Abstract
Bacterial cutinases are promising catalysts for the modification and degradation of the widely used plastic polyethylene terephthalate (PET). The improvement of the enzyme for industrial purposes is limited due to the lack of structural information for cutinases of bacterial origin. We have crystallized and structurally characterized a cutinase from Thermobifida fusca KW3 (TfCut2) in free as well as in inhibitor-bound form. Together with our analysis of the thermal stability and modelling studies, we suggest possible reasons for the outstanding thermostability in comparison to the less thermostable homolog from Thermobifida alba AHK119 and propose a model for the binding of the enzyme towards its polymeric substrate. The TfCut2 structure is the basis for the rational design of catalytically more efficient enzyme variants for the hydrolysis of PET and other synthetic polyesters.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Polyethylene terephthalate (PET) (Fig. S1) is probably the most well known and widespread synthetic polyester all over the world. Its usage comprises foils and bottles as well as fibres for textile industry (Herrero Acero et al. 2011). The current strategies to modify the properties of PET for different applications often involve harsh chemical and physicochemical conditions (Chen and McCarthy 1998; Riccardi et al. 2003). As a result, unwanted side reactions compromising the integrity of the final products are a serious problem in the industry (Brueckner et al. 2008). Furthermore, the used chemicals are often environmentally harmful. Therefore, a strategy to overcome these limitations is required. Enzymes offer a cleaner and often more gentle method to modify polymers (Guebitz and Cavaco-Paulo 2008). The family of cutinases, found in bacteria and fungi, is responsible for the degradation of cutin, a plant polyester (Chen et al. 2010a, b; Chen et al. 2008). It could be shown that some of these enzymes are able to cleave the ester bonds in PET (Fig. S1), which finally leads to the complete degradation of the plastic (Ronkvist et al. 2009). Moreover, it was shown that these enzymes can be adapted or are able to degrade other synthetic esters such as polycaprolactone, nylon and others (Araùjo et al. 2007; Kitadokoro et al. 2012; Thumarat et al. 2012). The currently best characterized polyester hydrolase is the fungal cutinase from Fusarium solani, for which numerous biochemical studies as well as several high-resolution structures are available (Longhi and Cambillau 1999). However, cutinases from thermophilic bacteria are of higher thermostability and show a greater resistance to detergents and organic solvents, which further enhance their potential for industrial processes (Chen et al. 2010a, b; Chen et al. 2008; Zimmermann and Billig 2011). The thermophilic bacterium Thermobifida fusca harbours two open reading frames for cutinases termed Tfu_0882 (TfCut1) and Tfu_0883 (TfCut2) (Chen et al. 2008). It has been shown that both proteins catalyze the breakdown of triolein and the artificial chromogenic substrate p-nitrophenylbutyrate (pNPB) with a temperature optimum around 60 °C (Chen et al. 2010a, b). TfCut2 was originally isolated from T. fusca KW3 with a high-PET hydrolytic activity (Herrero Acero et al. 2011). A detailed analysis of the substrate specificity shows that these cutinases prefer short chain substrates (Chen et al. 2010a, b). Both types of cutinases have a high tolerance towards different organic solvents including n-hexane or acetone, which makes them very well suitable for many industrial processes, for example, the production of biodiesel (Chen et al. 2010a, b). However, despite their high sequence identity of about 93 %, TfCut2 has a roughly two times higher activity towards PET compared to TfCut1 (Chen et al. 2010a, b). Recently, the first crystal structure of a bacterial cutinase Est 119 from Thermobifida alba was characterized (Kitadokoro et al. 2012). This enzyme belongs to the α/β-hydrolase superfamily with a typical chymotrypsin-like catalytic triad (Kitadokoro et al. 2012).
In this work, we present three crystal structures of TfCut2, in native as well as in an inactivated form with a phenylmethylsulfonyl fluoride (PMSF) moiety-bound to the active serine. Together with molecular modelling studies, we analyse the structural determinants for substrate and product specificity as well as the enzyme thermostability. The high-resolution structure provides a suitable starting point for further structure-based enzyme engineering attempts.
Materials and methods
Expression and purification
TfCut2 (ENA: FR727681) from T. fusca KW3 (DSM6013) was recombinantly expressed and purified by immobilized metal ion chromatography (IMAC) in the same manner as previously described for a carboxylesterase from the same strain (Oeser et al. 2010). Briefly, Escherichia coli Bl21 (DE3) cells containing the codon-optimized gene of TfCut2 (ENA: HG939556) were grown at 37 °C to an OD600 of 1.5. Protein production was induced in the presence of 0.5 mM IPTG at 18 °C for 14 h. Cells were harvested by centrifugation at 4,500xg and 4 °C for 45 min. Cells were resuspended in 50 mM phosphate buffer (pH 8) containing 300 mM NaCl and lysed by sonication. The cell debris was removed by centrifugation at 11,000xg for 30 min. The resulting supernatant containing soluble TfCut2 was applied on a Ni-NTA Superflow column (Qiagen, Hilden, Germany). The column was washed with the same buffer containing up to 50 mM imidazole. Afterwards, TfCut2 was eluted using a buffer containing 250 mM imidazole. TfCut2 containing fractions were pooled and stored at 4 °C until further usage.
Esterase activity assay
The esterolytic activity was assayed with para-nitrophenylbutyrate (pNPB) as substrate in a 96-well microplate format using the PowerWave™ XS universal microplate reader (BioTek Instruments Inc., Winooski, USA) for detection. Eighty microlitres of 100 mM phosphate buffer (pH 8) and 10 μL of enzyme solution were mixed, and the reaction was started by adding 10 μL of 5 mM pNPB dissolved in absolute ethanol to each well. The absorbance change of each sample was monitored at 405 nm over a time course of 5 min. The activity was determined by the formation of para-nitrophenol (pNP) using an empirically determined extinction coefficient of 17,000 M−1 cm−1 for pNP at pH 8.
The thermal stability of TfCut2 was assayed by incubating the enzyme at temperatures ranging from 25 to 79 °C at a concentration of 250 μg/mL in 100 mM phosphate buffer at pH 8 in a final volume of 100 μL for 1 h. Afterwards, the samples were cooled on ice for 15 min before the esterolytic activity was determined in triplicate as described above. All data for TfCut2 were normalized to 100 % activity at 25 °C.
Circular dichroism spectroscopy
Circular dichroism spectra (CD spectra) of TfCut2 were recorded using a Jasco J-715 spectropolarimeter (JASCO, Easton, USA). Protein samples of 10 μM were prepared in 50 mM sodium borate buffer (pH 8.5) in quartz cuvettes with a path length of 2 mm (Hellma, Jena, Germany) and measured as described (Haack et al. 2008). The raw CD signal was converted into mean residue ellipticity in deg cm2/dmol as described before (Greenfield 2004). The thermal denaturation process was monitored by the ellipticity changes at 220 nm as a function of temperature from 20 to 100 °C in 0.1 °C steps. The apparent melting point (T m ) was estimated by nonlinear fitting using a double Boltzman fit as implemented in Origin (Origin Labs).
Molecular docking
The docking program GOLD version 5.1 (Cambridge Crystallographic Data Centre, Cambridge, UK) (Jones et al. 1997) was used to explore the possible binding mode for a PET substrate. A model substrate called ‘2PET’ consisting of 2 repeating units of PET polymer was prepared using MOE (Chemical Computing Group, Montreal, Canada). The central ester bond of 2PET was constrained in the oxyanion hole formed by Met131 and Tyr60 with the correct orientation to form the tetrahedral intermediate. The other atoms of 2PET were allowed to be flexible for a reasonable conformation docked to the rigid protein structural models by a genetic algorithm (Jones et al. 1997). The top-ranked docking conformations of 2PET based on the default scoring function of GOLD were selected for further investigation.
Molecular dynamics simulation
Molecular dynamics (MD) simulation was carried out using GROMACS version 4.6 package (Groningen University, Groningen, Netherlands) employing Amber99SB force fields (Hornak et al. 2006). The crystal structures of TfCut2 and T. alba Est119 (protein data bank (PDB)-ID: 3VIS) were centred in a cubic box containing explicit water molecules with a distance of ≥1.0 nm from the box edge as the starting structures. The steepest descent method was applied to perform the energy minimization until the maximum force (F max) of less than 1,000 kJ/mol/nm was reached. The system was equilibrated for 100 ps by a position-restrained simulation at desired temperatures in the isothermal-isobaric (NPT) ensemble. The isotropic pressure coupling using the Berendsen algorithm was applied with a reference pressure of 1.0 bar (Berendsen et al. 1984). Each simulation was carried out three times under the same condition as that for equilibration for 50 ns in steps of 2-fs at 298 K (25 °C) and 353 K (80 °C), respectively. To analyse the thermostability of TfCut2 and T. alba Est119, the time course of root mean square deviation (rmsd) of the backbone structures and the root mean square fluctuation (rmsf) of each amino acid residue were calculated using the GROMACS 4.6 package.
Crystallization
Prior to the crystallization, the protein buffer was exchanged by size exclusion chromatography against 10 mM Tris–HCl (pH 7.2). The monodispersity of the sample was assayed using dynamic light scattering. Initial crystallization conditions were found using in-house custom-made crystallization screens similar to commercially available screens. Crystallization setups were performed by mixing equal amounts of protein with reservoir solution in 96-well CrystalQuick plate (Greiner Bio-One). Plates were stored at 292 K and examined regularly for crystal formation. Optimization of initial hits was carried out in 24-well crystallization plates using the hanging drop vapour diffusion method. Best crystals were grown in 0.1 M sodium acetate at pH 4.0–4.6, 0.2 M ammonium sulphate, and 25–35 % PEG MME 2000.
Data collection, processing and refinement
Data were collected on a Rayonix MX225 detector at 100 K using synchrotron radiation at beamline 14.1 at BESSY (Berlin) and processed using XDS (Kabsch 2010) or MOSFLM/SCALA (Evans 2006; Leslie 1992). The phase problem was solved by molecular replacement using PHASER (Vagin and Teplyakov 2010). An initial solution was found using a poly-Ala model of Streptomyces exofoliatus lipase as deposited in the protein data bank (PDB-ID: 1JFR (Wei et al. 1998)). Room temperature data collection was carried out on a BrukerMicrostar rotating anode equipped with a Mar345 image plate system. Data were processed as described for the data collected at the synchrotron.
Refinement was carried out using REFMAC (Murshudov et al. 2011) or PHENIX (Adams et al. 2010) in conjunction with manual rebuilding and real space refinement in COOT (Emsley et al. 2010). The quality of the final models was evaluated with MOLPROBITY (Chen et al. 2010a, b), WHATIF (Hooft et al. 1996) and PROCHECK (Laskowski et al. 1993). All relevant data collection and refinement statistics are found in Table 1. The final structures and maps are deposited in the PDB under the accession codes 4cg1, 4cg2 and 4cg3.
Molecular graphics
Molecular figures of the structural assembly were prepared using PYMOL (Schrödinger, LLC, Munich, Germany). The analysis and visualization of the substrate-protein interaction was carried out with LIGPLOT (Wallace et al. 1995).
Results
Overall structure
The structure of TfCut2 was determined using molecular replacement with a poly-Ala model of S. exofoliatus lipase as deposited in the protein data bank (PDB-ID: 1JFR, 63 % sequence identity) (Wei et al. 1998). This structure was the most similar structure available at the time of structure determination. Strong difference density was observed for the missing side chains which allowed an unambiguous assignment of the whole sequence. In addition, up to two amino acids from the C-terminal tag extension are visible in the density. The enzyme forms a classical α/β-hydrolase fold with a central nine-stranded β-sheet flanked by 11 α-helices on both sides. The catalytic triad, comprising S130, D176 and H208, is located in a crevice on the surface of the enzyme (Fig. 1a).
a Cartoon representation of TfCut2 with α-helices in red and β-strands in green. The active site residues are shown in stick representation. b Structure-based superposition of TCut2 in fawn and Est119 in green. The catalytic triad and the characteristic disulphide bridge are shown in stick representation. c Structure-based superposition of TfCut2 (fawn) and the Fusarium solani cutinase (brown). For both cutinases, the active site residues are shown in stick representation. The characteristic disulphide bond of TfCut2 is shown as sticks as well (Colour figure online)
Thermal stability
The most desirable property of cutinases from thermostable bacteria is their outstanding thermostability. This offers an important advantage for an industrial application, as many processes dealing with PET are performed at elevated temperatures (Liu et al. 2009). Thus, enzymes with an enhanced thermal stability are of high economic interest. We have determined the apparent melting temperature of TfCut2 to be 70 °C using temperature-dependent CD spectroscopy (Fig. 2a). However, activity measurements indicate a loss of activity already at 61 °C (Fig. 2b).
Thermostability of TfCut2. a Thermal denaturation curve of TfCut2 measured by temperature-dependent CD spectroscopy. The respective fit curve for the determination of the melting point is shown in red. b Residual activity of TfCut2 towards pNPB after 1-h incubation at different temperatures. The error bars represent the standard deviation of triplicate measurements (Colour figure online)
To identify determinants for the high-temperature stability of TfCut2, we analysed the atomic mobility of a structure determined at room temperature and refined with phenix ensemble refinement (Burnley et al. 2012). In addition, molecular dynamics calculations were employed to study the protein dynamics of TfCut2. The overall structure of TfCut2 showed very low B-factors, indicating a high rigidity of the structure and a low positional disorder of the protein in the crystal. An exception is a highly flexible region around the amino acids 245 to 247 (Fig. 3a), which made it necessary to fit multiple conformations in the ensemble refinement (Fig. 3b). The same region showed also a high flexibility in the molecular dynamics simulations (Fig. 3c).
Atomic mobility of TfCut2. a Tube representation of the B-factor distribution of TfCut2. The thickness of the tube and the colour of the backbone represent the B-factor distribution of the main-chain atoms of TfCut2. b Tube representation of TfCut2 visualizing the different conformations found by an ensemble refinement with phenix.refine. The thickness of the tube represents the number of models used to fit the data and therefore reflects the flexibility of the structure. Residues which differ in sequence between TfCut2 and 1 are shown in stick representation. The active site residues S130 and H208, defining the location of the active site, are also shown in yellow. c Visualization of the rmsf of the MD simulations for TfCut2 (black) and T. alba Est119 (cyan). Both structures show a similar pattern of flexible hot spots, but the variability is lower for TfCut2. The curves represent the mean of three independent simulations (Colour figure online)
Inhibitor complex
In order to get a deeper insight into enzyme-substrate interactions, we tried to modify the enzyme using different types of covalent binding inhibitors like PMSF, phosphonates and phosphinates. So far, except for PMSF, no complex with a phosphonate or phosphinate resembling the more natural polymeric substrate could be obtained. However, the PMSF complex revealed covalent modification of S130 and binding of one of the oxygens of the tetrahedral adduct in the oxyanion hole formed by the main chain nitrogens of M131 and Y60 (Fig. 4a). The phenyl ring of the PMSF moiety is mainly stabilized by hydrophobic interactions with I178 and to a lesser extent with Y60 (Fig. 4b).
Crystal structure of the TfCut2xPMS complex. a View in to the active site with PMS bound to the catalytic serine. The protein backbone is shown as cartoon. The catalytic triad as well as the oxyanion-hole-forming residues are shown in stick representation. The respective interactions are shown as dotted lines. The observed electron density map (2Fo-Fc) for the PMSF-S130 adduct is shown in mesh representation contoured at 1σ. b Scheme of the interactions between the covalently bound PMS moiety and interacting residues
Discussion
Structural similarity
A DALI (Holm and Rosenström 2010) search for homologous structures showed that TfCut2 has a high similarity to the recently structurally characterized thermostable cutinase EST119 from T. alba (PDB-ID: 3VIS, 82 % sequence identity), which also exhibits activity with several polymeric substrates (Thumarat et al. 2012) (Fig. 1b). However, the activity towards PET remains elusive. A high similarity is also shown for the S. exofoliatus lipase (PDB-ID: 1JFR, 63 % sequence identity), which was used as a molecular replacement model to avoid model bias (Wei et al. 1998). Numerous other esterases and lipases share a high structural similarity due to the conserved α/β-hydrolase fold (Table 2). No fungal cutinase was found to be homologous to TfCut2 in the DALI search, although these enzymes also have an α/β-hydrolase fold and are classified as cutinases due to their shared catalytic properties. The most pronounced differences in contrast to structurally characterized cutinases, such as the F. solani cutinase (PDB-ID: 1AGY (Longhi et al. 1997)), are located in the N- and C-terminal regions as well as a disulfide bridge only shared by bacterial cutinases, whereas the core fold and the catalytic triad is conserved (Fig. 1c).
Thermostability
The observed thermostability of TfCut2 outperforms all characterized cutinases so far. A higher thermostability was only reported for the fungal cutinase from Humicola insolens, which shows significant activity even at 85 °C (Baker et al. 2012), whereas the closest structural homolog, T. alba EST119, was inactivated within 1 h at 50 °C (Thumarat et al. 2012). In contrast to the T. alba Est119 (Thumarat et al. 2012), no sign for a metal dependence of the thermostability was found for TfCut2 (Thumarat et al. 2012; Chen et al. 2010a, b). As for the Est119 structure, no bound metal ion could be detected in the crystal structure of TfCut2, and a characteristic sequence motif for metal binding was not present. The unusual metal-dependent stability of the T. alba Est119 is so far unique and could not be explained by the structural data obtained. In our MD simulations, TfCut2 showed lower root mean square fluctuation (rmsf) values for the backbone compared to EST119 at 80 °C (Fig. 3c). Especially, the N-terminal region has a higher atomic mobility compared to TfCut2. Despite a fourfold increase in the thermostability the Est119_S219P mutant still did not outrival the thermostability of TfCut2 (Thumarat et al. 2012). In general, the secondary structure content, the content of hydrophobic amino acids, the number of disulfide bonds, the number salt bridges and the number of hydrogen bonds are thought to be responsible for the additional stability of thermostable proteins (Chakravarty and Varadarajan 2002). The disulfide bridge between Cys241 and Cys259 in TfCut2 is located in close vicinity to the highly flexible region around the amino acids 245 to 247 (Fig. 3) and may stabilize this region, thus preventing denaturation at higher temperatures. The stabilization of proteins through disulfide bonds is a common feature in thermophilic organisms and is already successfully implemented in strategies to enhance the thermostability of proteins (Han et al. 2009; Kim and Ishikawa 2013; Porcelli et al. 2012). However, this disulfide bond is also present in T. alba Est119 and, therefore, this cannot be the only reason for the difference in their thermostability. The regions forming the boundary of the active site, mainly, residues 55 to 65, 175 to 180 and 208 to 211, showed also a higher flexibility compared to the overall structure, which may allow some induced fit motions necessary for catalysis. Such flexible hot spots might be prone to local unfolding and might explain the different apparent melting temperatures obtained by CD and activity assays. Other factors like secondary structure content or amino acid composition do not differ significantly between both cutinases. However, specific sequence alterations like the point mutation S219P in EST119 can have a high impact on the thermostability, probably due to a stabilization of the local structure. Indeed the equivalent residue in TfCut2 is already a proline (P180), proofing the importance of this mutation for the thermostability. Interestingly, despite the higher intrinsic rigidity of proline, no differences were observed in the molecular dynamics simulations for this region. Overall TfCut2 showed a slightly improved aliphatic index (86.8) compared to Est119 (74.5), which might result in a stabilized structure (Ikai 1980). Despite having only a marginally higher aliphatic index of 75.2, Est119_A68V/S218P shows a significantly higher thermal stability at 60 °C (Thumarat et al. 2012). Furthermore, an optimized hydrogen bond network within TfCut2 (138 internal hydrogen bonds) in comparison to T. alba Est119 (129 internal hydrogen bonds) might be also an important contributing factor to the higher stability of TfCut2.
Substrate specificity
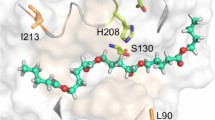
Currently, all studies on the binding mode of the polymeric substrates are based on docking studies to models of cutinases, based on an acetylhydrolase from the mesophilic S. exofoliatus (PDB-ID: 1JFR) (Wei et al. 1998). The proposed binding of PET involves regions found to be important for PET hydrolysis based on mutagenesis studies and, therefore, the chain was extended on the surface towards these sites (Herrero Acero et al. 2011). To get a more detailed understanding of the enzyme-substrate interactions, we docked a substrate composed of two PET monomers into the final model of TfCut2 (Fig. 5). The docking placed the PET dimer in a position superimposable with the PMSF moiety on the donor subsites and in a position suitable for a nucleophilic attack of the serine on the carbon of the scissile bond. The subsites are formed mainly by slightly hydrophobic and small polar residues, well suited to interact with the amphiphilic substrate. The same alternating pattern is found for the substrate groove of Est119 and might be a general feature of cutinases with activity towards PET (Kitadokoro et al. 2012). The active site groove shows a kink, which brings the acceptor subsites in an angle of roughly 130° in respect to the donor subsites. A prominent aromatic clamp, formed by Y60 and W155, is part of the first acceptor subsite +1. Due to the rigidity of the substrate ester bond, the docked molecule remains in an extended conformation, not following the observed kink in the substrate groove. It remains elusive if the hydrophobic clamp has a similar role like sugar tongs in glycosylhydrolases by sandwiching the terephthalic acid moiety on the acceptor subsite, thereby destabilizing the ground state of the substrate (Bozonnet et al. 2007). Further additional binding sites are difficult to predict due to the rather flat surface of TfCut2 with no pronounced binding cleft for the linear substrate. Hence, it is difficult to attribute the observed sequence differences, which are not located close to the active site to specific interactions with the substrate. Some of the amino acids different in the two T. fusca cutinases are facing the interior of the protein and may rather influence protein dynamics than specific surface characteristics. An influence of some mutations in the interior of the protein on the dynamic behaviour was also suggested by Herrero Acero et al. (2013) for the Thermobifida cellulosilytica isoenzymes.
Surface representation of TfCut 2 with a docked 2PET moiety (blue carbon atoms, three independently docked conformations are shown) within the substrate groove in stick representation. The PMS inhibitor from the TfCut2xPMS co-crystal structure is superimposed in yellow. The position of the active serine is marked on the surface in red (Colour figure online)
One of the remaining questions for bacterial cutinases from different Thermobifida strains is the cause for the pronounced differences in the activity and affinity towards PET, despite their high sequence similarity (Fig. S2). The differing amino acids are mostly not located in close vicinity to the active site (Fig. 3b). However, nearly all these amino acids cluster around the flexible N-terminal region or are in secondary structure elements in close vicinity (Fig. 3b). A second cluster, the amino acids 182 and 186, is in a helix directly following the loop comprising amino acids 175 to 180, which is a part of the active site boundary. Indeed, mechanistic studies on the two isoenzymes from T. cellulosilyticus have shown that mutations in the N-terminal region are important for proper substrate interactions, whereas the mutations around the 180-loop enhance the reaction but do not influence the interaction with the substrate (Herrero Acero et al. 2013). Variants of the T. alba Est119 with improved activity, generated by random mutagenesis, showed also a distinct pattern with the mutations mainly located in the N-terminal region (Thumarat et al. 2012). The most active variant of Est119, A68V, resembles the wild-type sequence of TfCut2 (V29). An improved hydrophobic pattern might alter the interaction pattern within the beta-sheet structure and thereby influence the internal dynamics of the enzyme leading to a more active enzyme (Herrero Acero et al. 2013; Thumarat et al. 2012).
In summary, we have determined the structure of the thermostable bacterial cutinase TfCut2 able to degrade PET with high efficiency. Our analysis suggests that an optimized sequence pattern, the optimized hydrogen bond network and probably the disulfide bond are important factors in TfCut2 for the thermostability of the enzyme. A complex with PMSF highlights the key active site residues and serves as a suitable starting point for further docking and studies to get a deeper insight in key elements responsible for the outstanding activity of TfCut2 towards PET. Furthermore, the structure opens up the potential for a rational design of the enzyme towards different polymeric substrates, which could lead to a family of designer enzymes for industrial plastic modification and degradation.
References
Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy A, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH (2010) PHENIX: a comprehensive python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66(Pt 2):213–221. doi:10.1107/S0907444909052925
Araùjo R, Silva C, O’Neill A, Micaelo N, Guebitz G, Soares CM, Casal M, Cavaco-Paulo A (2007) Tailoring cutinase activity towards polyethylene terephthalate and polyamide 6,6 fibers. J Biotechnol 128(4):849–857. doi:10.1016/j.jbiotec.2006.12.028
Baker PJ, Poultney C, Liu Z, Gross R, Montclare JK (2012) Identification and comparison of cutinases for synthetic polyester degradation. Appl Microbiol Biotechnol 93(1):229–240. doi:10.1007/s00253-011-3402-4
Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR (1984) Molecular dynamics with coupling to an external bath. J Chem Phys 81(8):3684–3690, http://link.aip.org/link/?JCP/81/3684/1
Bozonnet S, Jensen M, Nielsen M, Aghajari N, Jensen M, Kramhoft B, Willemoes M, Tranier S, Haser R, Svensson B (2007) The ‘pair of sugar tongs’ site on the non-catalytic domain c of barley-amylase participates in substrate binding and activity. FEBS J 274(19):5055–5067, PM:17803687
Brueckner T, Eberl A, Heumann S, Rabe M, Guebitz GM (2008) Enzymatic and chemical hydrolysis of poly(ethylene terephthalate) fabrics. J Polym Sci A Polym Chem 46(19):6435–6443. doi:10.1002/pola.22952
Burnley BT, Afonine PV, Adams PD, Gros P (2012) Modelling dynamics in protein crystal structures by ensemble refinement. Elife 1:e00311. doi:10.7554/eLife.00311
Chakravarty S, Varadarajan R (2002) Elucidation of factors responsible for enhanced thermal stability of proteins: a structural genomics based study. Biochemistry 41(25):8152–8161
Chen W, McCarthy TJ (1998) Chemical surface modification of poly(ethylene terephthalate). Macromolecules 31(11):3648–3655. doi:10.1021/ma9710601
Chen S, Tong X, Woodard RW, Du G, Wu J, Chen J (2008) Identification and characterization of bacterial cutinase. J Biol Chem 283(38):25854–25862
Chen S, Su L, Billig S, Zimmermann W, Chen J, Wu J (2010a) Biochemical characterization of the cutinases from Thermobifida fusca. J Mol Catal B Enzym 63:121–127, http://www.sciencedirect.com/science/article
Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC (2010b) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66(Pt 1):12–21. doi:10.1107/S0907444909042073
Emsley P, Lohkamp B, Scott WG, Cowtan K (2010) Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66(Pt 4):486–501. doi:10.1107/S0907444910007493
Evans P (2006) Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr 62(Pt 1):72–82
Greenfield NJ (2004) Analysis of circular dichroismdata. Methods Enzymol 383:282–317. doi:10.1016/S0076-6879(04)83012-X
Guebitz GM, Cavaco-Paulo A (2008) Enzymes go big: surface hydrolysis and functionalization of synthetic polymers. Trends Biotechnol 26(1):32–38. doi:10.1016/j.tibtech.2007.10.003
Haack M, Enck S, Seger H, Geyer A, Beck-Sickinger AG (2008) Pyridone dipeptide backbone scan to elucidate structural properties of a flexible peptide segment. J Am Chem Soc 130(26):8326–8336. doi:10.1021/ja8004495
Han Z-l, Han S-Y, Zheng S-Y, Lin Y (2009) Enhancing thermostability of a Rhizomucor miehei lipase by engineering a disulfide bond and displaying on the yeast cell surface. Appl Microbiol Biotechnol 85(1):117–126. doi:10.1007/s00253-009-2067-8
Herrero Acero E, Ribitsch D, Steinkellner G, Gruber K, Greimel K, Eiteljoerg I, Trotscha E, Wei R, Zimmermann W, Zinn M, Cavaco-Paulo A, Freddi G, Schwab H, Guebitz G (2011) Enzymatic surface hydrolysis of PET: effect of structural diversity on kinetic properties of cutinases from Thermobifida. Macromolecules 44(12):4632–4640. doi:10.1021/ma200949p
Herrero Acero E, Ribitsch D, Dellacher A, Zitzenbacher S, Marold A, Steinkellner G, Gruber K, Schwab H, Guebitz GM (2013) Surface engineering of a cutinase from Thermobifida cellulosilytica for improved polyester hydrolysis. Biotechnol Bioeng. doi:10.1002/bit.24930
Holm L, Rosenström P (2010) Dali server: conservation mapping in 3D. Nucleic Acids Res 38(Web Server issue):W545–W549. doi:10.1093/nar/gkq366
Hooft RW, Vriend G, Sander C, Abola EE (1996) Errors in protein structures. Nature 381(6580):272. doi:10.1038/381272a0
Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C (2006) Comparison of multiple amber force fields and development of improved protein backbone parameters. Proteins 65(3):712–725. doi:10.1002/prot.21123
Ikai A (1980) Thermostability and aliphatic index of globular proteins. J Biochem 88(6):1895–1898
Jones G, Willett P, Glen RC, Leach AR, Taylor R (1997) Development and validation of a genetic algorithm for flexible docking. J Mol Biol 267(3):727–748. doi:10.1006/jmbi.1996.0897
Kabsch W (2010) XDS. Acta Crystallogr D Biol Crystallogr 66(Pt 2):125–132, PM:20124692
Kim H-W, Ishikawa K (2013) The role of disulfide bond in hyperthermophilic endocellulase. Extremophiles 17(4):593–599. doi:10.1007/s00792-013-0542-8
Kitadokoro K, Thumarat U, Nakamura R, Nishimura K, Karatani H, Suzuki H, Kawai F (2012) Crystal structure of cutinase Est119 from Thermobifida alba AHK119 that can degrade modified polyethylene terephthalate at 1.76 Å resolution. Polym Degrad Stab 97(5):771–775, http://www.sciencedirect.com/science/article
Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK—a program to check the stereochemical quality of protein structures. J Appl Cryst 26:283–291
Leslie AGW (1992) Recent changes to the MOSFLM package for processing film and image plate data, Joint CCP4 + ESF-EAMCB Newsletter on Proteincrystallography 26
Liu Z, Gosser Y, Baker PJ, Ravee Y, Lu Z, Alemu G, Li H, Butterfoss GL, Kong X-P, Gross R, Montclare JK (2009) Structural and functional studies of Aspergillus oryzae cutinase: enhanced thermostability and hydrolytic activity of synthetic ester and polyester degradation. J Am Chem Soc 131(43):15711–15716. doi:10.1021/ja9046697
Longhi S, Cambillau C (1999) Structure-activity of cutinase, a small lipolyticenzyme. Biochim Biophys Acta 1441(2–3):185–196
Longhi S, Czjzek M, Lamzin V, Nicolas A, Cambillau C (1997) Atomic resolution (1.0 Å) crystal structure of Fusarium solani cutinase: stereochemical analysis. J Mol Biol 268(4):779–799. doi:10.1006/jmbi.1997.1000
Murshudov GN, Skubák P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA (2011) REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr 67(Pt 4):355–367. doi:10.1107/S0907444911001314
Oeser T, Wei R, Baumgarten T, Billig S, Föllner C, Zimmermann W (2010) High level expression of a hydrophobic poly(ethylene terephthalate)-hydrolyzing carboxylesterase from Thermobifida fusca KW3 in Escherichia coli BL21(DE3). J Biotechnol 146(3):100–104. doi:10.1016/j.jbiotec.2010.02.006
Porcelli M, De Leo E, Del Vecchio P, Fuccio F, Cacciapuoti G (2012) Thermal unfolding of nucleoside hydrolases from the hyperthermophilic archaeon Sulfolobus solfataricus: role of disulfidebonds. Protein Pept Lett 19(3):369–374
Riccardi C, Barni R, Selli E, Mazzone G, Massafra MR, Marcandalli B, Poletti G (2003) Surface modification of poly(ethylene terephthalate) fibers induced by radio frequency air plasma treatment. Appl Surf Sci 211:386–397, http://www.sciencedirect.com/science/article
Ronkvist M, Xie W, Lu W, Gross RA (2009) Cutinase-catalyzed hydrolysis of poly(ethylene terephthalate). Macromolecules 42(14):5128–5138. doi:10.1021/ma9005318
Thumarat U, Nakamura R, Kawabata T, Suzuki H, Kawai F (2012) Biochemical and genetic analysis of a cutinase-type polyesterase from a thermophilic Thermobifida alba AHK119. Appl Microbiol Biotechnol 95(2):419–430. doi:10.1007/s00253-011-3781-6
Vagin A, Teplyakov A (2010) Molecular replacement with MOLREP. Acta Crystallogr D Biol Crystallogr 66(Pt 1):22–25
Wallace AC, Laskowski RA, Thornton JM (1995) LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng 8(2):127–134
Wei Y, Swenson L, Castro C, Derewenda U, Minor W, Arai H, Aoki J, Inoue K, Servin-Gonzalez L, Derewenda ZS (1998) Structure of a microbial homologue of mammalian platelet-activating factor acetylhydrolases: Streptomyces exfoliatus lipase at 1.9Å resolution. Structure 6(4):511–519
Zimmermann W, Billig S (2011) Enzymes for the biofunctionalization of poly(ethylene terephthalate). Adv Biochem Eng Biotechnol 125:97–120. doi:10.1007/10_2010_87
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 517 kb)
Rights and permissions
About this article
Cite this article
Roth, C., Wei, R., Oeser, T. et al. Structural and functional studies on a thermostable polyethylene terephthalate degrading hydrolase from Thermobifida fusca . Appl Microbiol Biotechnol 98, 7815–7823 (2014). https://doi.org/10.1007/s00253-014-5672-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-014-5672-0