Abstract
Digital image analysis of high time resolution video microscopy was used to investigate hyphal growth dynamics in different Candida albicans strains. The effects of the quorum sensing molecules tyrosol and farnesol, the deletion of the fungus specific protein phosphatase Z1 CaPPZ1), and the hypha-specific cyclin (HGC1) genes were analyzed by this method. Our system monitored cell growth in a CO2 incubator under near-physiological conditions and measured three major parameters under the following stringent conditions: (a) the time of yeast cell adherence, (b) the time of hyphal outgrowth, and (c) the rate of hyphal growth. This method showed that hyphal extension of wild-type SC5314 cells was accelerated by tyrosol and inhibited by farnesol. Hyphal growth rate was moderately lower in cappz1 and strongly reduced in hgc1 mutants. In addition, tyrosol treatment caused a firm adherence, while farnesol treatment and hgc1 mutation prevented the adherence of yeast cells to the surface of the culture flask. Transition from yeast-to-hyphal state was faster after tyrosol treatment, while it was reduced in farnesol-treated cells as well as in the cappz1 and hgc1 mutants. Our data confirm the notion that the attachment of yeast cells, the yeast-to-hyphal transition, and hyphal growth rate are closely related processes. Time-lapse video microscopy combined with image analysis offers a convenient and reliable method of testing chemicals, including potential drug candidates, and genetic manipulations on the dynamic morphological changes in C. albicans strains.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Beside the characteristic yeast and filamentous forms, different populations of Candida albicans show various types of morphologies. The regulatory mechanism that switches among these forms is considered to be a critical factor in virulence, as hyphae and pseudohyphae are more invasive in terms of tissue penetration during the early stage of infection than the yeast form promoting dissemination in the bloodstream (Sudbery et al. 2004). Signaling pathways that transduce environmental clues into morphological changes have been studied in C. albicans (Whiteway 2000; Liu 2001; Brown 2002). The hyphal form has an important role in causing disease by invading epithelial cells and causing tissue damage (Sudbery 2011). Adherence to mucosal epithelia and concomitant morphological switching are crucial elements of the C. albicans’ invasion (Calderone et al. 1984; Naglik et al. 2011; Sudbery 2011; Jacobsen et al. 2012). C. albicans can also adhere to inert abiotic surfaces such as plastics routinely used in medicine (Kiremitçi–Gümüşderelioğlu and Pesmen 1996; Radford et al. 1999; Ramage et al. 2006; von Fraunhofer and Loewy 2009; Busscher et al. 2010). Adherence and morphological transitions are likely coupled processes (Tronchin et al. 1988, 1989; Radford et al. 1998; Bulad et al. 2004).
Although the cell wall structure and composition change dynamically during the adherence and germination (Tronchin et al. 1989), and the appearances of adhesion molecules are clearly morphology dependent (Gow and Hube 2012; de Groot et al. 2013; Mayer at al. 2013), studies that could monitor these events simultaneously are scarce. Investigations related to the adherence of C. albicans cells in different human epithelia in the late 70s and early 80s (Kimura and Pearsall 1978, 1980; King et al. 1980; Sobel et al. 1981) proved to be more static than dynamic. Environmental conditions affecting cellular morphology have also been monitored (Madhani and Fink 1998; Brown and Gow 1999), yet hyphal development from the unbudded yeast form of C. albicans to its adhesion and hyphal growth has not been demonstrated using dynamic methods. More recently, time-lapse microscopy and imaging methods have been tracing hyphal compartments (Barelle et al. 2003), host-pathogen interactions (Tam et al. 2011; Brothers et al. 2013), and the formation of biofilms (Kaneko et al. 2013). These approaches were unable to distinguish between different cell forms and at the same time to monitor (a) the adhesion and budding of mother cells, (b) the appearance of individual hyphae colonies, and (c) the speed of hyphae elongation over longer periods of time.
An obstacle in in vitro experiments of this kind is the often unsuitable conditions in which the yeast cells are grown in, raising questions as to the physiological relevance of these measurements. To overcome these difficulties, we have recently developed a computer-based image analysis of time-lapse video microscopy that allows visualization and analysis of mammalian and fungal cell growth up to 2 weeks (Nagy et al. 2010; Banfalvi et al. 2012). Low phototoxicity, low light dispersion, near-infrared illumination, constant temperature, and pH of this system serves the purpose of maintaining cells as close as possible to physiological conditions.
This paper describes the application of long-term scanning microscopy for the analysis of adherence and growth patterns of different C. albicans strains. We investigated the morphological effects of (a) quorum-sensing molecules tyrosol and farnesol on the wild-type SC5314 strain, (b) the genetic deletions of the fungus-specific protein phosphatase Z1 (CaPPZ1), and the hypha-specific cyclin (HGC1) genes. The results demonstrate the usefulness of our system in studying external effectors and genetic changes on C. albicans’ hyphal morphogenesis and its potential clinical impact.
Materials and methods
Culturing and growth conditions for C. albicans strains
The C. albicans strains used in the present study are summarized in Table 1. The suitability of our method was tested with chemicals and genetic mutations that affect germination and hyphal growth. The opportunistic pathogen C. albicans SC5314 wild-type strain (deposited as ATCC MYA-2876) was used to demonstrate the effects of quorum-sensing molecules, namely farnesol (Hornby et al. 2001) and tyrosol (Chen et al. 2004). The SC5314 strain was grown in Sabouraud medium (Sabouraud, 1910) for 18 h at 30 °C without shaking. Aliquots (1 ml) of the culture were centrifuged for 5 min at 2,500 g and washed twice with 1-ml phosphate-buffered saline (PBS) (Gibco, Life Technologies, Budapest, Hungary). Cells (1–2 × 105) were transferred into T-25-cm2 ventilated culture flasks (VWR-International, Debrecen, Hungary) containing 5 ml of the filamentation medium in Roswell Park Memorial Institute (RPMI) (PAA Laboratories GmbH, Pasching, Austria) plus 10 % fetal bovine serum (FBS) (PAA Laboratories) alone (Lo et al. 1997) or supplemented with 0.5-mM tyrosol, 2-(4-hydroxyphenyl)ethanol (Sigma-Aldrich, Budapest, Hungary) (Alem et al. 2006). Farnesol treatment took place in a similar way with the exception that the cells were preincubated for 2 min in the medium containing 1-mM trans,trans-farnesol (Sigma-Aldrich, Budapest) before the addition of 10 % final concentration of FBS (Alem et al. 2006).
The hgc1 mutant (WYZ12.2) and its genetically matched control strain (WYZ12.1) were cultured similarly to SC5314 (Zheng and Wang 2004). The cappz1 mutant and its appropriate control strain (QMY23) were grown in Sabouraud medium for 18 h at 37 °C without shaking (Adam et al. 2012). The culture (1 ml) was centrifuged for 5 min at 2,500 g, the pellet was washed two times with 1-ml PBS, and 1–2 × 105 cells were transferred into the T-25 culture flask containing 5-ml RPMI supplemented with 10 % fetal bovine serum.
During the analysis, all of the C. albicans cells were incubated at 37 °C in a CO2 incubator (Lo et al. 1997) in ventilated T-25-cm2 flasks (VWR-International) for 5–6 h.
Time-lapse imaging
The description of the system and protocol for time-lapse scanning (TLS) is given below:
-
(a)
Incubator. A SANYO MCO18-AC (Wood Dale, IL, US) CO2 incubator was used with a back-side instrument port. Its chamber was modified to host four microscopes.
-
(b)
Microscopes. Olympus (Tokyo, Japan) upright microscopes were modified for inverted usage, and revolver turrets were installed to replace the original illumination. Charge-coupled device (CCD) camera boards were placed under the turrets, using the monocular adapter lower parts of Olympus Tokyo as housing. Specimen tables were unmodified, but the slide orientation mechanisms were removed. Ocular sockets were used for illuminators.
-
(c)
Illumination. Diodes emitting light at 940 nm (LED: 5-mm diameter; 1.2 V and 50 mA, driven at 5 V using a serial 82 Ohm resistor) were used to illuminate cells while minimizing heat and phototoxicity. The longer wavelength offered a deeper penetration (up to 3-mm thickness) and less light dispersion through the culture medium and the wall of the T-flask. The theoretical limit of resolution under our conditions using a 940-nm wavelength at 1.25 numerical aperture was 1.88 nm based on the Abbe equation. The original 5-mm spherical light-emitting diode (LED) head was used as a condenser for a better reproducibility of the setup. Illuminators were centered and fixed with glue in microscope ocular tubes. The distance between the upper surface of the T-25 culture flask and the spherical ballhead of the LED was 120 mm. Cells were only illuminated during image acquisition periods (~5 s per timepoint).
-
(d)
Microscope objectives. Carl Zeiss (Jena, Germany) plan achromatic objectives (×10: 0.25 NA) were used to enable a broad field of view to be imaged.
-
(e)
Cameras. Custom-modified 2 megapixel UVC USB 2.0 camera boards (Asus Computer International, Fremont, CA, USA) were modified by removing the camera housing, objectives, and infrared-cut filters. Status indicator LEDs and attached resistors were desoldered, and driver circuit terminals were elongated with wires for near-infrared illumination.
-
(f)
Image capture and display. Ten images were collected, each within a 5-s interval and averaged to minimize noise. The collection of images within a 5-s interval is regarded as high time resolution. Images from the four cameras were displayed simultaneously on screen to show time-matched morphological changes between wild-type and mutant C. albicans cells. Examples of movies showing the growth and mobility of different strains of C. albicans cells are provided in the figure legends.
Data of videos (S1–S7) of time-lapse imaging are given in Table S1 in the supplementary material.
Motion analysis of hyphae extension
Stack preparation
Image sequences of hyphae growth were converted from color (24-bit red, green, and blue (RGB)) .bmp files to 16-bit grayscale .tiff files using a GraphicConverter (Lemke Software, Piene, Germany). Movies were recorded at 1600 × 1200 pixels with a 0.434-μm/pixel spatial dimension. Temporal sampling was at an average of 85.73 ± 0.27 s per frame (n = 7), corresponding to a frequency of 0.117 Hz. Movies of hyphae growth were captured for 5 h. The .tiff images were imported as a stack into custom-written software (Volumetry G8a, GWH) and the timestamp in the filename extracted (Fig. 1a). Variations in illumination throughout the stack were normalized by finding a region of interest (ROI) where no hyphae were observed during the movie and calculating the average intensity in that region (ROI ~ 30 × 30 μm in size). The difference in intensity of each frame compared to the average intensity throughout the entire stack was subtracted. More advanced antihalo filters (see below) were not needed for spatio-temporal map and temporal accumulation analyses.
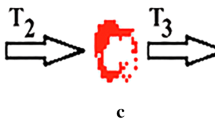
Methods of image amalysis. a–b Spatio-temporal map method. a Shows a frame from a movie displaying hyphae attachment points (bright white dots) with processes extending outwards (white lines with black edges). The spatio-temporal map (ST map) method requires regions of interest (ROIs) to be drawn around suitable extending processes (see red, green and blue ROIs in a; b/i–iii). The average intensity perpendicular to the process was calculated along the ROI in each frame. Resulting lines of average pixel intensities were stacked to create ST maps. b 3ST maps (b/i, b/ii, and b/iii) from different hyphae showing the growth of hyphae processes over time. A line was manually drawn over the edge created by the extending hyphae process in the ST map and the slope calculated to determine the velocity (see b/iii). The rate of hyphae process extension was essentially linear, although curve-linear analysis may have improved the accuracy of velocity analysis in some situations by a very small amount. The black and white horizontal streaks on the right hand side of the ST maps are due to secondary processes extending out perpendicularly from the original process (b/i, b/iii). c–f Temporal accumulation method. During manual selection, individual cells with sufficient empty spaces around them were selected for the analysis. c The temporal accumulation method does not require the manual selection of individual hyphae, but uses a series of routines to isolate and measure the average movement of the ends of all hyphae processes in the field of view. After normalizing average intensity, the movies were differentiated (16 bit: [(frame(t+∆t)−frame(t)) + 2^15]) to isolate only those processes that had extended between the chosen frames. d The resulting extension “patches” were thresholded and filtered for size and roundness. e A temporal accumulation routine was used overlap extension patches from individual frames to create an image of hyphal growth filaments, with time encoded within each filament as a color. f After separating non-continuous filaments using a temporal gradient filter, erosion, and size filtering, filaments were used to calculate hyphae extension velocity (using an initial sprouting position as reference) or instantaneous extension velocities (moving references), as well as a number of other parameters including coherence (average ∆ instantaneous velocities) and changes in angle of the extending hyphae processes
Spatio-temporal method
A user-directed spatio-temporal map (ST map) method was used to calculate hyphae extension velocity in stacks in which hyphae had poor adhesion to the growth surface evidenced by floating particles or hyphae extensions that had significant to-and-fro motion in the x, y, or z axis. Only those individual hyphae that remained in the same focal plane with minimal to-and-fro motion were chosen for analysis. To begin, the final frame of the movie was selected which showed the full extent of hyphae growth during the experiment and ROIs drawn in parallel over those hyphae processes that showed linear extension (see Fig. 1a: rectangular ROIs, i–iii). The average intensity of each perpendicular pixel column along the length of the ROIs was calculated in each frame of the movie to construct ST maps (Fig. 1b), with time represented in the x axis and distance in the y axis. Extension velocities were calculated manually by superimposing a line over the leading edge of the extending process and calculating the slope (see Fig. 1b iii: orange line). Repetitive calculations on the same extending process show that slight differences in the way the line was drawn accounted for velocity changes less than 0.1 nm.s−1 and were more reliable than using automated edge detection methods.
Temporal accumulation method
To isolate only the growing tip of the hyphae, the stack was differentiated using a suitable time step (∆t = 429 s [five frames]). This time separation created a circular growth “patch” in the differentiated movies, while removing any static information. To remove pixel noise, a Gaussian blur algorithm was used throughout the stack (5 × 5 pixels, SD 1.5; Fig. 1c). After applying an intensity threshold to outline the growth patches, a particle filter was employed that filtered out any particles that did not meet size and perimeter/area ratio criteria (size between 9.4–47 μm2, perimeter/area ratio <0.4). The remaining particles are a representative of extending hyphae growth with side-to-side movements were filtered out (Fig. 1d). A temporal accumulation routine was then used (starting at the end of the stack) that overlaid thresholded particles into a single image, coloring them based on their position (time) in the stack (8-bit spectrum LUT used). This created “trails” or filaments of the hyphae extensions (Fig. 1e). Towards the end of the recordings when secondary branches spawned from an initial filament, the trails overlapped slightly. Multiple filaments originating from a single cell were separated based on the temporal gradient in a 3 × 3 pixel square. If the gradient exceeded 429 s [five frames], the 3 × 3 pixel square was colored black. An erode routine was used to separate filaments further (3 × 3, 1 iteration). Finally, a size filter was applied to filter out any filaments that had an area less than 47 μm2 (Fig. 1f).
The start and finish time of each filament was calculated based on the encoded max/min colors. Both the instantaneous velocity and the extension velocity of the filament were calculated by thresholding those regions in the filament that corresponded to particular points in time. For instantaneous velocity, two regions in the filament separated by 171 s (2 frames) were thresholded and midpoint distance between the regions calculated. For extension velocity, the midpoint of the start region of a filament was used as a static reference to which midpoint of other regions were compared (advancing in 171-s [two frames] increments). To measure the coherence of motion, the change in instantaneous velocity was calculated (SD of the absolute ∆ velocity between timepoints). In conjunction with calculations of distance between two selected growth patches, the angle and ∆angle over the course of the growth of a hypha was also calculated.
There was no significant difference between the results of the ST map and the temporal accumulation methods using control (SC5314) movies. Temporal accumulation methods could only be reliably used in movies that showed better adhesion and faster growth rates; however, it allowed calculation of more motion parameters than the ST map method. Hyphae extension velocities presented in Fig. 2 were all calculated using the ST map method.
Hyphal growth rates of different C. albicans strains. a The wild-type SC5314 control strain was investigated alone and after treatment with tyrosol or farnesol. b The QMY23 control strain was compared to the phosphatase-deficient cappz1 mutant, and c the WYZ12.1 control was compared to the hypha-specific G1 cyclin-related hgc1 mutant strain (WYZ12.2). In the three series of experiments (a, b, c), each set was tested relative to its own appropriate control. Statistical analysis *p < 0.05, **p < 0.01
Digital image processing and analysis of adherence, transitions, and motion of non-adhered cells
Numbered image sequences were transferred to a custom-built computer (Biological Image Processing Station, BIPS) running a barebone Windows 7 operating system. Image processing was done by the National Health Institute ImageJ software bundle (http://rsbweb.nih.gov/ij/docs/install/windows.html), using custom-developed plugins and macros. Image sequences were first deflickered using a sequence stack histogram to avoid transient brightness changes between separate frames. Every sequence contains an empty frame at the beginning and at the end representing the field of view without a specimen as a reference image for background. The average of these frames resulted in a background image representing the error caused by uneven illumination (bulb-effect), slight differences of CCD sensor manufacturing, or dust particles in the light path. This background image was subjected to a Gaussian blur of radius 2 pixels, which was subtracted from each frame of the sequence. Background was further reduced by employing a fast Fourier-transform bandpass filtering to exclude large structures down to 40 pixels and filtering small structures up to 3 pixels in size and a background extraction process using a rolling ball at a radius of 50 pixels. The RGB channels of the 24-bit bitmap images were then merged (10 % blue, 40 % green, 50 % red) resulting in an 8-bit grayscale image. Contrast and brightness were equalized based on the stack (sequence) histogram at 0.4 % of the pixels saturated. Image sequences were then thresholded using a stack histogram. The consequent binary image sequence was carefully scrutinized, and non-flickering particles with a solid outline were selected for analysis avoiding overlaps of hyphae. Analysis was done by measuring the thresholded areas of particles in subsequent frames. The image sequences were digitally processed using image enhancement algorithms available as plugins for the Java language-based NIH ImageJ (FiJi) open-source software bundle. Parameters measured using ImageJ include time of adherence, time of transition of yeast-to-hyphal growth, and motion of non-adhered yeast cells.
Statistical analysis
Standard deviation was calculated as a measure of dispersion of data from its mean and the t test, one-way analysis of variance (ANOVA) for statistical significance. Data were presented as mean ± SD. *p < 0.05 was considered statistically significant and **p < 0.01 as statistically highly significant.
Results
Hyphae extension velocities
The movement, changes in the shape, and growth of individual C. albicans cells was followed by time-lapse video microscopy under near-physiological conditions in a carbon dioxide incubator. Real-time imaging of each experiment was carried out with four simultaneously working time-lapse scanning (TLS) microscopes. Only those TLS experiments were chosen for analysis that met rigorous criteria, including (a) contrast and sharpness of images, (b) low cell number with distinct dispersion of cells, (c) low interframe oscillations, and (d) negligible brightness differences between neighboring frames. Based on these criteria, seven experimental sets have been selected for further in-depth analysis of hyphae extension velocity data (Fig. 2). The average hyphal growth rate in wild-type SC5314 C. albicans strain was 7.31 ± 1.34 nm.s−1. Tyrosol (0.5 μM), that is known to accelerate hyphae extension, increased the velocity only moderately to 8.32 ± 1.24 nm.s−1, while farnesol (1 μM), an inhibitor of hyphal growth, reduced the speed of hyphal growth to 3.03 ± 0.56 nm.s−1. The hyphal growth rate of the second control QMY23 was 8.51 ± 0.75 nm.s−1, while its phosphatase-deficient cappz1 mutant counterpart showed a moderate but significantly reduced extension velocity of 6.98 ± 1.43 nm.s−1. The third control WYZ12.1 strain exhibited an initially lower 4.90 ± 0.48 nm.s−1 hyphal growth rate that was drastically reduced in the hgc1 cyclin mutant WYZ12.2 to 1.33 ± 0.47 nm.s−1.
Distribution of hyphae extension velocities
Besides the average velocity values, the distribution of the hyphae extension velocities were found relatively uniform (most between 5 and 10 nm.s−1) in the wild-type strain SC5314 (upper left panel, Fig. 3). Upon facilitating the hyphal growth of the SC5314 strain by tyrosol treatment, the distribution of instantaneous velocities narrowed slightly and shifted to the right (6–12 nm.s−1) indicating a more uniform growth pattern at a higher velocity (lower left panel, Fig. 3). The instantaneous velocities of hyphae extension from the QMY23 strain showed a relatively skewed distribution (most between 5 and 11 nm.s−1; upper right panel, Fig. 3). An uneven and broad instantaneous velocity distribution of hyphal growth was observed with the phosphatase-deficient cappz1 mutant of QMY23 (most between 3 and 15 nm.s−1) with a relatively sharp peak between 7–9 nm.s−1 (lower right panel, Fig. 3) indicating that different genetic backgrounds of the control strains (Table 1) may influence both the extension velocities’ values and their distributions.
Yeast-to-hyphal transitions
Hyphal growth of the SC5314 wild type strain
Time-lapse video microscopy was adapted to measure the time of adherence of yeast cells of different C. albicans cells to the growth surface. During the tracking of the overall motion of the wild-type SC5314 cells, the changes in hyphae appearance (between 40 and 100 min) could be divided into the following four subphases (Fig. 4a):
Yeast cell attachment and hyphae outgrowth of wild-type SC5314 C. albicans in the absence and in the presence of tyrosol or farnesol. From the adhesion time to the beginning of the next motion (originating from initial hypha growth) was taken as the time of hypha outgrowth. a Subphases (i–iv) of movement intensities of wild-type SC5314 C. albicans cells. b Changes in movement after tyrosol treatment of SC5314 strain. c Movement of SC5314 cells upon farnesol treatment. The overall movement intensity of particles (cells, hyphae) is expressed in arbitrary units extracted from square pixels reflecting changes between two consecutive frames and is depicted as a function of cultivation time. Black arrows indicate the end of sedimentation of yeast cells (end of phase i), white arrows show the time points of 50 % hyphae appearance (end of phase ii). These time points have been extracted from actual videos belonging to the curves. Blue lines indicate all movements within the visual field. As a rule, 10–20 cells were in the visual field of view to avoid the overlapping of growing hyphae. Branching hyphae have been excluded from the measurements. Red lines summarize the averages of three consecutive frames to filter out mechanical oscillations
-
(i)
shows the overall motion of yeast cells right after they have been suspended in the medium,
-
(ii)
during the sedimentation phase, a 10-min sedimentation time was allowed to exclude gravitation effects from the molecular motion of the sedimented but not yet adhered yeast cells. Cell particles gradually settled down and attached to the surface of the tissue flask reducing their overall motion to near zero.
-
(iii)
after 40 min, initial hyphal growth started on the surface of the flask causing an expansion of the cell/hyphae surface area on the flask surface that was followed by
-
(iv)
active hyphal growth/movement outside the focal plane.
Quorum-sensing molecules affecting cell adhesion and hyphal outgrowth
Upon tyrosol treatment, SC5314 yeast cells showed a slightly biphasic sedimentation curve (Fig. 4b). After the sedimentation period, hyphal growth took place almost exclusively parallel to the attachment surface, in contrast to control SC5314 cells in which hyphae appeared to extend vertically out of the focal plane towards the end of the recording (Fig. 4a, b). Farnesol treatment of SC5314 strain caused an even faster sedimentation of yeast cells, but prevented their attachment to the surface of the T-flask (Fig. 4c). Hyphal growth without attachment to the basement is characterized by a considerable movement frequency of individual particles.
The results of 3–6 independent measurements are summarized in Fig. 5a. It is seen that tyrosol reduced, while farnesol increased the time required for both the adhesion of the cells and the initiation of 3D hyphae outgrowth. The time of adherence in the wild-type SC5314 strain was 35.75 ± 1.58 min (100 %). Tyrosol (29.67 ± 1.51 min, 83 %) reduced moderately, while farnesol (47 ± 6.08 min, 131 %) increased significantly the time of attachment of yeast cells to the focal plane.
Time of adhesion and hyphal outgrowth after sedimentation of yeast cells of different C. albicans strains. The time of adherence to the growth surface of the standard T-25 flask is indicated by the dark gray columns. The adherence is expressed in time elapsed from the sedimentation to the 90 % standstill of Brownian particle movement. Hyphal outgrowth generating 3D movement of hyphae is shown by the light gray boxes on the top of the dark gray columns. In the three series of experiments (a, b, c), each set was tested relative to its own appropriate control. Statistical analysis *p < 0.05, **p < 0.01
Effect of mutations on cell adhesion and hyphal outgrowth
Similarly to the wild-type SC5314, we have characterized the QMY23 control strain that showed early focal attachment, followed by hyphal growth at or near the attachment sites. Knocking out the CaPPZ1 gene resulted in a multiphasic sedimentation profile with a delayed yeast-to-hyphal transition and a prolonged movement of individual particles. QMY23 exhibited the fastest adherence (27 ± 1.55 min, 100 %), the phosphatase-deficient cappz1 (33.67 ± 6.12 min, 125 %) exhibited a moderately increased time of adhesion (Fig. 5b). WYZ12.1 (32 ± 2.00 min, 100 %) showed a rapid attachment, while the adhesion time of its hgc1 hypha-specific G1 cyclin-related mutant WYZ12.2 (73.67 ± 6.03 min, 230 %) indicated a weak, if any, attachment to the growth surface. The WYZ12.2 exhibited the slowest attachment rate, the lack of binding to the growing surface, and a poor hyphal growth. The comparison of the duration of yeast cell adhesion (Fig. 5) with the hyphae extension velocity data (Fig. 2) indicates a reciprocal relationship between the two processes.
Hyphal extension velocities
A further process followed by time-lapse video microscopy beside yeast cell adhesion and hypae extension velocity was the time elapsed between adhesion and hyphae outgrowth. The temporal order of increasing time of hyphae outgrowth are indicated by the three experimental setups and are depicted as white boxes in Fig. 5.
-
(i)
SC5314 + tyrosol (3.83 ± 0.75 min, 68 %) < SC5314 (5.63 ± 1.06 min, 100 %), < SC5314 + farnesol (12.67 ± 11.06 min, 225 %),
-
(ii)
QMY23 (3.83 ± 2.23 min, 100 %) < cappz1KO (5.67 ± 1.03 min, 148 %),
-
(iii)
WYZ12.1 (9.2 ± 1.10 min, 100 %) < WYZ12.2 (22.0 ± 4.36 min, 239 %).
Although the time intervals follow the same order as the adherence times (gray bars in Fig. 5), the experimental deviations do not allow us to draw statistically significant correlations from these data.
Discussion
A new method has been developed and tested, based on digital image analysis of high time resolution video microscopy, serving the investigation of hyphal growth dynamics in different C. albicans strains. We have demonstrated that (a) the time of adherence of C. albicans yeast cells to the culture dish, (b) the time of appearance of hyphae, and (c) the hyphal growth rate can be followed by time-lapse image analysis. These studies dynamically document that C. albicans undergoes a dramatic morphological transition in response to various stimuli and show that the time-lapse method can be used to characterize different yeast mutants.
In the first set of experiments, we have tested the effects of quorum-sensing molecules, namely tyrosol and farnesol. C. albicans cells are similar to bacteria in that they use quorum-sensing to coordinate certain behaviors including virulence and antibiotic resistance (Miller and Bassler 2001). The autoregulatory tyrosol is released into the medium during the growth of C. albicans, accelerates the formation of germ tubes, and stimulates the formation of the filamentous protrusions. Farnesol is known for its potential to inhibit germ-tube formation of C. albicans (Chen et al. 2004). In agreement with earlier publications, we found that tyrosol accelerated yeast-to-hyphal transition. It also caused a stronger attachment of cells to the growth surface suggesting that germ-tube formation depends on the strength of the attachment. Farnesol treatment had the opposite effect; C. albicans cells did not show any tendency to attach through focal points to the basement of the tissue flasks and consequently prevented germ-tube formation and hyphal extension.
In the second set of experiments, we tested well-characterized gene deletions. Due to the importance of yeast-to-hyphal transition for virulence of C. albicans, the correlation between the morphological changes and infection rates are of particular interest (Sudbery 2011). We reported earlier that the deletion of CaPPZ1 resulted in a reduced rate of germination (Adam et al. 2012). Here, we confirm that the deletion of the CaPPZ1 reduces the rate of hyphal growth. Yeast-to-hyphal transition of the cappz1 mutant strain was slower as compared to QMY23 control strain, suggesting that the lack of the phosphatase results in a poor attachment, a delayed hyphal growth, and significantly reduced hyphae extension velocities. HGC1 plays a specific and critical role in C. albicans hyphal morphogenesis (Zheng et al. 2004), as its deletion caused defective hyphal growth. We confirmed that the hyphal growth velocities were drastically reduced in the hgc1 mutant WYZ12.2 strain. The absence of the adherence of the WYZ12.2 cells to focal points, which is a new phenotype not described in the original publication of Zheng and Wang (2004), could explain the poor and delayed hyphal outgrowth.
The presented data confirm the suitability of microscopic analysis to evaluate dimorphic changes in fungi that could be further improved. Our long-term scanning (LTS) microscopy system has several advantages: (a) it consists of four inverse microscopes allowing the simultaneous study of either identical parallel cultures or different treatments of the same culture beside the control sample, (b) each microscope is equipped with a high sensitivity video camera connected to the dual image acquisition computer system, (c) illumination was developed to minimize heat and phototoxicity, (d) exposure times were minimized to avoid phototoxicity, (e) transmission light microscopic images can be taken every minute up to a week, and (f) exposures can be converted to video films by speeding up the projection to 30 exposures/s. However, the continuous analysis of live cells would need the automation of cell breeding and experimental procedures. Such an integrated system does not exist. To reduce the limitations of the LTS system, the development of an LTS-perfusion platform would be useful. In such a combined system, the cell culture vessel could be firmly attached to the inverted microscope and the old medium replaced with a fresh one by avoiding physical contact with the cell culture. The elimination of direct contact with the cell culture would also prevent its infection and allow cell growth, analysis, and treatment for an unlimited time.
In summary, we have developed and tested the suitability of our time-lapse image analyzing system under a wide range of experimental conditions and demonstrated that this method is useful for quickly screening of C. albicans cell adherence, hyphal outgrowth, and hyphal growth rate in vitro. Our method can be adapted to a great variety of experiments for the detection of morphological changes elicited by effector molecules or specific mutations. Candida (spp.) can attach to plastic surfaces, intravenous devices, bladder catheters, and other prostheses routinely used in medicine (Klotz et al. 1985; Kiremitçi–Gümüşderelioğlu and Pesmen 1996; Radford et al. 1999; Ramage et al. 2006; von Fraunhofer and Loewy 2009; Busscher et al. 2010). Since adherence and morphological transitions are inherently coupled processes, the inhibition of adherence to such surfaces will greatly prevent hyphal growth. Antiadhesive treatments of devices combined with the inhibition of hyphal growth are expected to be an applicable approach to prevent iatrogenic Candida infections. Our method presented in this paper may help experts efficiently working on novel Candida control strategies.
References
Adam C, Erdei E, Casado C, Kovacs L, Gonzalez A, Majoros L, Petrenyi K, Bagossi P, Farkas I, Molnar M, Pocsi I, Arino J, Dombradi V (2012) Protein phosphatase cappz1 is involved in cation homeostasis, cell wall integrity and virulence of Candida albicans. Microbiology 158(Pt 5):1258–1267. doi:10.1099/mic.0.057075-0
Alem MA, Oteef MD, Flowers TH, Douglas LJ (2006) Production of tyrosol by Candida albicans biofilms and its role in quorum sensing and biofilm development. Eukaryot Cell 5(10):1770–1779. doi:10.1128/EC.00219-06
Banfalvi G, Sarvari A, Nagy G (2012) Chromatin changes induced by Pb and Cd in human cells. Toxicol In Vitro 26(6):1064–1071. doi:10.1016/j.tiv.2012.03.016
Barelle CJ, Bohula EA, Kron SJ, Wessels D, Soll DR, Schäfer A, Brown AJ, Gow NA (2003) Asynchronous cell cycle and asymmetric vacuolar inheritance in true hyphae of Candida albicans. Eukaryot Cell 2(3):398–410. doi:10.1128/EC.2.3.398-410.2003
Brothers KM, Gratacap RL, Barker SE, Newman ZR, Norum A, Wheeler RT (2013) NADPH oxidase-driven phagocyte recruitment controls Candida albicans filamentous growth and prevents mortality. PLoS Pathog 9(10):e1003634. doi:10.1371/journal.ppat.1003634
Brown AJP (2002) Morphogenetic signalling pathways in Candida albicans. In: Calderone R (ed) Candida and Candidiasis. American Society for Microbiology, Washington, DC, pp 95–106
Brown AJ, Gow NA (1999) Regulatory networks controlling Candida albicans morphogenesis. Trends Microbiol 7(8):333–338. doi:10.1016/S0966-842X(99)01556-5
Bulad K, Taylor RL, Verran J, McCord JF (2004) Colonization and penetration of denture soft lining materials by Candida albicans. Dent Mater 20(2):167–175. doi:10.1016/S0109-5641(03)00088-5
Busscher HJ, Rinastiti M, Siswomihardjo W, van der Mei HC (2010) Biofilm formation on dental restorative and implant materials. J Dent Res 89(7):657–665. doi:10.1177/0022034510368644
Calderone RA, Lehrer N, Segal E (1984) Adherence of Candida albicans to buccal and vaginal epithelial cells: ultrastructural observations. Can J Microbiol 30(8):1001–1007
Chen H, Fujita M, Feng Q, Clardy J, Fink GR (2004) Tyrosol is a quorum-sensing molecule in Candida albicans. Proc Natl Acad Sci U S A 101(14):5048–5052. doi:10.1073/pnas.0401416101
de Groot PW, Bader O, de Boer AD, Weig M, Chauhan N (2013) Adhesins in human fungal pathogens: glue with plenty of stick. Eukaryot Cell 12(4):470–481. doi:10.1128/EC.00364-12
Gillum AM, Tsay EY, Kirsch DR (1984) Isolation of the Candida albicans gene for orotidine-5’-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol Gen Genet 198(1):179–182. doi:10.1007/BF00328721
Gow NA, Hube B (2012) Importance of the Candida albicans cell wall during commensalism and infection. Curr Opin Microbiol 15(4):406–412. doi:10.1016/j.mib.2012.04.005
Hornby JM, Jensen EC, Lisec AD, Tasto JJ, Jahnke B, Shoemaker R, Dussault P, Nickerson KW (2001) Quorum sensing in the dimorphic fungus Candida albicans is mediated by farnesol. Appl Environ Microbiol 67(7):2982–2992. doi:10.1128/AEM.67.7.2982-2992.2001
Jacobsen ID, Wilson D, Wächtler B, Brunke S, Naglik JR, Hube B (2012) Candida albicans dimorphism as a therapeutic target. Expert Rev Anti Infect Ther 10(1):85–93. doi:10.1586/eri.11.152
Kaneko Y, Miyagawa S, Takeda O, Hakariya M, Matsumoto S, Ohno H, Miyazaki Y (2013) Real-time microscopic observation of Candida biofilm development and effects due to micafungin and fluconazole. Antimicrob Agents Chemother 57(5):2226–2230. doi:10.1128/AAC.02290-12
Kimura LH, Pearsall NN (1978) Adherence of Candida albicans to human buccal epithelial cells. Infect Immun 21(1):64–68
Kimura LH, Pearsall NN (1980) Relationship between germination of Candida albicans and increased adherence to human buccal epithelial cells. Infect Immun 28(2):464–468
King RD, Lee JC, Morris AL (1980) Adherence of Candida albicans and other Candida species to mucosal epithelial cells. Infect Immun 27(2):667–674
Kiremitçi-Gümüşderelioglu M, Pesmen A (1996) Microbial adhesion to ionogenic PHEMA, PU and PP implants. Biomaterials 17(4):443–449. doi:10.1016/0142-9612(96)89662-1
Klotz SA, Drutz DJ, Zajic JE (1985) Factors governing adherence of Candida species to plastic surfaces. Infect Immun 50:97–101
Liu H (2001) Transcriptional control of dimorphism in Candida albicans. Curr Opin Microbiol 4(6):728–735. doi:10.1016/S1369-5274(01)00275-2
Lo HJ, Köhler JR, DiDomenico B, Loebenberg D, Cacciapuoti A, Fink GR (1997) Nonfilamentous C. albicans mutants are avirulent. Cell 90(5):939–949. doi:10.1016/S0092-8674(00)80358-X
Madhani HD, Fink GR (1998) The control of filamentous differentiation and virulence in fungi. Trends Cell Biol 8(9):348–353. doi:10.1016/S0962-8924(98)01298-7
Mayer FL, Wilson D, Hube B (2013) Candida albicans pathogenicity mechanisms. Virulence 4(2):119–128. doi:10.4161/viru.22913
Miller MB, Bassler BL (2001) Quorum sensing in bacteria. Annu Rev Microbiol 55:165–199. doi:10.1146/annurev.micro.55.1.165
Mitrovich QM, Tuch BB, Guthrie C, Johnson AD (2007) Computational and experimental approaches double the number of known introns in the pathogenic yeast Candida albicans. Genome Res 17(4):492–502. doi:10.1101/gr.6111907
Naglik JR, Moyes DL, Wächtler B, Hube B (2011) Candida albicans interactions with epithelial cells and mucosal immunity. Microbes Infect 13(12–13):963–976. doi:10.1016/j.micinf.2011.06.009
Nagy G, Pinter G, Kohut G, Adam AL, Trencsenyi G, Hornok L, Banfalvi G (2010) Time-lapse analysis of cell death in mammalian and fungal cells. DNA Cell Biol 29(5):249–259. doi:10.1089/dna.2009.0980
Radford DR, Challacombe SJ, Walter JD (1998) Adherence of phenotypically switched Candida albicans to denture base materials. Int J Prosthodont 11(1):75–81
Radford DR, Challacombe SJ, Walter JD (1999) Denture plaque and adherence of Candida albicans to denture-base materials in vivo and in vitro. Crit Rev Oral Biol Med 10(1):99–116. doi:10.1177/10454411990100010501
Ramage G, Martinez JP, Lopez-Ribot JL (2006) Candida biofilms on implanted biomaterials: a clinically significant problem. FEMS Yeast Res 6(7):979–986. doi:10.1111/j.1567-1364.2006.00117.x
Sabouraud R (1910) Les teignes. Masson et Cie, Paris, France
Sobel JD, Myers PG, Kaye D, Levison ME (1981) Adherence of Candida albicans to human vaginal and buccal epithelial cells. J Infect Dis 143(1):76–82. doi:10.1093/infdis/143.1.76
Sudbery PE (2011) Growth of Candida albicans hyphae. Nat Rev Microbiol 9(10):737–748. doi:10.1038/nrmicro2636
Sudbery P, Gow N, Berman J (2004) The distinct morphogenic states of Candida albicans. Trends Microbiol 12(7):317–324. doi:10.1016/j.tim.2004.05.008
Tam JM, Castro CE, Heath RJ, Mansour MK, Cardenas ML, Xavier RJ, Lang MJ, Vyas JM (2011) Use of an optical trap for study of host-pathogen interactions for dynamic live cell imaging. J Vis Exp 53:e3123. doi:10.3791/3123
Tronchin G, Bouchara JP, Robert R, Senet JM (1988) Adherence of Candida albicans germ tubes to plastic: ultrastructural and molecular studies of fibrillar adhesins. Infect Immun 56(8):1987–1993
Tronchin G, Bouchara JP, Robert R (1989) Dynamic changes of the cell wall surface of Candida albicans associated with germination and adherence. Eur J Cell Biol 50(2):285–290
von Fraunhofer JA, Loewy ZG (2009) Factors involved in microbial colonization of oral prostheses. Gen Dent 57(2):136–143; ibid. 144–135
Whiteway M (2000) Transcriptional control of cell type and morphogenesis in Candida albicans. Curr Opin Microbiol 3(6):582–588. doi:10.1016/S1369-5274(00)00144-2
Zheng X, Wang Y (2004) Hgc1, a novel hypha-specific G1 cyclin-related protein regulates Candida albicans hyphal morphogenesis. EMBO J 23(8):1845–1856. doi:10.1038/sj.emboj.7600195
Acknowledgements
Thanks are due to Prof. Alexander Johnson (Department of Biochemistry and Biophysics, University of California, San Francisco, USA) for the QMY23 and to Prof. Yue Wang (Institute of Molecular and Cell Biology, Singapore, Singapore) for the WYZ12.1 and WYZ12.2C. albicans strains. This work was supported by the OTKA grant K108989 and by the UD Faculty of Medicine Research Fund (Bridging Fund 2012) to VD, TÁMOP 4.2.1/B-09/1/KONV-2010-0007 to IP, as well as by TÁMOP 4.2.4. A/2-11-1-2012-0001 National Excellence Program that was subsidized by the European Union and co-financed by the European Social Fund to GN and KP. GWH is supported by grants from the National Center for Research Resources (5P20RR018751-09) and the National Institute of General Medical Sciences (8 P20 GM103513-09) from the National Institutes of Health.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 24 kb)
(AVI 29,865 kb)
(AVI 30,469 kb)
(AVI 29,044 kb)
(AVI 51,190 kb)
(AVI 20,806 kb)
(AVI 29,909 kb)
(AVI 22,989 kb)
Rights and permissions
About this article
Cite this article
Nagy, G., Hennig, G.W., Petrenyi, K. et al. Time-lapse video microscopy and image analysis of adherence and growth patterns of Candida albicans strains. Appl Microbiol Biotechnol 98, 5185–5194 (2014). https://doi.org/10.1007/s00253-014-5696-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-014-5696-5