Abstract
Agaricus subrufescens is one of the most important culinary-medicinal cultivable mushrooms with potentially high-added-value products and extended agronomical valorization. The development of A. subrufescens-related technologies is hampered by, among others, the lack of suitable molecular tools. Thus, this mushroom is considered as a genomic orphan species with a very limited number of available molecular markers or sequences. To fill this gap, this study reports the generation and analysis of the first set of expressed sequence tags (EST) for A. subrufescens. cDNA fragments obtained from young sporophores (SP) and vegetative mycelium in liquid culture (CL) were sequenced using 454 pyrosequencing technology. After assembly process, 4,989 and 5,125 sequences were obtained in SP and CL libraries, respectively. About 87 % of the EST had significant similarity with Agaricus bisporus-predicted proteins, and 79 % correspond to known proteins. Functional categorization according to Gene Ontology could be assigned to 49 % of the sequences. Some gene families potentially involved in bioactive compound biosynthesis could be identified. A total of 232 simple sequence repeats (SSRs) were identified, and a set of 40 EST-SSR polymorphic markers were successfully developed. This EST dataset provides a new resource for gene discovery and molecular marker development. It constitutes a solid basis for further genetic and genomic studies in A. subrufescens.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Mushrooms have always been a part of the human diet since ancient times, due to their gourmet interest but also to their nutraceutical and medicinal properties (Chang and Wasser 2012). Among the cohort of edible mushrooms, the cultivated species belonging to Agaricus genera hold an important place. Agaricus bisporus (Lange) Imbach, the button mushroom, is one of the most widely consumed mushrooms throughout the world. In recent years, an increasing attention has been paid to another Agaricus species, Agaricus subrufescens Peck (syn. A. blazei Murrill sensu Heineman, Agaricus rufotegulis Nauta or Agaricus brasiliensis Wasser, M. Didukh, Amazonas and Stamets), popularly known as the almond mushroom (Largeteau et al. 2011). It is a tropical species that can rarely be found in temperate countries (Zhao et al. 2011), and as leaf litter secondary decomposers, it is cultivated on composts prepared from various agricultural wastes, following the general cultivation processes developed for A. bisporus (Llarena-Hernandez et al. 2013). Due to its particular fragrance and taste, this basidiomycete is appreciated as a gourmet mushroom, but its main interest lies in its potential health benefits and other potential valorizations.
Indeed, A. subrufescens has been reported to produce various bioactive compounds that have potential medicinal applications for the prevention and treatment of cancer, diabetes, hyperlipidemia, arteriosclerosis, chronic hepatitis, and stimulation of the immune system (Firenzuoli et al. 2008; Wisitrassameewong et al. 2012). These compounds are mainly polysaccharides or polysaccharide-enriched fractions corresponding to riboglucans (Cho et al. 1999), β-glucans (Mizuno et al. 1990; Fujimiya et al. 1998; Gonzaga et al. 2005), and glucomannans (Hikichi et al. 1999). Other bioactive molecules providing medicinal benefits in A. subrufescens have been identified such as ergosterol (Takaku et al. 2001), sodium pyroglutamate (Kimura et al. 2004), or lectin (Kawagishi et al. 1990). Aromatic hydrazins, especially agaritin, and its derivatives also abound in A. subrufescens (Nagaoka et al. 2006). The agaritin extracted from A. subrufescens showed an in vitro antitumor activity (Endo et al. 2010; Akiyama et al. 2011). Blazein is a steroid derivative found in A. subrufescens and is supposed to exert an anticancer activity (Itoh et al. 2008). All these examples illustrate the extensive literature available on biomolecules with potential beneficial properties that can be found in A. subrufescens. But surprisingly, whereas numerous metabolic pathways have been highlighted, no study dedicated to identify underlying molecular mechanisms and genes has been reported yet.
Beyond the medicinal issue, other agronomic valorizations of A. subrufescens mushroom have been proposed (for review, see Largeteau et al. 2011). As well as A. bisporus, such a leaf litter decomposer fungus is able to convert lignocellulosic waste materials into highly nutritious food. In this way, it contributes to a more sustainable agriculture, and industrial production of lignocellulolytic enzymes by A. subrufescens appears promising. The culture of this species offers a new market niche and a potential source of diversification for the Agaricus growers. Due to the higher optimal temperature required for its production when compared to the button mushroom, A. subrufescens represents an interesting agricultural option for tropical emerging countries, and it could be a good seasonal alternative in western region during summer (Largeteau et al. 2011).
Despite its importance as a culinary-medicinal cultivable mushroom with potentially high-added-value products and extended outlets, the expansion of A. subrufescens-related biotechonologies is hampered by several bottlenecks, among which a lack of knowledge on its ecology, reproductive biology, biodiversity, and genetics (Largeteau et al. 2011). The almost total absence of adequate molecular tools and genomics resources greatly contributes to impede research in those fields. In order to fill this gap, we have initiated a sequencing approach of A. subrufescens expressed sequence tagged (EST) fragments. The generation of large-scale EST dataset has proved useful in addressing a large array of biological questions, through comparative genomics, functional genomics, gene discovery, and marker development. This approach has been broadly applied in numerous plant, animal, and fungal species (Soanes et al. 2002; Parkinson and Blaxter 2009), but EST-sequencing projects in cultivated and edible mushrooms are scarce (Ospina-Giraldo et al. 2000; Lee et al. 2002; Joh et al. 2007, 2009; Chum et al. 2008; Ramirez et al. 2011). The recent advances in sequencing technologies provide now rapid and cost-effective means to access to transcriptomic and genomic resources of a given species, particularly for nonmodel organisms (Vera et al. 2008; Varshney et al. 2009; Ward et al. 2012).
The present study describes the development and possible applications allowed by the deciphering of the first EST dataset available for A. subrufescens: we first reported in detail the EST assembly and functional annotation processes obtained from 454 pyrosequencing in the light of the genomic data available for the congeneric species A. bisporus (Morin et al. 2012); second, we explored in silico the assembled contig sequence data to identify putative genes involved in bioactive compound production pathways or secondary metabolic engineering; third, we developed and characterized a set of EST-SSR markers. Finally, we assessed the reliability and usefulness of such an approach for further genomic and genetic applications in A. subrufescens studies.
Materials and methods
Fungal tissue source
One A. subrufescens genotype, the hybrid strain CA487-100 × CA454-3 (Llarena-Hernandez et al. 2013), was used in this study. This strain, together with the two homokaryotic parental strains CA487-100 and CA454-3, is maintained in the “Collection du Germplasm des Agarics à Bordeaux” (CGAB), INRA-Bordeaux (Callac et al. 2002) (http://www6.bordeaux-aquitaine.inra.fr/mycsa/Des-ressources-biologiques-uniques/La-collection-d-Agarics-CGAB). In order to obtain the widest range of expressed genes, two contrasted conditions were analyzed. For the sporophore (SP) condition, the strain was grown on commercial compost at INRA facilities as described by Peter-Valence et al. (2011). Samples of tissue (from five mushrooms) were excised from young nonmature SPs (equivalent to stage 3 in A. bisporus according to Hammond and Nichols 1976) and immediately frozen in liquid nitrogen. For the liquid culture (CL) condition (static CL), fungal biomass was produced using 100-mL flasks containing 50 mL of liquid medium (cristomalt 15 g/L, bio-polytone 1.5 g/L) in three replicates. The flasks were incubated at 25 °C for 10 days. The mycelium was recovered by filtration and rinsed with cold sterile water before freezing in liquid nitrogen.
RNA isolation and cDNA preparation
For each condition, total RNA was extracted by using the RNeasy mini kit (Qiagen) according to the manufacturer’s protocol with an additional RNase-free DNase (Qiagen, Hilden, Germany) treatment. RNA quality was checked on agarose gel electrophoresis, and the quantification was assessed using an ND-1000 spectrophotometer (NanoDrop, Wilmington, USA). Five micrograms of total RNA from each condition was sent to GATC Biotech SARL (Mulhouse, France) for library construction, tagging and subsequent high-throughput sequencing according to GS-FLX Titanium chemistry procedures (Roche Applied Science, Penzberg, Germany).
EST sequencing, assembly, and functional annotation
A one-quarter-plate run on a 454 GS-FLX machine (Roche Applied Science, Penzberg, Germany) was dedicated to our samples. Raw sequence reads were cleaned of adapters, primers, and low sequence quality. Then, the trimmed sequences were first clustered with the CD-HIT suite (Li and Godzik 2006) using cd-hit-est algorithm configured with a sequence identity cutoff of 90 %. The assembly into contigs and singletons was performed with Newbler V2.3 software (Margulies et al. 2005) (module gsAssembler–cdna option) with the following parameters: 40-bp min overlapping length; 90 % min overlapping identity; 100-bp all contig threshold, 500-bp large contig threshold.
A. subrufescens sequences were mapped to A. bisporus scaffolds V2.0 (http://genome.jgi-psf.org/Agabi_varbisH97_2/Agabi_varbisH97_2.home.html) using BLAT (Kent 2002) with parameters for EST mapping to genome across species (six translated frames for both contigs and genome). A score of 60 % which corresponds to the product of the first quartile value for alignment coverage (75 %) and identity (80 %) was used as threshold. A. subrufescens contigs/A. bisporus genome scaffold mapping coordinates were then compared to A. bisporus gene coordinates using SMART package (Schultz et al. 1998).
Blastx analyses (Altschul et al. 1990) were performed between A. subrufescens EST sequences and three protein databanks: nonredundant databank, Swissprot, and A.bisporus-predicted proteins (e value < 1e−10). The longest open reading frame (lORF) was inferred using GETORF from EMBOSS package (Rice et al. 2000) and considered as candidate CDS. The corresponding translated protein sequence was annotated using several programs including InterProScan V4.8 (Quevillon et al. 2005) for domain/motif search, RPS-Blast (Altschul et al. 1997) for conserved domain search. Blastp for similarity search (against swissprot, kegg, and pdb databanks (e value cutoff ≤1e-6), Signalp (Petersen et al. 2011), tmhmm (Sonnhammer et al. 1998), and targetP (Emanuelsson et al. 2007) for predicting subcellular localization and targeting. Gene ontology annotations were performed using Blast2GO (Conesa et al. 2005) on Blastx and iprscan results.
All annotation features and analysis results were stored in the Chado or Bio::SeqFeature::Store schema (gmod.org) according to the need (speed access or genericity). GBrowse (Stein et al. 2002) and the Genome Report System (GRS, unpublished) were, respectively, set up to provide graphical and textual interfaces to provide users with comprehensive categories of reports (functional domains, Blast hits) (https://urgi.versailles.inra.fr/gb2/gbrowse/agasynte; https://urgi.versailles.inra.fr/grs/index_agasynte.html).
Genes specifically expressed in one condition or in the other were identified by sequence comparison of the two EST datasets using all-against-all blastn (Altschul et al. 1990) searches (% identity > 90 %; e value <1e-30). CL sequences that do not match against SP sequences were considered as being specific to the CL condition and reciprocally. The result of mapping against A. bisporus genome was also used, and we identified homologs of A. bisporus overlapping with sequences specific to either condition. The resulting subsets of sequences were then compared to transcriptomic data available for A. bisporus in mycelium grown on culture media and fruiting bodies (Morin et al. 2012).
EST-SSR identification, primer design, and marker validation
Microsatellite detection was performed using the Perl script program MISA (http://pgrc.ipk-gatersleben.de/misa/). All perfect SSRs were searched for with a minimum repeats of five for dinucleotide to hexanucleotide. Primer pairs were designed using Primer3 software (Rozen and Skaletsky 2000). Primer pairs were designed to amplify products between 80 and 330 bp in order to facilitate further multiplexing reaction. To avoid some cross-priming, the specificity of each primer pairs was checked by blastn (Altschul et al. 1990) against the sequence datasets.
The EST-SSR primer pairs were tested for their usefulness as potential genetic markers on the 14 genotypes of A. subrufescens previously used for the characterization of genomic SSR (Foulongne-Oriol et al. 2012). PCR amplification, multiplexing optimization, and electrophoresis on ABI3130 sequencer (Applied Biosystems, Life technologies, Carlsbad, CA) were performed as recommended in Foulongne-Oriol et al. (2012).
For each EST-SSR locus, genetic parameters were assessed with Powermarker V3.25 (Liu and Muse 2005).
Results
Generation of ESTs
After trimming and filtering, a total of 97,218 (31.7 Mb) and 81,512 (27.1 Mb) read sequences were generated from SP and CL libraries, respectively. Table 1 summarizes the sequence output data for each library, at each step of the assembly process. The average read lengths (325 and 332 pb) were similar between the two libraries. The assembly resulted in a total of 2,162/2,180 contigs and 17,716/17,683 singletons from SP/CL libraries, respectively. The portion of reads assembled into contigs is about 36 %. Regarding the SP library dataset, the majority of contigs (61.7 %) were derived from less than 5 reads, and 0.2 % of the contigs were composed of more than 50 reads. Most of the contigs were longer than 500 bp (86.9 %) with an average length of 889 bp. Distributions of contig length and number of reads per contig are shown on Fig. 1. For singletons, the average length was 330 bp with 16 % of the sequences showing size equal or longer than 500 bp. Same order of magnitude was observed for the features of the second library (Table 1). For both SP and CL libraries, singleton sequences with length less than 500 bp were excluded for further analyses. This cutoff has been chosen in order to optimize further molecular marker development. Therefore, a set of 4,989 (2,162 contigs and 2,827 singletons) and 5,125 (2,180 contigs and 2,945 singletons) sequences in SP and CL libraries, respectively, were selected for annotation and marker development. The sequence dataset generated in this study has been deposited in the GenBank (Sequence Read Archive accession ID SRP028187, Transcriptome Shotgun Assembly project ID GBEJ00000000).
Functional annotation and classification of ESTs
A. subrufescens sequences mapping against A.bisporus scaffolds showed that 66 and 67.4 % of CL and SP sequences, respectively, were highly similar to A. bisporus sequences and could be mapped on the genome (Table 2). About 97 % of the A. subrufescens-mapped sequences overlapped with A. bisporus gene annotations. The number of A. bisporus genes overlapped by A. subrufescens sequences was estimated between 25.6 and 26.9 % of the 10,438 predicted gene models. An example of alignment of A. subrufescens sequence on A. bisporus scaffolds is illustrated on Supplemental Fig. S1A.
Using the BlastX program, 4,324 (86.7 %) and 4,535 (88.5 %) sequences from the SP and CL libraries, respectively, have significant hits with A. bisporus-predicted proteins (Table 3). Against the nonredundant (nr) protein database, the numbers of sequences per library that found significant hits were slightly lower but still high (80 and 78.7 % for CL and SP sequences, respectively). Polypeptides translated from the lORF were annotated using InterProScan: 2,605 (50.8 %) CL and 2,338 (46.8 %) SP sequences contained at least one domain identified. Conserved domains (CDDs) were found using rpsBlast in 41.3 % of CL and 38 % of CP polypeptides. Classifications according to Gene Ontology performed using Blast2GO (using BlastX results and domain annotations as input) allowed the assignation of GO terms to 2,576 sequences from CL library (50.3 %) and 2,382 sequences from SP library (47.7 %). As expected, the annotation process was less successful for short sequences than for long ones and consequently for singletons than for contigs (Table 3). All the results of the annotation processes are available in supplementary material (Supplemental Table S1) or through the user-friendly web-based tool Agasynte genome browser (Supplemental Fig. S1B). The request system based on either A. bisporus scaffold position or on A. subrufescens sequence id allowed reciprocal searches.
Figure 2 shows the distribution of GO terms among annotated sequences for each category. Molecular function is the most represented category of the GO tree with a total of 2,359 CL sequences and 2,187 SP sequences annotated. This analysis shows that 25 % of sequences annotated have an evidence of hydrolase activity. The other most popular GO assignations concern ion binding, transferase activity, nucleic acid binding, oxyreductase activity, and protein binding (more than 12 % of annotated sequences). The second category is the biological process with 2,014 (CL) and 1,820 (SP) sequences annotated. The two dominant groups are primary metabolic process and cellular metabolic process with an assignation rate higher than 54 % of sequences. Moreover, macromolecule metabolic process (39 %), nitrogen compound metabolic process (36 %), and biosynthetic process (29 %) are also frequently assigned. Finally, cellular component branch assigned GO terms to 1,271 (CL) and 1,237 (SP) sequences and highlighted particularly intracellular term (77 %).
Bar plot representations of GO-annotation results (in percentage) of 2,382 sequences of SP library and 2,576 sequences of CL library. The total numbers of SP and CL library sequences annotated for each category are, respectively, 1,820 and 2,014 for biological process (a), 2,187 and 2,359 for molecular function (b), 1,237 and 1,271 for cellular component (c). Since some sequences have several GO terms, the total percentage of each category exceed 100 %
The all-against-all blastn analyses allowed the identification of 2,284 sequences that are common to both conditions (22.6 % of redundancy). Thus, 2,705 and 2,841 sequences were found to be specific to SP and CL libraries, respectively. Regarding the 5,511 A. bisporus gene model homologs of A. subrufescens sequences, 1,161 and 1,015 were found to be specifically overlapped by CL and SP sequences, respectively (26 % of redundancy) (Supplemental Table S1). The comparison with A. bisporus transcriptomic data showed that 15.6 % of the A. bisporus homologs of CL-specific A. subrufescens sequences were found significantly (p < 0.01) overexpressed in mycelium grown on solid culture media. In the same way, 28.1 % of the A. bisporus homologs of SP-specific A. subrufescens sequences were significantly (p < 0.01) overexpressed in fruiting bodies.
As an example of a potential application of the present EST dataset, some genes related to the bioactive compound biosynthesis or other interesting secondary metabolism pathway were searched for (Supplemental Table S1). Indeed, in our EST collection, among the cohort of genes involved in sucrose metabolic process, we found sequences annotated relating to glucan molecule biosynthesis such as β-(1-3) glucan synthase (cl_GSIH7AY04IIDFT, sp_GSIH7AY04JG4RC), glycosyle transferase (cl_GSIH7AY04IGKLH), or glycosyle hydrolase (sp_isotig00247). Partial sequences of genes potentially involved in aromatic amino acid biosynthesis and nitrogen metabolism were identified: shikimate 5-dehydrogenase (cl_isotig00906), transaminase (sp_isotig01515), or glutamate dehydrogenase (sp_isotig01693). Several sequences were annotated as genes encoding lignocellulolytic enzymes among which laccase (cl_isotig00050), cellulose 1,4-β-cellobiosidase (sp_GSIH7AY04JLYDM), or β-glucosidase (sp_GSIH7AY04IO3UE). Sequences encoding key enzymes in sterol and terpenoid biosynthesis pathways could be also pointed out such as terpene synthase (cl_GSIH7AY04H53WK), C-3 sterol deshydrogenase (sp_isotig00391), farnesyl transferase (sp_isotig00292), or squalene epoxydase (sp_isotig01858).
Identification of EST microsatellite markers
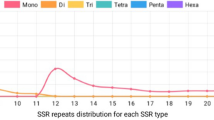
A total of 220 sequences containing 232 microsatellites have been identified from the 10,114 sequences (SP + CL dataset). The frequency of EST-SSR in A. subrufescens EST sequences is 2.3 %, and the distribution is 41 SSR per Mb of expressed sequences. The major types of identified repeats are dinucleotide (54.7 %) and trinucleotide (41.8 %). Microsatellite motifs exhibit an average of 5.4 repeats per locus. Of the 232 SSR loci, 141 (60.8 %) were eligible for primer design and amplified a unique PCR product. A subset of 65 EST-SSR primer pairs randomly chosen was selected for further characterization on 14 A. subrufescens genotypes. Forty-seven (72.3 %) provided scorable and reliable amplified products. Five markers (ESS04, ESS12, ESS15, ESS35, and ESS40) showed a size of the amplified product longer than the expected one, suggesting the presence of introns in the corresponding genomic regions. Forty (61.5 %) showed polymorphism among the 14 genotypes examined. The genetic parameters of these 40 polymorphic EST-SSR markers estimated on the 14 genotypes studies are summarized in Table 4. Altogether, these loci exhibited 159 alleles, ranging from 2 to 13 per locus with an average of 3.98. The polymorphic information content (PIC) ranged from 0.07 to 0.88, with an average of 0.44.
Discussion
A limited amount of sequence data was publicly available for A. subrufescens prior to the initiation of our studies, and this mushroom could be considered as a genomic orphan species. We have recently released a previous set of genomic data generated for the development of microsatellite markers, using the procedure of microsatellite-enriched library pyrosequencing (Foulongne-Oriol et al. 2012). However, the usefulness of this set based on gDNA sequence could be limited for functional characterization of genes, genome annotation, or comparative genomics. Therefore, in order to broaden the range of the genomic tools available for A. subrufescens, we have developed the first set of EST sequences dedicated to this medicinal mushroom using 454 pyrosequencing on samples from young SPs or vegetative mycelium in CL. This technology is a rapid, efficient, and cost-effective method to obtain large-scale DNA sequences, particularly for less-studied species (Ekblom and Galindo 2011). In fungi, it has been recently used to perform metagenomic studies (Buée et al. 2009; Opik et al. 2009; Kubartova et al. 2012), genome sequencing (Diguistini et al. 2009; Tedersoo et al. 2010; Forgetta et al. 2013), and transcriptomic analyses (Sato et al. 2009; Cornman et al. 2012; Wessling et al. 2012). To our knowledge, the present study is the first one describing an effective 454 pyrosequencing approach applied to an edible and cultivated mushroom.
Assembly and annotation
Using one-quarter plate pyrosequencing run for two tagged samples, we obtained about 30 Mb of raw data per sample. The number of generated sequences and their length are in the range of the expected ones for 454 technology (Metzker 2010). A relatively small portion of reads were assembled into contigs (36 %), and singletons were found predominant. Such a profile in high-throughput sequencing data is not unusual and probably due to a variety of causes such as sequencing error, effect of assembly algorithm, or insufficient coverage depth (Pop and Salzberg 2008). However, the assembly process resulted in a large percentage of contigs with length greater than 500 bp and good read coverage. The contig features described here are comparable to those observed in other studies (Zeng et al. 2010; Kaur et al. 2011). The high proportion of sequences, either contigs or singletons, which matched with known proteins using BLAST, illustrated the quality of the sequencing and the subsequent assembly. Half of the sequences were assigned to various gene ontology categories, illustrating the diversity of the transcripts. The remaining sequences without BLAST hits may correspond to additional genes not represented in databases and specific to A. subrufescens.
Preliminary transcriptomic data in A. subrufescens
Our primary objective was to produce a set of high-quality sequences available for gene discovery and coding sequence-based markers in A. subrufescens. Our results proved that the low-depth sequencing strategy used was sufficient and efficient to achieve this goal. Thus, the sequence data produced here represent a substantial new genic resource for the species, but they could not be considered as representative of the transcriptome. The estimation of the number of genes and level of transcript coverage represented in our EST collection is arduous to determine without a reference genome sequence. Assuming a similar number of genes as in the congeneric species A. bisporus (Morin et al. 2012), the sequences annotated in the present study are likely to reflect approximately 75 % of the gene space of A. subrufescens. Additional sequencing effort and dedicated approach would be needed to warrant a comprehensive transcriptome analysis in A. subrufescens (Vera et al. 2008; Wall et al. 2009; Ward et al. 2012).
The low-depth coverage of the EST sequences did not allow a quantitative analysis of differentially expressed genes between the two conditions (Vera et al. 2008). However, the reciprocal blast searches gave us a rough picture of the genes specifically expressed in one condition rather than in the other. In the subset of sequences specific to SP, several transcription factors (TF) were found (data not shown), with for example a homolog of hom1 which has been described as being upregulated in fruiting bodies compared to mycelium in A. bisporus (Morin et al. 2012) and Schizophyllum commune (Ohm et al. 2011). Sequences similar to G protein-coupled receptors, potentially involved in signal transduction network during fruiting body development (Xiang et al. 2014), were also found specifically in SP. In A. bisporus, the transcript that showed the highest level of overexpression in fruiting bodies compare to culture encoded for an hydrophobin (Morin et al. 2012) and one homolog of this gene in A. subrufescens was found uniquely in SP. This preliminary work is encouraging for future analyses of the regulation processes underlying fructification in this cultivable mushroom.
Synteny with A. bisporus
The alignment of the EST A. subrufescens sequences against the A. bisporus genome assembly revealed a strong homology between the compared sequences. Our analyses demonstrated that a large portion of the A. subrufescens sequences (87 %) showed best hits to proteins of A. bisporus. The level of homology between these two species is higher than between A. bisporus and other Agaricomycetes (Morin et al. 2012). The comparison with genes found preferentially expressed in quite comparable conditions in A. bisporus (Morin et al. 2012) suggested that the two species share common biological processes involved in the SP development. These results are promising to further promote comparative genomics and synteny analysis between these two species. They will, among others, facilitate investigation in genome evolution and dynamics (Stukenbrock 2013). Contrary to A. subrufescens (Largeteau et al. 2011), numerous genetic information are available for A. bisporus, such as quantitative trait loci for agronomic traits or useful marker-assisted selection (Savoie et al. 2013). The synteny highlighted by our results offers new opportunities for developing new strategies for breeding in A. subrufescens through the potential transferability between the two species (Duran et al. 2009). In this context, the web-based tool developed in this work will be very helpful. Despite the high level of synteny between the two species, 4 % of the A. subrufescens sequences encoded for lineage-specific genes. A deeper analysis of these genes could be interesting to define a genomic signature peculiar to A. subrufescens.
Putative gene discovery
The availability of EST sequences will allow the functional analysis of genes, especially those involved in metabolic pathways underlying the medicinal properties of A. subrufescens, as it has been successfully done in several plant of pharmacological interest (Zeng et al. 2010; Annadurai et al. 2012; Koo et al. 2013) or in traditional Chinese medicinal fungi (Luo et al. 2010; Xiang et al. 2014). It may help, for example, to decipher the biosynthesis of agaritine. The biochemical origin of this aromatic hydrazine molecule is still controversial. Several hypotheses have been described to explain its biogenesis. Agaritine has been firstly inferred to be a shikimate-derived metabolite (Stussi and Rast 1981). Baumgartner et al. (1998) postulated that agaritine is a fungal product metabolized from exogenic precursors resulting from lignin breakdown and bacterial diazotrophic activity. Several genes encoding enzymes of the shikimate pathway or involved in the nitrogen metabolism potentially related to the biosynthesis of agaritine and other hydrazine derivatives have been found among our sequences. Surprisingly, whereas terpenoids are known to be bioactive compounds of various plant and fungi (Kirby and Keasling 2009; Wawrzyn et al. 2012), the characterization of such molecules and their possible medicinal properties has not been investigated in Agaricus species. The genes identified in the present study as being involved in the terpenoid biosynthetic pathways offer also the opportunity to uncover new pharmaceutical compounds and establish new strategies for metabolic engineering. Other genes involved in secondary metabolic pathway of high biotechnological value were also highlighted as genes coding for lignocellulolytic enzymes (Dashtban et al. 2009). This set of A. subrufescens EST will support further molecular studies such as the cloning of gene of interest or the design of expression studies (Sharma and Sarkar 2013).
Marker development: example of EST-SSR
Another application of the EST sequences is the development of genic molecular markers. With the increasing amount of sequences available in public database together with the advance in next generation sequencing technologies, the development of such markers has become almost usual in various plant species (Varshney et al. 2007) and also in fungi (Feau et al. 2007; Luo et al. 2010). Such markers, based on gene sequence, are advantageous for several reasons (Varshney et al. 2007). First, they are supposed to be more conserved between species than markers based on noncoding sequences. Thus, they could be useful in taxonomy, population genetics, and evolutionary studies. Second, these markers might be linked to genes involved in the phenotypic expression of interesting traits. In this way, they could be highly informative in linkage mapping and association genetic studies to understand the control of agronomic or adaptive traits.
The data mining of the present A. subrufescens EST dataset allowed the identification of 232 SSR. The density of microsatellites found in A. subrufescens EST sequences (41 SSR/Mb) is comparable to that described in A. bisporus coding sequence (51 SSR/Mb) (Foulongne-Oriol et al. 2013) using the same bioinformatics methods. From our sequences, a total of 40 polymorphic EST-SSR markers were successfully developed. Since these new markers were characterized on the same sample than the one previously used for genomic SSR (gSSR) development (Foulongne-Oriol et al. 2012), we can readily compared the usefulness of EST-SSR versus gSSR in A. subrufescens. Thus, regarding genetic parameters, EST-SSRs were less slightly polymorphic (A = 3.9, PIC = 0.43, Ho = 0.27) than gSSRs (A = 4.159, PIC = 0.52, Ho = 0.33). This result is consistent with literature (Varshney et al. 2007) and reflects the fact that the mutation frequency of EST sequences is lower than for genomic sequences (Ellegren 2004; Li et al. 2004). The development of EST-SSR markers was particularly attractive because they are supposed to have a higher transferability rate across species than gSSR markers (Ellis and Burke 2007). Since we have already demonstrated the potential transferability of A. subrufescens gSSR to other Agaricus species (Foulongne-Oriol et al. 2012), we may reasonably expect a higher degree of transferability to related species with the present set of EST-SSR. Experiments to validate such a hypothesis are ongoing. Numerous other loci remain to be tested and could provide additional sources of polymorphic EST-SSR markers. In addition, the DNA resource reported here provides strong genomic basis to further SNP discoveries through forthcoming sequencing projects or targeted gene studies.
To conclude, the development of genomic resources and molecular markers in edible mushroom is a relatively new applied science as illustrated by recent advances in A. bisporus (Savoie et al. 2013) or Pleurotus ostreatus (Ramirez et al. 2011). The present work illustrates well these new trends in mushroom breeding and demonstrates also that they are not devoted to the major cultivable species. The method that we used, based on low-depth sequencing, has been proven to be efficient, rapid, and cost-effective to generate abundant specific sequences in a nonmodel organism. This strategy could be applied to other fungal species. The EST datasets presented here represent a significant contribution to the genomic resources available for the medicinal mushroom A. subrufescens. These ESTs provide solid bases for decoding the metabolic engineering and identifying genes potentially involved in medicinal biochemistry of this species. In addition, the development of molecular markers derived from these sequences, as illustrated by the present set of EST-SSR, will greatly facilitate further genetic analyses and molecular breeding. Such studies are ongoing in our laboratory. These EST collections offer also opportunities to initiate comparative genomic studies in Agaricus species and to a larger extent in basidiomycetes.
References
Akiyama H, Endo M, Matsui T, Katsuda I, Emi N, Kawamoto Y, Koike T, Beppu H (2011) Agaritine from Agaricus blazei Murrill induces apoptosis in the leukemic cell line U937. Biochim Biophys Acta 1810(5):519–525
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215(3):403–410
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST:a new generation of protein database search programs. Nucleic Acids Res 25(17):3389–3402
Annadurai RS, Jayakumar V, Mugasimangalam RC, Katta MA, Anand S, Gopinathan S, Sarma SP, Fernandes SJ, Mullapudi N, Murugesan S, Rao SN (2012) Next generation sequencing and de novo transcriptome analysis of Costus pictus D. Don, a non-model plant with potent anti-diabetic properties. BMC Genomics 13:663
Baumgartner D, Hoesch L, Rast DM (1998) The biogenesis of β-N-(gamma-glutamyl)-4-hydroxymethylphenylhydrazine (agaritine) in Agaricus bisporus. Phytochemistry 49(2):465–474
Buée M, Reich M, Murat C, Morin E, Nilsson RH, Uroz S, Martin F (2009) 454 Pyrosequencing analyses of forest soils reveal an unexpectedly high fungal diversity. New Phytol 184(2):449–456
Callac P, Theochari I, Kerrigan RW (2002) The germplasm of Agaricus bisporus: main results after ten years of collecting in France, in Greece and in North America. Acta Hortic 579:49–55
Chang ST, Wasser SP (2012) The role of culinary-medicinal mushrooms on human welfare with a pyramid model for human health. IntJ Med Mushrooms 14(2):95–134
Cho SM, Park JS, Kim KP, Cha DY, Kim HM, Yoo ID (1999) Chemical features and purification of immunostimulating polysaccharides from the fruiting bodies of Agaricus blazei. Korean J Mycol 27:170–174
Chum WW, Ng KT, Shih RS, Au CH, Kwan HS (2008) Gene expression studies of the dikaryotic mycelium and primordium of Lentinula edodes by serial analysis of gene expression. Mycol Res 112(Pt 8):950–964
Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M (2005) Blast2GO:a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21(18):3674–3676
Cornman RS, Bennett AK, Murray KD, Evans JD, Elsik CG, Aronstein K (2012) Transcriptome analysis of the honey bee fungal pathogen, Ascosphaera apis:implications for host pathogenesis. BMC Genomics 13:285
Dashtban M, Schraft H, Qin W (2009) Fungal bioconversion of lignocellulosic residues; opportunities and perspectives. Int J Biol Sci 5(6):578–595
Diguistini S, Liao NY, Platt D, Robertson G, Seidel M, Chan SK, Docking TR, Birol I, Holt RA, Hirst M, Mardis E, Marra MA, Hamelin RC, Bohlmann J, Breuil C, Jones SJ (2009) De novo genome sequence assembly of a filamentous fungus using Sanger, 454 and Illumina sequence data. Genome Biol 10(9):R94
Duran C, Edwards D, Batley J (2009) Genetic maps and the use of synteny. Methods Mol Biol 513:41–55
Ekblom R, Galindo J (2011) Applications of next generation sequencing in molecular ecology of non-model organisms. Heredity (Edinb) 107(1):1–15
Ellegren H (2004) Microsatellites:simple sequences with complex evolution. Nat Rev Genet 5(6):435–445
Ellis JR, Burke JM (2007) EST-SSRs as a resource for population genetic analyses. Heredity (Edinb) 99(2):125–132
Emanuelsson O, Brunak S, von Heijne G, Nielsen H (2007) Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc 2(4):953–971
Endo M, Beppu H, Akiyama H, Wakamatsu K, Ito S, Kawamoto Y, Shimpo K, Sumiya T, Koike T, Matsui T (2010) Agaritine purified from Agaricus blazei Murrill exerts anti-tumor activity against leukemic cells. Biochim Biophys Acta 1800(7):669–673
Feau N, Bergeron MJ, Joly DL, Roussel F, Hamelin RC (2007) Detection and validation of EST-derived SNPs for poplar leaf rust Melampsora medusae f. sp.deltoidae. Mol Ecol Notes 7(6):1222–1228
Firenzuoli F, Gori L, Lombardo G (2008) The medicinal mushroom Agaricus blazei Murrill:review of literature and pharmaco-toxicological problems. Evid Based Complement Alternat Med 5(1):3–15
Forgetta V, Leveque G, Dias J, Grove D, Lyons RJ, Genik S, Wright C, Singh S, Peterson N, Zianni M, Kieleczawa J, Steen R, Perera A, Bintzler D, Adams S, Hintz W, Jacobi V, Bernier L, Levesque R, Dewar K (2013) Sequencing of the Dutch elm disease fungus genome using the Roche/454 GS-FLX Titanium System in a comparison of multiple genomics core facilities. J Biomol Tech 24(1):39–49
Foulongne-Oriol M, Spataro C, Moinard M, Cabannes D, Callac P, Savoie JM (2012) Development of polymorphic microsatellite markers issued from pyrosequencing technology for the medicinal mushroom Agaricus subrufescens. FEMS Microbiol Lett 334(2):119–126
Foulongne-Oriol M, Murat C, Castanera R, Ramirez L, Sonnenberg AS (2013) Genome-wide survey of repetitive DNA elements in the button mushroom Agaricus bisporus. Fungal Genet Biol 55(1):6–21
Fujimiya Y, Suzuki Y, Oshiman K, Kobori H, Moriguchi K, Nakashima H, Matumoto Y, Takahara S, Ebina T, Katakura R (1998) Selective tumoricidal effect of soluble proteoglucan extracted from the basidiomycete, Agaricus blazei Murill, mediated via natural killer cell activation and apoptosis. Cancer Immunol Immunother 46(3):147–159
Gonzaga MLC, Ricardo NMPS, Heatley F, Soares SD (2005) Isolation and characterization of polysaccharides from Agaricus blazei Murill. Carbohydr Polym 60(1):43–49
Hammond JBW, Nichols R (1976) Carbohydrate metabolism in Agaricus bisporus(Lange) Sing.; changes in soluble carbohydrates during growth of mycelium and sporophore. J Gen Microbiol 93:309–320
Hikichi M, Hiroe E, Okubo S (1999) Protein polysaccharide 0041. European Patent 0939082
Itoh H, Ito H, Hibasami H (2008) Blazein of a new steroid isolated from Agaricus blazei Murrill (himematsutake) induces cell death and morphological change indicative of apoptotic chromatin condensation in human lung cancer LU99 and stomach cancer KATO III cells. Oncol Rep 20(6):1359–1361
Joh JH, Lee SH, Lee JS, Kim KH, Jeong SJ, Youn WH, Kim NK, Son ES, Cho YS, Yoo YB, Lee CS, Kim BG (2007) Isolation of genes expressed during the developmental stages of the oyster mushroom, Pleurotus ostreatus, using expressed sequence tags. FEMS Microbiol Lett 276(1):19–25
Joh JH, Kim KY, Lim JH, Son ES, Park HR, Park YJ, Kong WS, Yoo YB, Lee CS (2009) Comparative analysis of expressed sequence tags from Flammulina velutipes at different developmental stages. J Microbiol Biotechnol 19(8):774–780
Kaur S, Cogan NO, Pembleton LW, Shinozuka M, Savin KW, Materne M, Forster JW (2011) Transcriptome sequencing of lentil based on second-generation technology permits large-scale unigene assembly and SSR marker discovery. BMC Genomics 12:265
Kawagishi H, Kanao T, Inagaki R, Mizuno T, Shimura K, Ito H, Hagiwara T, Nakamura T (1990) Formolysis of a potent antitumor (1→6)-β-d-glucan protein complex from Agaricus blazei fruiting bodies and antitumor-activity of the resulting products. Carbohydr Polym 12(4):393–403
Kent WJ (2002) BLAT–the BLAST-like alignment tool. Genome Res 12(4):656–664
Kimura Y, Kido T, Takaku T, Sumiyoshi M, Baba K (2004) Isolation of an anti-angiogenic substance from Agaricus blazei Murill:its antitumor and antimetastatic actions. Cancer Sci 95(9):758–764
Kirby J, Keasling JD (2009) Biosynthesis of plant isoprenoids:perspectives for microbial engineering. Annu Rev Plant Biol 60:335–355
Koo HJ, McDowell ET, Ma X, Greer KA, Kapteyn J, Xie Z, Descour A, Kim H, Yu Y, Kudrna D, Wing RA, Soderlund CA, Gang DR (2013) Ginger and turmeric expressed sequence tags identify signature genes for rhizome identity and development and the biosynthesis of curcuminoids, gingerols and terpenoids. BMC Plant Biol 13:27
Kubartova A, Ottosson E, Dahlberg A, Stenlid J (2012) Patterns of fungal communities among and within decaying logs, revealed by 454 sequencing. Mol Ecol 21(18):4514–4532
Largeteau M, Llarena-Hernandez RC, Regnault-Roger C, Savoie JM (2011) The medicinal Agaricus mushroom cultivated in Brazil:biology, cultivation and non-medicinal valorisation. Appl Microbiol Biotechnol 92(5):897–907
Lee SH, Kim BG, Kim KJ, Lee JS, Yun DW, Hahn JH, Kim GH, Lee KH, Suh DS, Kwon ST, Lee CS, Yoo YB (2002) Comparative analysis of sequences expressed during the liquid-cultured mycelia and fruit body stages of Pleurotus ostreatus. Fungal Genet Biol 35(2):115–134
Li W, Godzik A (2006) Cd-hit:a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22(13):1658–1659
Li YC, Korol AB, Fahima T, Nevo E (2004) Microsatellites within genes:structure, function, and evolution. Mol Biol Evol 21(6):991–1007
Liu K, Muse SV (2005) PowerMarker:an integrated analysis environment for genetic marker analysis. Bioinformatics 21(9):2128–2129
Llarena-Hernandez RC, Largeteau ML, Farnet AM, Foulongne-Oriol M, Ferrer N, Regnault-Roger C, Savoie JM (2013) Potential of European wild strains of Agaricus subrufescens for productivity and quality on wheat straw based compost. World J Microbiol Biotechnol 29(7):1243–1253
Luo H, Sun C, Song J, Lan J, Li Y, Li X, Chen S (2010) Generation and analysis of expressed sequence tags from a cDNA library of the fruiting body of Ganoderma lucidum. Chin Med 5:9
Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, Dewell SB, Du L, Fierro JM, Gomes XV, Godwin BC, He W, Helgesen S, Ho CH, Irzyk GP, Jando SC, Alenquer ML, Jarvie TP, Jirage KB, Kim JB, Knight JR, Lanza JR, Leamon JH, Lefkowitz SM, Lei M, Li J, Lohman KL, Lu H, Makhijani VB, McDade KE, McKenna MP, Myers EW, Nickerson E, Nobile JR, Plant R, Puc BP, Ronan MT, Roth GT, Sarkis GJ, Simons JF, Simpson JW, Srinivasan M, Tartaro KR, Tomasz A, Vogt KA, Volkmer GA, Wang SH, Wang Y, Weiner MP, Yu P, Begley RF, Rothberg JM (2005) Genome sequencing in microfabricated high-density picolitre reactors. Nature 437:376–380
Metzker ML (2010) Sequencing technologies - the next generation. Nat Rev Genet 11(1):31–46
Mizuno T, Hagiwara T, Nakamura T, Ito H, Shimura K, Sumiya T, Asakura A (1990) Antitumor activity and some properties of water-soluble polysaccharides from “Himematsutake”, the fruiting body of Agaricus blazei murrill. Agric Biol Chem 54:2897–2906
Morin E, Kohler A, Baker AR, Foulongne-Oriol M, Lombard V, Nagy LG, Ohm RA, Patyshakuliyeva A, Brun A, Aerts AL, Bailey AM, Billette C, Coutinho PM, Deakin G, Doddapaneni H, Floudas D, Grimwood J, Hildén K, Kües U, Labutti KM, Lapidus A, Lindquist EA, Lucas SM, Murat C, Riley RW, Salamov AA, Schmutz J, Subramanian V, Wösten HA, Xu J, Eastwood DC, Foster GD, Sonnenberg AS, Cullen D, de Vries RP, Lundell T, Hibbett DS, Henrissat B, Burton KS, Kerrigan RW, Challen MP, Grigoriev IV, Martin F (2012) Genome sequence of the button mushroom Agaricus bisporus reveals mechanisms governing adaptation to a humic-rich ecological niche. Proc Natl Acad Sci U S A 109(43):17501–17506
Nagaoka MH, Nagaoka H, Kondo K, Akiyama H, Maitani T (2006) Measurement of a genotoxic hydrazine, agaritine, and its derivatives by HPLC with fluorescence derivatization in the Agaricus mushroom and its products. Chem Pharm Bull 54(6):922–924
Ohm RA, de Jong JF, de Bekker C, Wösten HAB, Lugones LG (2011) Transcription factor genes of Schizophyllum commune involved in regulation of mushroom formation. Mol Microbiol 81(6):1433–1445
Opik M, Metsis M, Daniell TJ, Zobel M, Moora M (2009) Large-scale parallel 454 sequencing reveals host ecological group specificity of arbuscular mycorrhizal fungi in a boreonemoral forest. New Phytol 184(2):424–437
Ospina-Giraldo MD, Collopy PD, Romaine CP, Royse DJ (2000) Classification of sequences expressed during the primordial and basidiome stages of the cultivated mushroom Agaricus bisporus. Fungal Genet Biol 29(2):81–94
Parkinson J, Blaxter M (2009) Expressed sequence tags:an overview. Methods Mol Biol 533:1–12
Petersen TN, Brunak S, von Heijne G, Nielsen H (2011) SignalP 4.0:discriminating signal peptides from transmembrane regions. Nat Methods 8(10):785–786
Peter-Valence F, Llarena-Hernandez C, Largeteau M, Savoie JM, Ruaudel F, Ziarelli F, Ferre E, Farnet AM (2011) Chemical characterization of the biomass of an edible medicinal mushroom, Agaricus subrufescens, via solid-state 13C NMR. J Agric Food Chem 59(16):8939–8943
Pop M, Salzberg SL (2008) Bioinformatics challenges of new sequencing technology. Trends Genet 24(3):142–149
Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R, Lopez R (2005) InterProScan:proteindomains identifier. Nucleic Acids Res 33(Web Server issue):W116–W120
Ramirez L, Oguiza JA, Perez G, Lavin JL, Omarini A, Santoyo F, Alfaro M, Castanera R, Parenti A, Muguerza E, Pisabarro AG (2011) Genomics and transcriptomics characterization of genes expressed during postharvest at 4 °C by the edible basidiomycete Pleurotus ostreatus. Int Microbiol 14(2):111–120
Rice P, Longden I, Bleasby A (2000) EMBOSS:the European Molecular Biology Open Software Suite. Trends Genet 16(6):276–277
Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132:365–386
Sato S, Feltus FA, Iyer P, Tien M (2009) The first genome-level transcriptome of the wood-degrading fungus Phanerochaete chrysosporium grown on red oak. Curr Genet 55(3):273–286
Savoie JM, Foulongne-Oriol M, Barroso G, Callac P (2013) Genetics and genomics of cultivated mushrooms, application to breeding of Agarics. In: Kempken F (ed) The Mycota, Agricultural Applications, vol 11. Springer, Berlin, pp 3–33
Schultz J, Milpetz F, Bork P, Ponting CP (1998) SMART, a simple modular architecture research tool:Identification of signaling domains. Proc Natl Acad Sci U S A 95:5857–5864
Sharma V, Sarkar IN (2013) Bioinformatics opportunities for identification and study of medicinal plants. Brief Bioinform 14(2):238–250
Soanes DM, Skinner W, Keon J, Hargreaves J, Talbot NJ (2002) Genomics of phytopathogenic fungi and the development of bioinformatic resources. Mol Plant Microbe Interact 15(5):421–427
Sonnhammer EL, von Heijne G, Krogh A (1998) A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol 6:175–182
Stein LD, Mungall C, Shu S, Caudy M, Mangone M, Day A, Nickerson E, Stajich JE, Harris TW, Lewis S (2002) The generic genome browser:a building block for a model organism system database. Genome Res 12(10):1599–1610
Stukenbrock EH (2013) Evolution, selection and isolation:a genomic view of speciation in fungal plant pathogens. New Phytol 199(4):895–907
Stussi H, Rast DM (1981) The biosynthesis and possible function of gamma-glutaminyl-4-hydroxybenzene in Agaricus bisporus. Phytochemistry 20(10):2347–2352
Takaku T, Kimura Y, Okuda H (2001) Isolation of an antitumor compound from Agaricus blazei Murill and its mechanism of action. J Nutr 131(5):1409–1413
Tedersoo L, Nilsson RH, Abarenkov K, Jairus T, Sadam A, Saar I, Bahram M, Bechem E, Chuyong G, Koljalg U (2010) 454 Pyrosequencing and Sanger sequencing of tropical mycorrhizal fungi provide similar results but reveal substantial methodological biases. New Phytol 188(1):291–301
Varshney RK, Mahendar T, Aggarwal RK, Börner A (2007) Genic molecular markers in plants:development and applications. In: Varshney RK, Tuberosa R (eds) Genomics-assisted crop improvment:genomics approaches and platforms, vol 1. Springer, Dordrecht, pp 13–29
Varshney RK, Nayak SN, May GD, Jackson SA (2009) Next-generation sequencing technologies and their implications for crop genetics and breeding. Trends Biotechnol 27(9):522–530
Vera JC, Wheat CW, Fescemyer HW, Frilander MJ, Crawford DL, Hanski I, Marden JH (2008) Rapid transcriptome characterization for a nonmodel organism using 454 pyrosequencing. Mol Ecol 17(7):1636–1647
Wall PK, Leebens-Mack J, Chanderbali AS, Barakat A, Wolcott E, Liang H, Landherr L, Tomsho LP, Hu Y, Carlson JE, Ma H, Schuster SC, Soltis DE, Soltis PS, Altman N, dePamphilis CW (2009) Comparison of next generation sequencing technologies for transcriptome characterization. BMC Genomics 10:347
Ward JA, Ponnala L, Weber CA (2012) Strategies for transcriptome analysis in nonmodel plants. Am J Bot 99(2):267–276
Wawrzyn GT, Bloch SE, Schmidt-Dannert C (2012) Discovery and characterization of terpenoid biosynthetic pathways of fungi. Methods Enzymol 515:83–105
Wessling R, Schmidt SM, Micali CO, Knaust F, Reinhardt R, Neumann U, Loren V, van Themaat E, Panstruga R (2012) Transcriptome analysis of enriched Golovinomyces orontii haustoria by deep 454 pyrosequencing. Fungal Genet Biol 49(6):470–482
Wisitrassameewong K, Karunarathna SC, Thongklang N, Zhao R, Callac P, Moukha S, Férandon C, Chukeatirote E, Hyde K (2012) Agaricus subrufescens: a review. Saudi J Biol Sci 19(2):131–146
Xiang L, Li Y, Zhu Y, Luo H, Li C, Xu X, Sun C, Song J, Shi L, He L, Sun W, Chen S (2014) Transcriptome analysis of the Ophiocordyceps sinensis fruiting body reveals putative genes involved in fruiting body development and cordycepin biosynthesis. Genomics 103(1):154–159
Zeng S, Xiao G, Guo J, Fei Z, Xu Y, Roe BA, Wang Y (2010) Development of a EST dataset and characterization of EST-SSRs in a traditional Chinese medicinal plant, Epimedium sagittatum (Sieb. Et Zucc.) Maxim. BMC Genomics 11:94
Zhao RL, Karunarathna S, Raspé O, Parra LA, Guinberteau J, Moinard M, De Kesel A, Barroso G, Courtecuisse R, Hyde K, Guelly AK, Desjardin DE, Callac P (2011) Major clades in tropical Agaricus. Fungal Divers 51:279–296
Acknowledgments
This work was financially supported by a research project funded by a bilateral cooperation between Mexico (project 115790 CONACYT) and France (ANR-09-BLAN-0391-01). We acknowledge the URGI platform and particularly people involved in the GnpIS information system development and system administration.
Author information
Authors and Affiliations
Corresponding author
Additional information
Joelle Amselem and Jean-Michel Savoie were joint senior authors on this work.
Rights and permissions
About this article
Cite this article
Foulongne-Oriol, M., Lapalu, N., Férandon, C. et al. The first set of expressed sequence tags (EST) from the medicinal mushroom Agaricus subrufescens delivers resource for gene discovery and marker development. Appl Microbiol Biotechnol 98, 7879–7892 (2014). https://doi.org/10.1007/s00253-014-5844-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-014-5844-y