating, the growth, development, and metabolic responses of microorganisms to the extreme environment of space are receiving increasing concerns. As streptomycetes have a complex life cycle and are the richest source of medically useful natural products, notably antibiotics, exploring the response of streptomycetes to microgravity environment not only allows for understanding the growth and development of filamentous microorganisms in the normal Earth gravity, but is also important for utilizing space resources (microgravity and radiation) to produce natural drugs. In this study, we investigated the effects of long-term (16.5 days) spaceflight on S. coelicolor subjected to the Shenzhou-8 space mission. It cannot be ignored that, during spaceflight, some factors other than microgravity could also have effects on microorganisms, such as radiation, vibration generated by rocket, and acceleration during launch and landing of the spacecraft; however, our experiment successfully eliminated the interference factors by using the simulated 1-g control in space (S-1 g). Due to technical limitations, on-orbit fixation was not carried out on microorganisms during the mission; so, we were not able to obtain direct evidence for the short-term effects of spaceflight on S. coelicolor. Nevertheless, before and after the mission, we analyzed the short-term (<8 days) and long-term (16.5 days) alterations in S. coelicolor A3(2) exposed to clinostat-SMG on ground, in consideration of the life cycle period of streptomycetes and in coordination with the spaceflight duration of Shenzhou-8 mission. Our results showed that the effects of spaceflight on S. coelicolor were largely in accordance with the SMG effects and revealed for the first time that microgravity accelerates morphological differentiation process and alters secondary metabolism of streptomycetes.
The cultural characteristics and microscopic morphological observations suggested that the process from aerial hyphae to mature spores (sporulation) of S. coelicolor on agar was accelerated under microgravity conditions. Moreover, the much more fragmented, thicker, and well-dispersed myceliumin liquid suspension cultures at μg position on-orbit was unique among all samples (Fig. 4), indicating that the size and shape of S. coelicolor mycelium in liquid medium were also affected by space microgravity.
The microarray data confirmed that morphological differentiation and development of S. coelicolor were susceptible to microgravity conditions. Spore pigmentation in S. coelicolor A3(2) depends on the production of a polycyclic aromatic polyketide (TW95a) during the maturation of the spores, which is specified by a complex locus whiE, and the whiE transcription is regulated positively by whiD, sigF, whiH, and whiI (Davis and Chater 1990; Kelemen et al. 1998). The whiH and whiI are “early-to-mid” whi genes required for aerial hyphae septum formation at the early stage of sporulation (Aínsa et al. 1999; Ryding et al. 1998), and the whiD, sigF, and whiE are “late” whi genes required for spore maturation and pigmentation at the late stage of sporulation (Kelemen et al. 1998; Molle et al. 2000). In our study, the upregulation of all these five gene loci during days 4–6 of solid cultivation under SMG conditions is consistent with the phenotypes of accelerated morphological differentiation and increased spore pigment accumulation. Moreover, the significant upregulation of whiD, sigF, and whiE on the 4th day in SMG clearly indicates that the development of S. coelicolor A3(2) has been switched into the late stage of sporulation ahead of normal schedule. It is reasonable that no differences were detected in the expression of the other four “early-to-mid” whi genes, whiG, whiJ, whiA, and whiB, which are even earlier than the whiH and whiI (Chater 1998), because S. coelicolor A3(2) had entered into late life cycle stages after 4 days (Fig. 1). A previous study also showed that whiG might be regulated posttranscriptionally and its transcription was constant in the whole development process (Kelemen et al. 1996). The upregulation of most of the other differentially expressed genes involved in the development of S. coelicolor in SMG conditions further supports at the gene transcriptional level that morphological differentiation of S. coelicolor A3(2) was accelerated under SMG. In the 16.5-day samples, the sporulation of S. coelicolor A3(2) had completed; thus, the whi genes showed no difference in expression (16.5-day μg vs S-1 g). The upregulation (whiA and whiG) and downregulation (sigF and whiEIII) of some whi genes in the case of 16.5-day μg versus 1 g, plus the upregulation of “early” genes of bldK, might indicate the initiation of another round of aerial hyphal growth on the old S. coelicolor colonies in space, possibly due to some other space environmental factors, for example, cosmic radiation. By integration of the morphological, chemical, microarray, and qRT-PCR data, we conclude that the complex life cycle of S. coelicolor A3(2) is shorten under both SMG and spaceflight conditions.
Our study indicated that the growth rate of S. coelicolor A3(2) cultured on yeast-starch agar was not influenced by either SMG or spaceflight, but the cell biomass of liquid cultures was increased under both conditions. These results are similar to the previous studies on E. coli and B. subtilis, which also showed that the latter two microorganisms grew faster and yielded more biomass in liquid suspension cultures during spaceflight (Kacena et al. 1999; Klaus et al. 1997; Mennigmann and Lange 1986), but exhibited no visible difference in growth rate in agar or semi-solid cultures (Kacena and Todd 1997; Kacena et al. 1997). It was speculated that the growth rate of strains was related to fluid mechanics and the distribution of liquid medium, rather than to cellular effects induced by microgravity environment (Kacena et al. 1997), so a potential explanation of our observation that S. coelicolor A3(2) liquid cultures grew faster in SMG and spaceflight is a reduced requirement for culture mixing to equalize nutrient levels throughout the culture volume.
The observation of stronger bacteriostatic activity against B. subtilis possibly indicates that S. coelicolor A3(2) could produce more bioactive substances under microgravity conditions. It has been reported that S. coelicolor A3(2) produces four anti-B. subtilis products, ACT (with weak activity) (Wright and Hopwood 1976), CDA (Lakey et al. 1983), MMY (Haneishi et al. 1974), and a yellow-pigmented antibiotic yCPK or its colorless precursor abCPK (Gottelt et al. 2010). Because both chemical and microarray analyses revealed that the biosynthesis of ACT was declined under SMG and spaceflight, and microarray analysis showed either no significant induction or downregulation of the cda and mmy gene clusters under the conditions, it is unlikely that the enhanced bioactivity was derived from ACT, CDA, or MMY. On the other hand, the significant upregulation of some CPK biosynthetic enzyme genes and a transporter gene (cpkF) at day 4 under SMG suggested that the enhanced bioactivity might be contributed by-products of the cpk gene cluster, yCPK, and/or abCPK, which are cryptic type I polyketides that emerge during mid-transition phase of growth and are difficult to detect (Gottelt et al. 2010; Pawlik et al. 2007). Furthermore, as we did not observe any yellow pigment during our experiments, we speculate that the enhanced activity of S. coelicolor A3(2) against B. subtilis under microgravity may be ascribed to the colorless abCPK, while other unknown bioactive compounds produced by S. coelicolor A3(2) cannot be excluded.
Our phenotypic and transcriptomic data indicate that microgravity conditions also alter secondary metabolism of S. coelicolor A3(2); however, the effects vary among different pathways. For example, the production of the only two type II polyketides, spore pigment and ACT, as well as the transcription of their gene clusters responded contrary to both spaceflight and SMG. It has been demonstrated that biochemical “cross-talk” between these two PKS (i.e., whiE-PKS and act-PKS) is normally prevented due to a differential spatial localization of expression of their gene sets (Yu and Hopwood 1995); so, the increase of whiE-PKS could not complement the decrease of its act-PKS homolog but consumed more substrates shared with the latter. In addition, as antibiotic biosynthesis in streptomycetes is also controlled by global regulators, the upregulation of the global regulatory gene sigQ, which works together with the TCS afsQ1/afsQ2, could inhibit the biosynthesis of ACT, RED, and CDA (Shu et al. 2009), while the downregulation of some other negative regulators, such as the TCS response regulator SCO0204 (Wang et al. 2009) under SMG (4-d, FC = −7.46; 6-d, FC = −1.53) and spaceflight microgravity (16.5-d μg vs S-1 g, FC = −1.64), might stimulate the antibiotic biosynthesis. Previous studies of streptomycete secondary metabolism under microgravity also showed that productions of some antibiotics decreased, such as cephalosporin C by S. clavuligerus and rapamycin by S. hygroscopicus in RWBs under SMG (Fang et al. 2000; Fang et al. 1997), while some antibiotics increased, such as actinomycin D by S. plicatus during spaceflight (Benoit et al. 2006; Lam et al. 2002). From this and previous studies, and considering the complex regulatory network of antibiotic production, it is likely that the alteration of microgravity to secondary metabolism of streptomycetes is pathway- and/or case-specific, lacking directed and consistent behavior.
As the clinostat-SMG on ground is not equal to the real microgravity in space and is based on the principle of the minimum response time (MRT) of organisms to the continuous changing direction of the gravity vector, the short-term effects of SMG in our results might not all be the same as the short-term effects of microgravity by spaceflight. Nevertheless, to our knowledge, this is the first report on the responses of morphological differentiation and secondary metabolism of filamentous bacteria, streptomycetes, to microgravity at both the phenotypic and whole transcriptome levels, which could pave the way for future microgravity studies on this kind of promising microorganisms, in well-equipped spacecrafts or the Space Station.
References
Aínsa JA, Parry HD, Chater KF (1999) A response regulator-like protein that functions at an intermediate stage of sporulation in Streptomyces coelicolor A3(2). Mol Microbiol 34:607–619. doi:10.1046/j.1365-2958.1999.01630.x
Benoit MR, Klaus DM (2007) Microgravity, bacteria, and the influence of motility. Adv Space Res 39:1225–1232. doi:10.1089/ast.2010.0536
Benoit MR, Li W, Stodieck LS, Lam KS, Winther CL, Roane TM, Klaus DM (2006) Microbial antibiotic production aboard the International Space Station. Appl Microbiol Biotechnol 70:403–411. doi:10.1007/s00253-005-0098-3
Bentley SD, Chater KF, Cerdeno-Tarraga AM, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D, Bateman A, Brown S, Chandra G, Chen CW, Collins M, Cronin A, Fraser A, Goble A, Hidalgo J, Hornsby T, Howarth S, Huang CH, Kieser T, Larke L, Murphy L, Oliver K, O’Neil S, Rabbinowitsch E, Rajandream MA, Rutherford K, Rutter S, Seeger K, Saunders D, Sharp S, Squares R, Squares S, Taylor K, Warren T, Wietzorrek A, Woodward J, Barrell BG, Parkhill J, Hopwood DA (2002) Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141–147. doi:10.1038/417141a
Bibb MJ (2005) Regulation of secondary metabolism in streptomycetes. Curr Opin Microbiol 8:208–215. doi:10.1016/j.mib.2005.02.016
Brungs S, Hauslage J, Hilbig R, Hemmersbach R, Anken R (2011) Effects of simulated weightlessness on fish otolith growth: clinostat versus rotating-wall vessel. Adv Space Res 48:792–798. doi:10.1016/j.asr.2011.04.014
Chater KF (1998) Taking a genetic scalpel to the Streptomyces colony. Microbiol-SGM 144:1465–1478. doi:10.1099/00221287-144-6-1465
Crabbé A, De Boever P, Van Houdt R, Moors H, Mergeay M, Cornelis P (2008) Use of the rotating wall vessel technology to study the effect of shear stress on growth behaviour of Pseudomonas aeruginosa PAO1. Environ Microbiol 10:2098–2110. doi:10.1111/j.1462-2920.2008.01631.x
Crabbé A, Pycke B, Van Houdt R, Monsieurs P, Nickerson C, Leys N, Cornelis P (2010) Response of Pseudomonas aeruginosa PAO1 to low shear modelled microgravity involves AlgU regulation. Environ Microbiol 12:1545–1564. doi:10.1111/j.1462-2920.2010.02184.x
Crabbé A, Schurr MJ, Monsieurs P, Morici L, Schurr J, Wilson JW, Ott CM, Tsaprailis G, Pierson DL, Stefanyshyn-Piper H, Nickerson CA (2011) Transcriptional and proteomic responses of Pseudomonas aeruginosa PAO1 to spaceflight conditions involve Hfq regulation and reveal a role for oxygen. Appl Environ Microbiol 77:1221–1230. doi:10.1128/Aem. 01582-10
Davis NK, Chater KF (1990) Spore color in Streptomyces coelicolor A3(2) involves the developmentally regulated synthesis of a compound biosynthetically related to polyketide antibiotics. Mol Microbiol 4:1679–1691. doi:10.1111/j.1365-2958.1990.tb00545.x
Demain AL, Fang A (2001) Secondary metabolism in simulated microgravity. Chem Rec 1:333–346. doi:10.1002/Tcr.1018
Eiermann P, Kopp S, Hauslage J, Hemmersbach R, Gerzer R, Ivanova K (2013) Adaptation of a 2-D clinostat for simulated microgravity experiments with adherent cells. Microgravity Sci Technol 25:153–159. doi:10.1007/s12217-013-9341-1
Fang A, Pierson DL, Mishra SK, Koenig DW, Demain AL (1997) Secondary metabolism in simulated microgravity: β-lactam production by Streptomyces clavuligerus. J Ind Microbiol Biotechnol 18:22–25. doi:10.1038/sj.jim.2900345
Fang A, Pierson DL, Mishra SK, Demain AL (2000) Growth of Streptomyces hygroscopicus in rotating-wall bioreactor under simulated microgravity inhibits rapamycin production. Appl Microbiol Biotechnol 54:33–36. doi:10.1007/s002539900303
Flardh K, Buttner MJ (2009) Streptomyces morphogenetics: dissecting differentiation in a filamentous bacterium. Nat Rev Microbiol 7:36–49. doi:10.1038/Nrmicro1968
Gao C, Hindra MD, Yin C, Elliot MA (2012) Crp is a global regulator of antibiotic production in Streptomyces. MBio 3:e00407–e00412. doi:10.1128/mBio. 00407-12
Gottelt M, Kol S, Gomez-Escribano JP, Bibb M, Takano E (2010) Deletion of a regulatory gene within the cpk gene cluster reveals novel antibacterial activity in Streptomyces coelicolor A3(2). Microbiol-SGM 156:2343–2353. doi:10.1099/Mic. 0.038281-0
Haneishi T, Kitahara N, Takiguch Y, Arai M, Sugawara S (1974) New antibiotics, methylenomycins A and B. I. Producing organism, fermentation and isolation, biological-activities and physical and chemical properties. J Antibiot 27:386–392. doi:10.7164/antibiotics.27.386
Hemmersbach R, Strauch SM, Seibt D, Schuber M (2006) Comparative studies on gravisensitive protists on ground (2D and 3D clinostats) and in microgravity. Microgravity Sci Technol 18:257–259. doi:10.1007/Bf02870423
Herranz R, Anken R, Boonstra J, Braun M, Christianen PC, de Geest M, Hauslage J, Hilbig R, Hill RJ, Lebert M, Medina FJ, Vagt N, Ullrich O, van Loon JJ, Hemmersbach R (2013) Ground-based facilities for simulation of microgravity: organism-specific recommendations for their use, and recommended terminology. Astrobiology 13:1–17. doi:10.1089/ast.2012.0876
Kacena MA, Todd P (1997) Growth characteristics of Escherichia coli and Bacillus subtilis cultured on an agar substrate in microgravity. Microgravity Sci Technol 10:58–62
Kacena MA, Leonard PE, Todd P, Luttges MW (1997) Low gravity and inertial effects on the growth of Escherichia coli and Bacillus subtilis in semi-solid media. Aviat Space Environ Med 68:1104–1108
Kacena MA, Merrell GA, Manfredi B, Smith EE, Klaus DM, Todd P (1999) Bacterial growth in space flight: logistic growth curve parameters for Escherichia coli and Bacillus subtilis. Appl Microbiol Biotechnol 51:229–234. doi:10.1007/s002530051386
Kang SG, Jin W, Bibb M, Lee KJ (1998) Actinorhodin and undecylprodigiosin production in wild-type and relA mutant strains of Streptomyces coelicolor A3(2) grown in continuous culture. FEMS Microbiol Lett 168:221–226. doi:10.1016/S0378-1097(98)00446-7
Kelemen GH, Brown GL, Kormanec J, Potuckova L, Chater KF, Buttner MJ (1996) The positions of the sigma-factor genes, whiG and sigF, in the hierarchy controlling the development of spore chains in the aerial hyphae of Streptomyces coelicolor A3(2). Mol Microbiol 21:593–603. doi:10.1111/j.1365-2958.1996.tb02567.x
Kelemen GH, Brian P, Flärdh K, Chamberlin L, Chater KF, Buttner MJ (1998) Developmental regulation of transcription of whiE, a locus specifying the polyketide spore pigment in Streptomyces coelicolor A3(2). J Bacteriol 180:2515–2521
Kirby R, Herron P, Hoskisson P (2011) Analysis of developmental gene conservation in the Actinomycetales using DNA/DNA microarray comparisons. Anton Leeuw Int J G 99:159–177. doi:10.1007/s10482-010-9473-x
Klaus DM (2001) Clinostats and Bioreactors. Gravit Space Biol Bull 14(2):55–64
Klaus D, Simske S, Todd P, Stodieck L (1997) Investigation of space flight effects on Escherichia coli and a proposed model of underlying physical mechanisms. Microbiol-SGM 143:449–455. doi:10.1099/00221287-143-2-449
Lakey JH, Lea EJA, Rudd BAM, Wright HM, Hopwood DA (1983) A new channel-forming antibiotic from Streptomyces coelicolor A3(2) which requires calcium for its activity. J Gen Microbiol 129:3565–3573. doi:10.1099/00221287-129-12-3565
Lam KS, Mamber SW, Pack EJ, Forenza S, Fernandes PB, Klaus DM (1998) The effects of space flight on the production of monorden by Humicola fuscoatra WC5157 in solid-state fermentation. Appl Microbiol Biotechnol 49:579–583. doi:10.1007/s002530051216
Lam KS, Gustavson DR, Pirnik DL, Pack E, Bulanhagui C, Mamber SW, Forenza S, Stodieck LS, Klaus DM (2002) The effect of space flight on the production of actinomycin D by Streptomyces plicatus. J Ind Microbiol Biotechnol 29:299–302. doi:10.1038/sj.jim.7000312
Lawal A, Jejelowo OA, Rosenzweig JA (2010) The effects of low-shear mechanical stress on Yersinia pestis virulence. Astrobiology 10:881–888. doi:10.1089/ast.2010.0493
Lawal A, Kirtley ML, van Lier CJ, Erova TE, Kozlova EV, Sha J, Chopra AK, Rosenzweig JA (2013) The effects of modeled microgravity on growth kinetics, antibiotic susceptibility, cold growth, and the virulence potential of a Yersinia pestis ymoA-deficient mutant and its isogenic parental strain. Astrobiology 13:821–832. doi:10.1089/ast.2013.0968
Liu G, Chater KF, Chandra G, Niu GQ, Tan HR (2013) Molecular regulation of antibiotic biosynthesis in Streptomyces. Microbiol Mol Biol Rev 77:112–143. doi:10.1128/Mmbr. 00054-12
Lynch SV, Brodie EL, Matin A (2004) Role and regulation of σS in general resistance conferred by low-shear simulated microgravity in Escherichia coli. J Bacteriol 186:8207–8212. doi:10.1128/Jb.186.24.8207-8212.2004
Lynch SV, Mukundakrishnan K, Benoit MR, Ayyaswamy PS, Matin A (2006) Escherichia coli biofilms formed under low-shear modeled microgravity in a ground-based system. Appl Environ Microbiol 72:7701–7710. doi:10.1128/Aem. 01294-06
Mennigmann HD, Lange M (1986) Growth and differentiation of Bacillus subtilis under microgravity. Naturwissenschaften 73:415–417. doi:10.1007/Bf00367283
Molle V, Palframan WJ, Findlay KC, Buttner MJ (2000) WhiD and WhiB, homologous proteins required for different stages of sporulation in Streptomyces coelicolor A3(2). J Bacteriol 182:1286–1295. doi:10.1128/Jb.182.5.1286-1295.2000
Nasir A, Strauch SM, Becker I, Sperling A, Schuster M, Richter PR, Weisskopf M, Ntefidou M, Daiker V, An YA, Li XY, Liu YD, Lebert M (2014) The influence of microgravity on Euglena gracilis as studied on Shenzhou 8. Plant Biol 16:113–119. doi:10.1111/plb.12067
Paulsen K, Tauber S, Goelz N, Simmet DM, Engeli S, Birlem M, Dumrese C, Karer A, Hunziker S, Biskup J, Konopasek S, Suh D, Hurlimann E, Signer C, Wang A, Sang C, Grote KH, Zhuang FY, Ullrich O (2014) Severe disruption of the cytoskeleton and immunologically relevant surface molecules in a human macrophageal cell line in microgravity-Results of an in vitro experiment on board of the Shenzhou-8 space mission. Acta Astronaut 94:277–292. doi:10.1016/j.actaastro.2013.06.007
Pawlik K, Kotowska M, Chater KF, Kuczek K, Takano E (2007) A cryptic type I polyketide synthase (cpk) gene cluster in Streptomyces coelicolor A3(2). Arch Microbiol 187:87–99. doi:10.1007/s00203-006-0176-7
Pietsch J, Ma X, Wehland M, Aleshcheva G, Schwarzwalder A, Segerer J, Birlem M, Horn A, Bauer J, Infanger M, Grimm D (2013) Spheroid formation of human thyroid cancer cells in an automated culturing system during the Shenzhou-8 Space mission. Biomaterials 34:7694–7705. doi:10.1016/j.biomaterials.2013.06.054
Prasad G, Jayaram S, Ward J, Gupta P (2004) SimBOX: a scalable architecture for aggregate distributed command and control of spaceport and service constellation. Proc SPIE Enabling Technol Simul Sci 5423(VIII):437–446. doi:10.1117/12.543058
Preu P, Braun M (2014) German SIMBOX on Chinese mission Shenzhou-8: Europe's first bilateral cooperation utilizing China's Shenzhou programme. Acta Astronaut 94:584–591. doi:10.1016/j.actaastro.2013.08.022
Ryding NJ, Kelemen GH, Whatling CA, Flardh K, Buttner MJ, Chater KF (1998) A developmentally regulated gene encoding a repressor-like protein is essential for sporulation in Streptomyces coelicolor A3(2). Mol Microbiol 29:343–357. doi:10.1046/j.1365-2958.1998.00939.x
Shu D, Chen L, Wang WH, Yu ZY, Ren C, Zhang WW, Yang S, Lu YH, Jiang WH (2009) afsQ1-Q2-sigQ is a pleiotropic but conditionally required signal transduction system for both secondary metabolism and morphological development in Streptomyces coelicolor. Appl Microbiol Biotechnol 81:1149–1160. doi:10.1007/s00253-008-1738-1
Thiel CS, Paulsen K, Bradacs G, Lust K, Tauber S, Dumrese C, Hilliger A, Schoppmann K, Biskup J, Golz N, Sang C, Ziegler U, Grote KH, Zipp F, Zhuang FY, Engelmann F, Hemmersbach R, Cogoli A, Ullrich O (2012) Rapid alterations of cell cycle control proteins in human T lymphocytes in microgravity. Cell Commun Signal 10:1–16. doi:10.1186/1478-811x-10-1
van Loon JJWA (2007) Some history and use of the random positioning machine, RPM, in gravity related research. Adv Space Res 39:1161–1165. doi:10.1016/j.asr.2007.02.016
Van Mulders SE, Stassen C, Daenen L, Devreese B, Siewers V, van Eijsden RG, Nielsen J, Delvaux FR, Willaert R (2011) The influence of microgravity on invasive growth in Saccharomyces cerevisiae. Astrobiology 11:45–55. doi:10.1089/ast.2010.0518
Viollier PH, Kelemen GH, Dale GE, Nguyen KT, Buttner MJ, Thompson CJ (2003) Specialized osmotic stress response systems involve multiple SigB-like sigma factors in Streptomyces coelicolor. Mol Microbiol 47:699–714. doi:10.1046/j.1365-2958.2003.03302.x
Wang WH, Shu D, Chen L, Jiang WH, Lu YH (2009) Cross-talk between an orphan response regulator and a noncognate histidine kinase in Streptomyces coelicolor. FEMS Microbiol Lett 294:150–156. doi:10.1111/j.1574-6968.2009.01563.x
Wilson JW, Ott CM, Ramamurthy R, Porwollik S, McClelland M, Pierson DL, Nickerson CA (2002a) Low-shear modeled microgravity alters the Salmonella enterica serovar typhimurium stress response in an RpoS-independent manner. Appl Environ Microbiol 68:5408–5416. doi:10.1128/Aem. 68.11.5408-5416.2002
Wilson JW, Ramamurthy R, Porwollik S, McClelland M, Hammond T, Allen P, Ott CM, Pierson DL, Nickerson CA (2002b) Microarray analysis identifies Salmonella genes belonging to the low-shear modeled microgravity regulon. Proc Natl Acad Sci U S A 99:13807–13812. doi:10.1073/pnas.212387899
Wilson JW, Ott CM, Bentrup KHZ, Ramamurthy R, Quick L, Porwollik S, Cheng P, McClelland M, Tsaprailis G, Radabaugh T, Hunt A, Fernandez D, Richter E, Shah M, Kilcoyne M, Joshi L, Neiman-Gonzalez M, Hing S, Parra M, Dumars P, Norwood K, Bober R, Devich J, Ruggles A, Goulart C, Rupert M, Stodieck L, Stafford P, Catella L, Schurr MJ, Buchanan K, Morici L, McCracken J, Allen P, Baker-Coleman C, Hammond T, Vogel J, Nelson R, Pierson DL, Stefanyshyn-Piper HM, Nickerson CA (2007) Space flight alters bacterial gene expression and virulence and reveals a role for global regulator Hfq. Proc Natl Acad Sci U S A 104:16299–16304. doi:10.1073/pnas.0707155104
Wright LF, Hopwood DA (1976) Actinorhodin is a chromosomally-determined antibiotic in Streptomyces coelicolor A3(2). J Gen Microbiol 96:289–297. doi:10.1099/00221287-96-2-289
Yu TW, Hopwood DA (1995) Ectopic expression of the Streptomyces coelicolor whiE genes for polyketide spore pigment synthesis and their interaction with the act genes for actinorhodin biosynthesis. Microbiol-SGM 141:2779–2791. doi:10.1099/13500872-141-11-2779
Yu TW, Shen YM, McDaniel R, Floss HG, Khosla C, Hopwood DA, Moore BS (1998) Engineered biosynthesis of novel polyketides from Streptomyces spore pigment polyketide synthases. J Am Chem Soc 120:7749–7759. doi:10.1021/Ja9803658
Acknowledgments
This work was supported by the China Manned Space Engineering Program (CMSE, 921-2). We thank Prof. Hua-Rong Tan (Institute of Microbiology, CAS) for kindly providing S. coelicolor A3(2) and M145, and Chunli Li and Jingnan Liang (Institute of Microbiology, CAS) for their help in SEM analyses. We are also grateful to German Aerospace Center’s (DLR) Space Administration, EADS (Astrium), the China Manned Space Engineering Office (CMSEO; now CMSA, China Manned Space Agency) and the General Establishment of Space Science and Application, Chinese Academy of Sciences (GESSA, CAS) for technical and logistical support to the SIMBOX-Shenzhou-8 space mission.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 489 kb)
Rights and permissions
About this article
Cite this article
Huang, B., Liu, N., Rong, X. et al. Effects of simulated microgravity and spaceflight on morphological differentiation and secondary metabolism of Streptomyces coelicolor A3(2). Appl Microbiol Biotechnol 99, 4409–4422 (2015). https://doi.org/10.1007/s00253-015-6386-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-015-6386-7