Abstract
The walnut (Juglans regia) has been extensively characterized for expressed sequence tags (EST) sequences and currently 6169492 ESTs are available in National Center for Biotechnology Information. Although this is a valuable resource for marker development, the redundancy in sequences makes the mining out of unique candidates for designing markers cumbersome. Keeping this in view, the present study was undertaken with the aim to remove the data redundancy in walnut ESTs and then to develop simple sequence repeats (SSRs) markers. The EST sequences were assembled into a non-redundant set of 85 contigs and 1584 singletons (total sequences 1699), indicating 16.55% reduction in data redundancy. These 1699 sequences were then used to mine out SSR motifs. 132 EST-SSRs were detected, with dinucleotide repeats being predominant (70.45%), followed by trinucleotide repeats (27.27%) and very less frequent hexanucleotides (2.27%). These markers were validated by designing primer pairs. 15 of these designed primers were tested on a group of 37 walnut genotypes. Out of which 7 markers gave robust amplification, generating polymorphism. These findings indicate the usefulness of EST-SSRs in genome analysis. This study further emphasizes the importance of assembly of the vast amounts of data submitted in public databases. Our results have generated a set of non redundant walnut ESTs which is of prime importance for development of marker systems without any repetition or overlapping.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Walnut (Juglans regia L.) is an important nut belonging to family Juglandaceae. It is commonly known as ‘Akhrot’, in India and almost all parts of which are used in one way or the other. Juglans regia—Persian walnut or English walnut is an indigenous species in Eurasia which is cultivated throughout the temperate regions of world for its high quality wood and edible nuts. Persian walnut is monoecious and heterodichogamous, with 2n chromosome number = 32. The mating system of Persian walnut is predominantly out crossing, as it is wind pollinated, although under particular environmental conditions self-pollination is also possible [1].
Cultivated varieties of walnut generally adapt well to climatic conditions of different production areas. Juglans regia, has an exceptionally wide natural distribution, it occurs from Carpathian Mountains of Eastern Europe, all through Western Asia, the Himalayan regions of Pakistan, India, Nepal, Bhutan and China. Wild trees of walnut are found commonly in mixed deciduous and coniferous forests at altitude ranging from 1550 to 3000 m. The wild walnuts are found in Kaghan valley, Ayubia National Park, Swat (in Pakistan). In some regions walnut trees grow to enormous sizes. The height of the largest tree ranges between 40 and 50 m. Juglans regia is a long lived species and even some are 1000 years old [2]. The nuts of the wild trees are smaller, rounder, and have a much thicker shell. There is an enormous variability in nut traits e.g., nut sizes (small to very large), shape, shell thickness (very thin to very thick), the degree of shell seal, the colour of kernel, and the taste and appearance of kernels.
Persian walnuts are the most common, their nutrient density and profile are significantly different from those of black walnuts. Unlike most nuts that are high in monounsaturated fatty acids, walnut oil is composed largely of polyunsaturated fatty acids, particularly alpha-linolenic acid and linoleic acid. They also contain triglycerides effective in reducing the risk of cardiovascular diseases and are also useful source of lipids. Nutritional value of walnut is gaining importance due to neuro- transmitter molecules, serotonin and melatonin which are used as nutraceuticals. Compared to certain other nuts, such as almonds, peanuts and hazelnuts; walnuts contain the highest spectrum of antioxidants, including free antioxidants and the antioxidants bound to fiber.
Markers are available in vast array for crop genome analysis in literature. Molecular markers detect the differences in DNA of individual plants. Among many types of molecular markers microsatellites, or simple sequence repeats (SSRs), have an edge because of polymorphic loci present in nuclear and organelle DNA, which consists of repeating units of 2–6 base pairs in length. They are hyper-variable, present throughout the genome of eukaryotes. Due to their abundance, codominant inheritance, distribution throughout the genome, multi-allelic variation, high reproducibility and high level of polymorphism, SSRs are considered powerful genetic markers. SSRs are mainly of two types Genomic SSRs and EST SSRs or genic SSRs. These markers often present high levels of inter- and intra-specific polymorphism, particularly when tandem repeats number ranges from 60 to 100. Genic- SSRs are specific regions of genome and are used to amplify the specific microsatellite repeat in a Polymerase Chain Reaction (PCR) reaction. SSR markers also have proven useful in the repository setting [3] to examine potential redundancies and propagation errors within collections [4, 5]. Keeping in view the above, present study was taken up with the objective to develop EST-SSRs for Juglans regia L..
Material and Methods
Source Plant Material and DNA Isolation
Source material was collected from plants of Juglans regia L. from field (Ochghat) of Fruit Science Department of Dr. Y.S. Parmar University of Horticulture and Forestry, Nauni, Solan (H.P.). Young and healthy leaves of thirty-seven genotypes of Juglans regia L. were excised from the plants in the field and transported to the laboratory and stored at − 80 °C till further use. Genomic DNA from the collected leaves of different plant species was isolated following CTAB method of Doyle and Doyle [6] followed by further purification.
Searching of Juglans regia L. EST Sequences from dbEST
Two thousand EST sequences of Juglans regia L. were obtained from NCBI website (www.ncbi.nih.gov) in FASTA format and saved as text file.
Clustering and Assembly of EST Sequences
EGassembler webserver [7] was used to produce a non-redundant dataset from 2000 redundant ESTs obtained from NCBI. The software masked the repetitive elements including small RNA pseudo genes, LINEs, SINEs, LTR elements, vector sequences, organelle and other interspersed repeat. The software automatically screens and cleans for various contaminants in the EST sequences. The sequences were clustered and assembled into contigs and singletons using CAP3 [8] by the server with the criterion of 80% overlap identity between one end of a default read to another end.
SSR Identification for the Assembled ESTs
Potential SSRs were detected in the assembled ESTs using SSR Identification Tool (SSRIT) [9]. The parameters for search of SSRs were maximum motif length of ten base pairs and at least five repeats of SSR motifs. The sequences were put as FASTA format into the software. The number of repeats along with their frequency was recorded from the SSR motifs obtained as output of SSRIT.
Primer Designing
Software PRIMER3 (www.frodo.wimit.edu/primer3) [10] was used for designing of SSR primers from EST-SSRs. The parameters for primer designing were as follows: primer size—20–22 bps, primer Tm—57–60 °C, GC content—40–61% and optimum primer Tm between 57 and 60 °C. The designed primer pairs were synthesized by Eurofins mwg/operon (Eurofins Genomics, Bangalore, India). 15 primer pairs were custom synthesized and were validated for their ability for amplification on a set of 37 walnut genotypes.
BLASTX Analysis
Putative functions of Juglans regia L. EST–SSRs were identified by comparing the EST-SSR sequences with UniProt database (http://www.uniprot.org/) using BLASTX tool. E value of <1E−5 was assumed as a significant criterion of homology.
PCR Amplifications and Gel Electrophoresis
PCR protocol was standardized for carrying out the amplification using EST-SSR primers. A mixture of 20 μl for PCR-SSR analysis was prepared using 10X PCR buffer, 2 mM MgCl2, 1 mM dNTPs, 0.3 μM each primer (forward and reverse), 0.3 U/μl Taq DNA Polymerase, 50 ng template DNA following a thermal profile as: 5 min of initial denaturation at 95 °C followed by 40 cycles of 1 min denaturation at 94 °C, annealing varied with Tm of each primer for 1 min and extension of 2 min at 72 °C, further followed by final extension of 5 min at 72 °C. The amplified DNA was mixed thoroughly with 6X loading dye (0.25% bromophenol blue, 40% sucrose) and then electrophoresed in 2% agarose gel in 1X TAE buffer (40 mM Tris–acetate, 1.0 mM EDTA). The gel was run at constant voltage at the rate of 5 V/cm for about 3 h. Ethidium bromide at rate of 0.5 μg/ml was incorporated in the gel.
Data Analysis
Primers which gave polymorphism with walnut (Juglans regia L.) genotypes were screened out. Genetic diversity, defined as polymorphism information content (PIC) [11], was used to measure allelic diversity at each SSR locus. PIC values were calculated as follows:
where pi was the frequency of the ith allele in the set of genotypes analyzed. Then the percentage of polymorphism were calculated. Binary code i.e. 0 and 1 was used to show the absence and presence of bands, respectively. Jaccard’s similarity coefficient matrix was obtained through NTSYSpc Version 2.02h software. Dendrograms were created for the results obtained and compared for the efficiency of generation of polymorphism by EST-SSRs.
Results and Discussion
Data Mining for dbEST-SSRs by Using EGassembler
Out of the 2000 ESTs, 85 contigs were assembled and 1584 singletons were recorded which showed no overlap with any ESTs. The whole dataset was reduced to 1669 sequences after assembly showing 16.55% of data redundancy as shown in Table 1. Similarly 2000 EST sequences of Prunus persica were obtained from NCBI website (www.ncbi.nih.gov/nucest) by Kaur et al. [12].
Use of SSR Identification Tool (SSRIT)
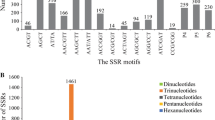
SSRIT search was carried out for contig and singleton data, which resulted in the detection of 139 SSRs, out of which 3 EST-SSRs were reported by contigs and rest 95 were from singletons. Analysis of the detected SSRs revealed that all of them represented di-, tri- and hexanucleotide repeats. It was found that dinucleotide SSR is the dominant repeat type (70.45%) followed by trinucleotide (27.27%) and hexanucleotide were less frequent (2.27%) shown in Table 2. Similarily, the SSRIT was used by Kaur et al. [12] for Prunus persica and in Malus by Vaidya et al. [13] (Tables 2, 3).
Primer Designing
Ninety-eight primers were designed using PRIMER3 software out of which fifteen were custom synthesized and used for further studies enlisted in Table 4. Similarily, primers were designed by Zhang et al. [14] in Juglans regia.
Sequence Identification by Using BLASTX Analysis
For ninety-eight custom synthesized primers, annotation was performed. Based on this analysis, a putative function could be assigned to 98 of the potential EST-SSR markers (95 singletons and 3 contigs) assuming a threshold value of < 1E−5. The annotation results indicated that 98 EST-SSR sequences showed highest homology with Juglans regia and lowest homology with Aphanomyces astaci. Similarly, BLASTX software was used by Zhang et al. [14] for the development of Juglans regia SSR Markers by Data Mining of the EST Database (Table 5).
Marker Validation by PCR Amplification
The 15 primer pairs were used to study genetic polymorphism in a set of 37 walnut genotypes. Total 7 primers were amplified out of which only, 6 being polymorphic and one is monomorphic. The rest 8 primers were not amplified. PIC values were calculated for all the polymorphic primers. The PIC was found to range from 0.23 to 0.57 (Fig. 1). Jaccard’s similarity matrix coefficient was obtained through NTSYSpc. The similarity coefficient values ranged from 0.53 to 1.00. The generated dendrogram revealed a wide genetic base of the walnut germplasms collection studied. The dendrogram generated for EST-SSR markers divided the genotypes into two main clusters A and B (Fig. 2). Group A could be further classified into subgroup A1 and A2. Subgroup A1 was comprised of ‘AKSU#71’, ‘GOBIND’, ‘PHAGLI-UP (SOLI)’, ‘CHICO’, ‘MEYLANNAISE’, ‘INDER AKHROT 1’, ‘PARSIENNE’, ‘NETAR AKHROAT 1’, ‘ZHANG LIN #3’, ‘BLACKMORE’, ‘CHAMBA SELECTION- 60’, ‘XIN ZHENG ZHU’, ‘AKSU#417’, ‘JOGINDER SELECTION-2’, ‘SCHARSCH FRANQUETTE’, ‘ROOPA AKHROT 1’, ‘JAUNAJI SELECTION-12’, ‘CHAMBA SELECTION-20’, ‘JOGINDER NAGAR SELECTION-61’, ‘XIN ZAD FEN’, ‘PLACENTIA’, ‘SHINREI’, ‘HOWARD’, ‘(PHAGLI-UP)-CHEYAMMA’, ‘SOLDING SELECTION’, ‘ROOPA AKHROT 2’, ‘INDER AKHROT 3’, ‘RATAN AKHROT’, ‘NETAR AKHROT 2’, ‘LUXMI AKHROT’, ‘HARTLEY’, ‘DAULAT RAM SELECTION’, ‘RONDE DE MONTIGNAC’, ‘JOGINDER NAGAR SELECTION-39’, and ‘PLANT NO.-46’. Subgroup A2 was found to contain only one genotype, i.e. ‘INDER AKHROT’ and Group B was also found to contain only one genotype, i.e. ‘LAKE ENGLISH’. Similarly, Ahmad [15] reported that the similarity coefficient values ranged from 0.28 to 1.00.
An example of an SSR banding pattern obtained from primer 4 in 37 genotypes of walnut L = DNA ladder 1 kb, 1 = AKSU#71, 2 = MEYLAINNAISE, 3 = ZHANG LIN#3, 4 = XIN ZAD FEN, 5 = PARSIENNE, 6 = PLACENTIA, 7 = SHINREI, 8 = AKSU#417, 9 = CHICO, 10 = CHAMBA SELECTION-60, 11 = DAULAT RAM SELECTION, 12 = SCHARSCH FRANQUETTE, 13 = JAUNAJI SELECTION-12, 14 = HOWARD, 15 = XIN ZHANG ZHU, 16 = JOGINDER NAGAR SELECTION-39, 17 = JOGINDER SELECTION-2, 18 = PLANT NO. 46, 19 = RONDE DE MONTIGNAC, 20 = BLACKMORE, 21 = CHAMBA SELECTION-20, 22 = JOGINDER NAGAR SELECTION-61, 23 = NETAR AKHROT, 24 = (PHAGLI- UP)-CHEYAMMA, 25 = SOLDING SELECTION, 26 = INDER AKHROT 1, 27 = ROOPA AKHROT 1, 28 = HARTLEY, 29 = GOBIND, 30 = INDER AKHROT, 31 = RATTAN AKHROT, 32 = NETAR AKHROT, 33 = INDER AKHROT, 34 = LUXMI AKHROT, 35 = PHAGLI-UP (SOLI), 36 = ROOPA AKHROT 2, 37 = LAKE ENGLISH
Inspite of extensive variation prevalent in the native walnut germplasm, there has been no systemic work on genetic characterization of indigenous and exotic germplasm of walnut in India. Proposed work on genetic characterization of walnut germplasm using EST-SSR markers will facilitate their use as identified genetic stocks in future breeding programmes. Accurate estimation, of distances between different genotypes of the germplasm, can provide useful data to breeders for optimizing sampling strategies in walnut cultivars which can be used in crop improvement programmes. But because of limited number of EST sequences available at NCBI website, the number of primers available is also less which prevents the applicability of this technology. Thus it is the need of the hour to develop more sequences and let them available publicly, so that molecular marker work can be employed at large scale.
Conclusion
The present study attempts to ascertain the frequency and distribution of SSRs in the walnut EST database and develops those EST-SSRs for use in genetic studies. The authors demonstrated the utility of computational approaches for mining SSRs from ever increasing repertoire of publicly available plant EST sequences present in different data-bases. The resulting EST-SSR set is a valuable tool for further genetic and genomic applications. The developed 98 EST-SSR markers have a high rate of PCR amplification and can be used in walnut breeding and genetic studies. The use of these markers would reduce the cost and therefore facilitate cultivar identification, genetic distance assessments, gene mapping and possible marker-assisted selection (MAS). The functional categorization of these markers corresponded to many genes with biological, cellular and molecular functions, thus providing an opportunity to investigate the consequences of SSR polymorphisms on gene function.
References
Pollegioni P, Major A, Bartoli S, Ducci F, Proiett I, Malvolti ME (2008) Application of microsatellite and dominant molecular markers for the discrimination of species and interspecific hybrids in genus Juglans. Acta Hortic 705:191–197
Khan WM, Khan IA, Ahmad H, Ali H, Ghafoor S, Afzal M, Khan FA, Shah M, Afridi SG (2010) Estmation of genetic diversity in walnut. Pak J Bot 42(3):1791–1796
Mitchell SE, Kresovich CA, Jester CA, Hernandez CJ, Szewc MAK (1997) Application of multiplex PCR and fluorescence-based, semi-automated sizing technology for genotyping plant genetic resources. Crop Sci 37:617–624
Phippen WB, Kresovich S, Candelas FG, McFerson JR (1997) Molecular characterization can discriminate and partition variation among gene bank holdings: a case study with phenotypically similar interspecific comparisons. Mol Ecol 5:99–110
Dangl GS, Mendum ML, Prins BH, Walker MA, Meredith CP, Simon CJ (2001) Simple sequence repeat analysis of a clonally propagated species: a tool for managing a grape germplasm collection. Genome 44:432–438
Doyle JJ, Doyle JJ (1987) A rapid DNA isolaton procedure from small quantities of fresh leaf tissues. Phytochem Bull 19:11–15
Masoudi-Nejad A, Koichiro T, Shuichi K, Yuki M, Masanori S, Masumi I (2006) EGassembler: online bioinformatics service for large-scale processing, clustering and assembling ESTs and genomic DNA fragments. Nucleic Acids Res 34:459–462
Huang X, Madan A (1999) CAP3: a DNA sequence assembly program. Genome Res 9:868–877
Temnykh S, Clerk G, Lukashova A, Lipovich L, Cartinhour S, McCouch SR (2001) Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): frequency, length variation, transposon associations, and genetic marker potential. Genome Res 11:1441–1452
Rozen S, Skaletsky HJ (2000) Primer 3 on the www for general users and for biologist programmers. In: Krawetz S, Misener S (eds) Bioinformatics methods and protocols: methods in molecular biology. Humana Press, Totowa, pp 365–386
Anderson JA, Churchill GA, Autrique JE, Tanksley SD, Sorrells ME (1993) Optimizing parental selection for genetic linkage maps. Genome 36:181–186
Kaur R, Shilpa VE, Kumar K (2015) Development, characterization and transferability of peach genic SSRs to some Rosaceae species. Adv Res 3(2):165–180
Vaidya E, Kaur R, Kumar K, Sharma N (2015) Exploitation of Malus ESTs for development of SSR markers after in silico analysis. J Appl Bot Food Qual 88:164–169
Zhang R, Zhu AD, Wang XJ, Yu J, Zhang HR, Gao JS (2010) Development of Juglans regia SSR markers by data mining of the EST database. Plant Mol Biol Rep 28:646–653
Ahmad NS (2013) Genetic analysis of plant morphology in bambara groundnut (Vigna subterranea (L.) Verdc.). Ph.D. Thesis, University of Nottingham
Acknowledgements
The authors thank Dr. Y. S. Parmar University Of Horticulture and Forestry, Nauni, Solan.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interest among them.
Additional information
Significance statement The developed 98 EST-SSR markers have a high rate of PCR amplification and can be used in walnut breeding and genetic studies. The use of these markers would reduce the cost and therefore facilitate cultivar identification, genetic distance assessments, gene mapping and possible marker-assisted selection.
Rights and permissions
About this article
Cite this article
Shailja, S., Kaur, R., Shilpa, C. et al. Exploitation of Walnut (Juglans regia L.) Expressed Sequence Tags for Development of SSR Markers After In Silico Analysis. Proc. Natl. Acad. Sci., India, Sect. B Biol. Sci. 89, 267–274 (2019). https://doi.org/10.1007/s40011-017-0941-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40011-017-0941-1