Abstract
Malonyl-CoA is the essential building block of natural products such as fatty acids, polyketides, and flavonoids. Engineering the biosynthesis of fatty acids is important for biofuel production while that of polyketides provides precursors of medicines and nutritional supplements. However, microorganisms maintain a small amount of cellular malonyl-CoA, which could limit production of lipid and polyketides under certain conditions. Malonyl-CoA concentration is regulated by multiple pathways and signals, and changes in intracellular malonyl-CoA often lead to complex alterations in metabolism. In the present work, overexpression of a plant malonyl-CoA synthetase gene (AAE13) in Saccharomyces cerevisiae resulted in 1.6- and 2.4-fold increases in lipid and resveratrol accumulation simultaneously. We also demonstrated that AAE13 partially complemented the temperature-sensitive acc1 mutant, replacing this key enzyme in central metabolism. Mechanistic analysis by CoA quantification and transcriptomic measurement suggested that increases in malonyl-CoA concentration were coupled with drastic reductions in other major CoA compounds and clear suppression of tricarboxylic acid cycle-related genes. These results suggest that malonyl-CoA is a critical target for fatty acid and polyketide engineering and that overexpression of malonyl-CoA synthetic enzymes needs to be combined with upregulation of CoA synthesis to maintain metastasis of central metabolism.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Malonyl-CoA is a ubiquitous metabolic intermediate in all living organisms. It is the main elongation unit during fatty acid biosynthesis. In both prokaryotic and eukaryotic cells, the fatty acid synthase complex can repeatedly incorporate malonyl-CoA into the elongating fatty acid chain (Tehlivets et al. 2007). Most fatty acids can be converted into triacylglycerides through the Kennedy pathway, after which they serve as the building blocks of lipid bilayers.
Malonyl-CoA has other biological functions and can be formed within the mitochondria, peroxisomes, and cytosol of eukaryotic cells (Saggerson 2008). In mammals, malonyl-CoA plays an important role in regulating ketogenesis and body fatty acid metabolism. This regulation is especially active in the hypothalamus, heart, and liver, where it influences calorie intake, myocardial infarction, and steatosis (Folmes and Lopaschuk 2007).
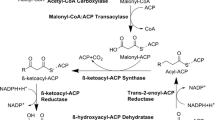
As the main generator of cellular malonyl-CoA, acetyl-CoA carboxylases (ACCs) are the rate-limiting enzymes in fatty acid biosynthesis in microbes, plants, and animals (Ekhterae et al. 1996; Schneiter et al. 2000; Shintani et al. 1997). ACCs use a covalently linked biotin cofactor to bind a carboxyl anion and transfer it to acetyl-CoA, producing malonyl-CoA (Fig. 1). Structural biology and extensive small molecule inhibitor studies have provided many insights into the catalytic mechanisms of ACCs (Tong 2005). In yeast, cytoplasmic ACC1 is encoded by a single gene as a 2233 amino acid-long product, and ACC1 deletion is lethal and cannot be rescued by external long-chain fatty acids (Hasslacher et al. 1993). HFA1, comprising a mitochondrial targeting signal, encodes a specific mitochondrial acetyl-CoA carboxylase providing malonyl-CoA for intra-organellar fatty acid synthesis. HFA1 with the ACC1 promoter complemented ACC1-defective yeast mutants to wild-type levels (Hoja et al. 2004).
Pathway of malonyl-CoA-related metabolism and resveratrol biosynthesis in engineered yeast. The key enzymes are shown in red. AAE13 and 4CL were isolated from Arabidopsis thaliana, and STS from Vitis vinifera. PDC, pyruvate decarboxylase; ALDH, aldehyde dehydrogenase; ACS, acetyl-CoA synthetase; ACL, ATP:citrate lyase; ACC1, acetyl-CoA carboxylase; AAE13, malonyl-CoA synthetase; FAS, fatty acids synthase; 4CL, 4-coumarate:CoA ligase; STS, stilbene synthase
Plants, in addition to ACCs, contain the enzyme malonyl-CoA synthetase, which ligates malonic acid and CoA to generate malonyl-CoA directly (Fig. 1). Chen et al. (2011) first reported the cloning and functional characterization of this enzyme from Arabidopsis (AAE13). Recombinant malonyl-CoA synthetase showed high activity against malonic acid but little or no activity against other dicarboxylic or fatty acids. Removal of excessive malonic acid is critical to plants. Since aae13-null mutants grew poorly and accumulated malonic acid, AAE13 was thought to be involved in the detoxification of short chain organic acids (Chen et al. 2011).
Additionally, malonyl-CoA is involved in the biosynthesis of many defensive phytochemicals. One major group of such compounds in all higher plants is the polyketides. These compounds are formed by polyketide synthase through repeated addition of various numbers of malonyl-CoA to a starter molecule. For example, flavonoids are formed by adding three molecules of malonyl-CoA to a coumaroyl-CoA starter, which is catalyzed by chalcone synthase and forms naringenin chalcone. Within the plant kingdom, a few families evolved chalcone synthase into stilbene synthase. Stilbene synthase condenses the same starting material, one coumaroyl-CoA and three malonyl-CoA, into resveratrol (Fig. 1) (Halls and Yu 2008).
Flavonoids, resveratrol, and fatty acids are important metabolic engineering targets. Flavonoids are potent antioxidants. Resveratrol is a compound that can boost longevity of many organisms (Wang et al. 2010). Additionally, fatty acids and related compounds are important nutraceuticals and biofuels. Many efforts have been dedicated to increase the yield of these compounds in engineered microorganisms. Among the different approaches, increasing cellular malonyl-CoA concentration was thought to be critical (Fowler et al. 2009; Miyahisa et al. 2005; Xu et al. 2011, 2013; Zha et al. 2009). However, results from overexpression of ACC have been inconsistent (Davis et al. 2000; Ruenwai et al. 2009; Shin et al. 2012). Although overexpression of ACC genes of different origins mostly led to insignificant changes in fatty acid accumulation, there were a few positive reports. Overexpression of the Mucor rouxii ACC1 in the non-oleaginous yeast showed an overall enhancement in fatty acid production, where the highest accumulation resulted in a 40 % increase in total fatty acid content (Ruenwai et al. 2009). In Escherichia coli, overexpressing ACC resulted in a 100-fold increase in ACCase activity and a 6-fold increase in the rate of fatty acid synthesis (Davis et al. 2000). In these reports, despite significantly increased ACCase activity, fatty acid content remained low. There may be feedback inhibition of the ACC enzyme by palmitoyl-CoA, the product of fatty acid synthase in yeast.
Recently, bacterial malonyl-CoA synthetase has been used in engineering fatty acid and flavonoid biosynthesis. For example, Leonard et al. (2008) demonstrated that overexpression of malonyl-CoA synthetase (MatB) and malonate carrier protein (MatC) with feeding malonate increased intracellular levels of malonyl-CoA and increased the titer of flavonoids in E. coli. Similarly, Park et al. (2011) used the same combination of the MatB/MatC genes in Streptomyces venezuelae and increased the production of heterologous flavonoids by up to sevenfold. Clearly, increasing malonyl-CoA supply may lead to enhanced polyketide accumulation, even though the detailed mechanism has not been analyzed.
Here, we report that overexpression of plant malonyl-CoA synthetase gene, AAE13, in Saccharomyces cerevisiae led to significant increases in both heterologous resveratrol accumulation and endogenous triacylglycerol accumulation. For the first time, the results show plant malonyl-CoA synthetase complementing a yeast ACC1 mutant. To delineate the mechanisms of these observations, the transcriptome and intracellular CoA distribution were comprehensively analyzed. We discovered that yeast cells maintained the total intracellular CoA at a constant level. We hypothesize that redirecting the CoA pool toward one particular pathway may deplete the CoA supply of other pathways. These results may provide an explanation for the previous inconsistent malonyl-CoA engineering results and suggest a new direction for engineering fatty acid biofuel biosynthesis in microorganisms.
Materials and methods
Chemicals
Standard chemicals including coenzyme A, acetyl-CoA, malonyl-CoA, succinyl-CoA and n-propionyl-CoA, N,N-dimethylbutyl amine (DMBA), diethyl ether and methanolic HCl (hydrochloric acid in methanol), and 3 N were purchased from Sigma-Aldrich (St Louis, MO, USA). Acetonitrile (HPLC grade) was purchased from EMD Chemicals (Gibbstown, NJ, USA). Trichloroacetic acid (10 % TCA) was purchased from BioWorld (Dublin, OH, USA). All enzymes were from New England Biolabs (Ipswich, MA, USA).
Strains and cultures
S. cerevisiae WAT11 (Urban et al. 1997) was used as the host strain. The temperature-sensitive mutant acc1-7-1 was kindly provided by Dr. Alan M. Tartakoff at the Case Western Reserve University (Cleveland, USA). WAT11 cells were grown at 30 °C in a shaker at 250 rpm in synthetic dextrose (SD) dropout medium containing 1.7 g/L Bacto-yeast nitrogen base without amino acids and ammonium sulfate, 20 g/L d-glucose, 5 g/L ammonium sulfate, and 1.4 g/L yeast synthetic drop-out medium supplements (Sigma-Aldrich, St Louis, USA). Depending on the nutrient requirement of strains, 20 mg/L histidine, 100 mg/L leucine, 50 mg/L tryptophan, and 40 mg/L uracil were added into medium. Plasmid vectors were transformed into yeast with the lithium acetate method using YeastmakerTM II (Clontech, Mountain View, CA, USA).
Plasmid constructions
The yeast expression vectors used to express AAE13 was constructed as follows: AAE13 (At3g16170) was amplified from the vector pENTR-AAE13 (Chen et al. 2011) with the AAE13-specific primers AAE13 forward (5′-ATGGAAGTGTTTAAAGCAGCTTTTTC-3′) and AAE13 reverse (5′-TTATTCTTGATTTTCCAGAGATTTC-3′). The amplified product was cloned into pCR®8⁄GW⁄TOPO® (Life Technologies, Carlsbad, USA). The resulting plasmid was combined with the Advanced Gateway destination vectors pAG305GPD-ccdB, pAG306GPD-ccdB, and pAG426GPD-ccdB (Addgene, Boston, USA) in an attL × attR recombination reaction to generate the vectors pAG305GPD-AAE13, pAG306GPD-AAE13, and pAG426GPD-AAE13. The vectors pAG305GPD-AAE13 and pAG306GPD-AAE13 contain an integrative recombination site and an expression cassette under the control of a constitutive promoter (GPD). The integrating vector containing AAE13 was introduced into WAT11, the empty vector pAG305GPD-ccdB was also transformed into WAT11 as control, and pAG426GPD-AAE13 and pAG426GPD-ccdB were transformed into the acc1-7-1 mutant by using the above-mentioned transformation kit.
For resveratrol biosynthesis, pAG306GPD-AAE13 and a previously constructed vector containing the 4CL::STS gene for resveratrol synthesis (Wang et al. 2011b) were co-transformed into WAT11 competent cells. Cells containing pAG304GPD-4CL::STS and empty vector pAG306GPD-ccdB were used as the control.
Nile Red staining
Nile Red, a marker of lipid bodies, was used to stain yeast cells as described (Kamisaka et al. 1999). Yeast cells cultured at 30 °C for 4 days were collected by centrifugation at 10,000 × g for 10 s and then were incubated in Nile red (0.5 μg/mL) dissolved in 10 mM phosphate buffer (pH 7.0) and 0.15 M potassium chloride (KCl) at 30 °C for 1 min. After staining, yeast cells were washed twice with phosphate buffer. Yeast cells that were stained with Nile Red were imaged using a Zeiss LSM 510 META confocal microscope (Jena, Germany) attached to a digital camera using 488 nm excitation and 500-530 BP detection, and 543 nm emission and 560 LP detection at Donald Danforth Plant Science Center. The scanning was done using a × 40 magnification water immersion lens.
Lipid extraction and quantification
The lipid was extracted as described previously (Kamisaka et al. 2006). The total fatty acid content in yeast cells was determined by gas chromatography after derivatization with 10 % methanolic HCl and methylene chloride. The lipid content (%) was expressed as total fatty acid amount (mg) per dry cell weight (mg) × 100. Samples (50 mL) of the yeast cultures were collected by centrifugation at 700 × g for 5 min. The pellets were washed once with distilled water, dried at 105 °C for 3 h, and then dissolved in 1 mL of 10 % methanolic HCl, 0.5 mL of methylene chloride, and 100 μL of C17:0 fatty acid (10 mg/mL) as an internal standard. After incubation at 60 °C for another 3 h, the mixture was supplemented with 2 mL of saturated NaCl and 1 mL of hexane. The recovered fatty acid methyl esters in the hexane layer were determined by performing quantitative gas chromatography (GC) analysis. GC samples were analyzed on a 15 m × 0.25 mm (inside diameter) AT-WAX Alltech column (Alltech, Deerfield, IL, USA) using the following programs: initial temperature, 190 °C for 2 min; ramp increase at 10 °C/min to 230 °C; final temperature hold, 4 min.
For separation of polar and neutral lipids, 3 mL of 2:1 methanol/chloroform was added into the dry yeast cells (about 100 mg), and the cells were ground with a glass stir rod. The tubes were covered with aluminum foil and incubated at room temperature for 30 min, and then 1 mL of chloroform and 1 mL of water were added and mixed by shaking. The extraction was centrifuged for 5 min at 513 × g in a swinging bucket centrifuge. The organic bottom layer was transferred to a new glass test tube. A 3-mL silica solid phase column (Thermo Scientific, Waltham, MA, USA) was prepared with 2 mL of chloroform by gravity flow. For neutral lipid collection (TAG), the organic phase samples were loaded onto the column using a pastor pipette and collected in a new glass tube. Once a sample had flowed through, the column was washed with 2 mL of chloroform (for a total of 10 mL), 5 mL of 80:20 (chloroform/acetone), and 7 mL of acetone before separating the next sample. For polar lipid collection (PC), the same column was charged with 5 mL of 100:50:40 (methanol/chloroform/water), while the organic phase sample was mixed with 1.3 mL of water and 1.3 mL of chloroform, and centrifuged at 513 × g for 10 min. The bottom layer was transferred to a new glass tube. The TAG and PC samples were completely dried under nitrogen gas. The extraction and GC analysis was the same as the above-mentioned methods.
Acyl-CoA extraction
Acyl-CoA standards were prepared as 1 mg/mL stock in 10 mM ammonium formate and stored at −80 °C. A 4-μL aliquot of 400 μg/mL n-propionyl-CoA was added to the frozen sample as the internal standard. The acyl-CoA extraction was performed as described with modifications (Perera et al. 2009). Briefly, the transgenic and control yeast were cultivated in 300 mL of SD dropout medium in 1-L Erlenmeyer flasks at 30 °C on a shaker at 250 rpm for different time intervals. A 50-mL aliquot of cell culture was centrifuged at 700 × g, 4 °C for 5 min. The pellets were pulverized in liquid nitrogen with mortar and pestle, and 500 μL of 10 % (v/v) ice-cold TCA was added immediately and vigorously vortexed for 3 min in cold room. The TCA supernatant was collected by centrifugation at 14,000 × g for 10 min at 4 °C. The pellet was resuspended in 300 μL of 10 % ice-cold TCA by vortexing and centrifuged at 4 °C. The supernatants were combined with the previous supernatant and extracted two more times with 400 μL of ice-cold diethyl ether. The aqueous extract was lyophilized in a SpeedVac (Thermo Scientific). The pellets were dissolved with 100 μL of ice-cold 10 mM ammonium formate, and then centrifuged for 30 min at 4 °C prior to liquid chromatography–mass spectrometry (LC/MS) analysis.
Acyl-CoA LC-MS/MS analysis
All samples were analyzed using a 4000 QTRAP LC-MS/MS system (Applied Biosystems, Foster City, CA, USA) by multiple reaction monitoring (MRM) in the negative ionization mode. Separation of (10 μL) samples was achieved by using a Gemini-NX 5u C18 HPLC column (Phenomenex, 150 mm × 3.00 mm) equipped with an analytical C18 guard cartridge system (Phenomenex, Torrance, CA, USA). The samples were maintained at 4 °C inside the closed chamber of the autosampler. Solvent A of the LC mobile phases consisted of 5 mM NH4OAc and 5 mM DMBA, finally solvent A was adjusted to pH 5.6 with acetic acid. Solvent B contained 0.1 % formic acid in acetonitrile. The third pump delivered pure acetonitrile isocratically as a post-column mixing solvent. Routinely, 10 μL of the sample was injected per analysis. At initial stage, the LC was running at a flow rate of 0.5 mL/min with 2.5 % solvent B. Total flow rate (pump A + B) was described as follows: 0–2.5 min, 0.5 mL/min; 2.5–12 min, 0.2 mL/min; 12–20 min, 0.5 mL/min. The elution gradient was as follows: 0–2 min, 2.5 % B; 2–2.5 min, 2.5–10 % B; 2.5–10 min, 10–50 %; 10–12 min, 50–100 % B; 12–12.1 min, 100–2.5 %; 12.1–20 min, 2.5 % B. Pump C was used to deliver 100 % acetonitrile at 0.125 mL/min to mix with the LC eluent post-column before MS analysis. During LC-MS/MS analysis, the electrospray ion source of the mass spectrometer was operated at 600 °C in negative ion mode to generate [M-H]− ions of the acyl-CoAs. Quantification was determined in the MRM mode in MS/MS based on monitoring of specific MS-MS fragments ion for each acyl-CoA. MRM-selected ions for free CoA, acetyl-CoA, malonyl-CoA, succinyl-CoA, and n-propionyl-CoA were 766/419, 808/408, 852/808, 866/408, and 822/408, respectively. For acyl-CoA quantification, four standards were used to create a calibration curve for each acyl-CoA. All calibration curves were linear, with R 2 ≥ 0.99.
Resveratrol analysis
Yeast cells containing pAG304GPD-4CL::STS and pAG306GPD-AAE13 or co-transformed with pAG306GPD-ccdB were cultured at 30 °C in 250 mL SD dropout liquid medium (Clontech, Mountain View, CA, USA) containing 2 % (w/v) glucose. In the time course assay, the samples were collected at 2, 4, 6, and 7 days. Aliquots of the cultures (400 μL) were extracted with 800 μL of ethyl acetate. Extracts were evaporated to dryness with an Eppendorf Vacufuge (Eppendorf Scientific, Hauppauge, NY, USA) at room temperature and redissolved in 200 μL of 80 % (v/v) methanol. The metabolites were extracted with ethyl acetate using previous methods (Ralston et al. 2005). Aliquots of the above extracts were analyzed on an Agilent 1100 series HPLC system (Agilent Technologies, Santa Clara, CA, USA) using a Gemini-NX 5u C18 HPLC column (Phenomenex, 5 μm; 150 mm × 3.00 mm) following previous methods (Wang et al. 2011b). Retention time and ultraviolet (UV) spectra were compared to those of authentic standard.
Extraction and measurement of malonic acid in yeast cells
The extraction and measurement of malonic acid were performed as described previously (Broeckling et al. 2005; Chen et al. 2011) with minor modifications. The extracted metabolites were derivatized with 100 μL of N,O-bis(trimethylsilyl)trifluoroacetamide (BSTFA) + trimethylchlorosilane (TMCS) (99:1) (Supelco, Bellefonte, PA, USA) for 30 min at 50 °C. The sample was subsequently transferred to a 100-μL glass insert and analyzed by GC-MS. The ribitol was added as internal standard.
Microarray analysis
Yeast cells were cultured in SD drop out medium at 30 °C under shaken conditions and harvested at mid-log phase. Yeast total RNA was isolated from yeast cells using a Master-Pure yeast RNA purification kit (EPICENTRE Biotechnologies, Madison, WI, USA) according to the manufacturer’s instructions. Total RNA quality was determined by Agilent 2100 bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) according to manufacturer’s recommendations. First-strand cDNA was generated by oligo-dT primed reverse transcription utilizing the Superscript Plus Indirect labeling system (Life Technologies, Carlsbad, CA, USA). Oligo-dT primers were utilized in which two additional nucleotides following the 20 dT residues allow the primer to attach to only the 5′ end of the poly (A) tail of mRNA. For cDNA synthesis, 2 μL of anchored oligo-dT primer is added to 10 μg of total RNA and the solution is incubated at 70 °C for 5 min, then cooled on ice for 1 min. To each sample, RNase inhibitor (Life Technologies) (1 μL), 5× first-strand buffer (6 μL), dNTP mix (dATP, dCTP, dGTP, dTTP, one aminoallyl-modified nucleotide, and one aminohexyl-modified nucleotide) (1.5 μL), 0.1 M dithiothreitol (DTT) (1.5 μL), and Superscript III RNase H- Reverse Transcriptase (2 μL) were added. Reverse transcription was carried out at 46 °C for 3 h. The reaction was terminated by adding 1 N NaOH (15 μL) and incubating at 70 °C for 10 min, then neutralized with 1 N HCL (15 μL). First-strand cDNA was purified with Zymo Research Clean and Concentrator 5 clean-up columns according to manufacturer’s instructions (Irvine, CA, USA). First-strand cDNA was dried in a speed vac at medium heat until the volume was reduced to 3 μL. Alexa Fluor 555 and 647 reactive dyes were prepared for coupling reaction by adding dimethyl sulfoxide (DMSO) (2 μL) directly to each dye pack. Coupling buffer (2×; 5 μL) and either DMSO-resuspended Alexa Fluor 555 or Alexa Fluor 647 reactive dye (2 μL) were then added to each appropriate cDNA sample. Sample/dyes were then incubated at room temperature in the dark for 2 h. Coupling reactions were then quenched with 3 M sodium acetate (20 μL). Fluorescently labeled cDNAs were purified with Zymo Research Clean and Concentrator 5 columns. The cDNAs were quantitated via spectrophotometry to confirm recovery. Samples were then paired and balanced by mass.
The paired and balanced cDNAs were suspended in Agilent 2× Gene Expression buffer (25 μL), Agilent 10× Blocking agent (5 μL), and water to a total volume of 50 μL. This hybridization solution was applied to Agilent Yeast v2 8x15K microarrays. Hybridization was carried out at 65 °C for 20 h. Washing procedures were carried out according to Agilent gene expression protocols.
Slides were scanned on an Axon 4000B scanner (Molecular Devices, Sunnyvale, CA, USA) to detect Cy3 and Cy5 fluorescence. Laser power was kept constant for Cy3/Cy5 scans but photomultiplier tube (PMT) was varied for each experiment based on optimal signal intensity with lowest possible background fluorescence. A low PMT setting scan was also performed to recover signal from saturated elements. Gridding and analysis of images was performed using Genepix v6.1 (Molecular Devices, Sunnyvale, CA, USA). Microarray data have been deposited with the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession number GSE52701.
Real-time RT-PCR analysis
For real-time RT-PCR, total RNA was treated with TURBO DNA-free kit (Ambion, Austin, TX, USA) prior to cDNA synthesis. The first-strand cDNA synthesis was synthesized from 1 μg of DNase I-treated total RNA with the anchored oligo-dT primer following the manufacturer’s protocol (Biolabs). Fifteen pairs of specific primers for the 15 amino acid biosynthesis and TCA cycle-related genes in S. cerevisiae strain WAT11 were designed using primer 5 software (Table S1 in the Supplementary Material). Real-time RT-PCR was performed with SYBR Advantage qPCR Premix (Clontech) using StepOneplus Real-time PCR system (Life Technologies). A serial dilution of the cDNA was used as standard curve to optimize the amplification efficiency with each primer pair. Relative expression levels were normalized using actin gene, ACT1 (Accession Number NP_116614), as an internal control in parallel reactions. All qRT-PCR (rea-time quantitative reverse transcription PCR) reactions were carried out with the following program: initiation with a 10-min denaturation at 95 °C, followed by 45 cycles of amplification with 20 s denaturation at 95 °C, 20 s annealing for 55 °C, 20 s extension at 68 °C; 15 s at 95 °C, melting curve was performed from 55 to 95 °C (1 s hold per 0.3 °C increasing) to check the specificity of the amplified product. Samples were analyzed in quadruplicates using independent RNA samples and were quantified using the comparative threshold cycle method.
Complementation of acc1-7-1 mutant with Arabidopsis AAE13
The yeast expression vector pAG426GPD-AAE13 was transformed into the acc1-7-1 temperature-sensitive mutant (Schneiter et al. 1996). The empty pAG426GPD-ccdB vector (harboring the selective URA3 gene) was transformed into WAT11 yeast (positive control) or mutant acc1-7-1 yeast (negative control). Transformants were selected on plates lacking uracil at 25 °C for 4 days. For temperature-sensitive mutant complementation, the colonies that grew at 25 °C were selected and grown on SD with all amino acids (SD) or SD minus uracil (SD-URA) with or without the anti-metabolite 5-fluoroorotic acid (5-FOA), and at 25 °C (permissive temperature) or 37 °C (restrictive temperature) to determine if growth and the selected phenotype were plasmid dependent.
Results
Determination of substrate concentration
To confirm appropriate substrate concentration to add to yeast cultures, control and AAE13 transgenic yeast were cultured on SD minus leucine medium (SD-LEU) containing different concentrations of malonic acid, 0, 0.2, 0.5, 1, and 2 mM, to measure total fatty acid content. Yeast cells expressing AAE13 had higher fatty acid content compared to that in the control cells at all substrate concentrations (Fig. 2). When malonic acid concentration was 0.2 or 0.5 mM, the average fatty acid content was the highest. When 1 mM malonic acid was supplemented, fatty acid content began to decrease, which indicated that too much malonic acid is toxic to yeast. Therefore, 0.2 mM malonic acid was used in the media for further experiments.
Effect of malonic acid concentration on yeast lipid content. The yeast cells containing pAG305GPD-AAE13 (AAE13) and control yeast cells expressing pAG305GPD-ccdB (Control) were cultured at 30 °C for 4 days in 50 mL SD dropout medium. Lipid content is expressed as total fatty acids (mg) per dry cell weight (mg) × 100. Results are means ± SD (n = 3) for a 50-mL sample of the culture
AAE13 expression increases lipid accumulation
Fluorescent staining of lipid bodies by Nile Red showed that yeast cells expressing AAE13 had lipid bodies that were relatively larger than those on control yeast cells (Fig. 3). Total fatty acid measurement by GC at 5 time points indicated that AAE13 yeast had significantly increased lipid content, although cellular growth was decreased (Fig. 4, Fig. S1 in the Supplementary Material). Lipid profiling showed that the fatty acid composition in AAE13 transgenic yeast was changed (Fig. 5). In both AAE13 transgenic and control yeast, palmitoleic acid (16:1) was the major component, occupying 41 and 43 % of the total lipids, respectively. Another fatty acid with similar levels in AAE13 transgenic and control yeast was palmitic acid (16:0), which represented 18 and 22 %, respectively, of the total lipids. Together, 16 carbon fatty acids comprised approximately 60 % of total cellular lipids. Among 18 carbon fatty acids, stearic acid (18:0) was present in much lower concentrations, at ca. 5 % whereas oleic acid (18:1) was a predominant one. The AAE13 transformants had increased percentage of oleic acid, with a concomitant small decrease in palmitic acid and palmitoleic acid, compared to the control. The AAE13 transformants averaged 32.4 % oleic acid compared with the control mean of 23 % (P < 0.01). The fatty acid composition of WAT11 was consistent with previous reports in Saccharomyces carlsbergensis (Hayashi et al. 1976) and S. cerevisiae (Kamisaka et al. 2006).
Staining of yeast cells carrying pAG305GPD-AAE13 (AAE13) and control yeast cells containing pAG305GPD-ccdB (Control) with Nile Red. Yeast cells cultured in SD dropout medium for 4 days were stained with Nile Red (0.5 μg/mL). Above in each panel is a fluorescent image, middle is white image, and below is differential interference contrast image. Bar = 5 μm
Total lipid content and growth of yeast cells carrying pAG305GPD-AAE13 (AAE13) and control yeast cells containing pAG305GPD-ccdB (Control). Yeast cells were cultured at 30 °C for 2, 3, 4, 5, and 7 days in 400 mL medium. A 50-mL sample was taken at every time point. Lipid content is expressed as total fatty acids (mg) per dry cell weight (mg) × 100. Results are means ± SD (n = 3) for a 50-mL sample of the culture
Fatty acids composition of total cell lipids between AAE13 and control yeast cells containing pAG305GPD-AAE13 (AAE13) and pAG305GPD-ccdB (Control), respectively. The AAE13 and control yeast cells were cultured at 30 °C for 4 days in 50 mL SD dropout medium. Results are means ± SD (n = 3) for a 50-mL sample of culture. **P < 0.01 shows extremely significant difference
We also determined the contents of neutral and polar lipids in dried yeast cells. As shown in Fig. S2 in the Supplementary Material, the content of neutral lipid increased drastically due to the overexpression of AAE13 gene whereas the content of polar lipids remained almost constant between the transgenic and the control cells. This result is in agreement with the observation of Nile Red staining of neutral lipid droplets (Fig. 3). Thus, the overproduced fatty acids were mainly deposited into neutral lipids as a storage form. Neutral lipids located in lipid bodies can later be mobilized for polar lipid biosynthesis or for energy production (Daum and Paltauf 1980).
Overexpression of AAE13 leads to significant increases in resveratrol accumulation
Transgenic WAT11 expressing 4CL::STS fusion gene alone or in combination with AAE13 was analyzed for resveratrol and lipid content. After 4 days of fermentation with feeding 4-coumaric acid (12 mg/L), resveratrol and total lipid were extracted using ethyl acetate and 10 % methanol HCl, respectively, from the culture and yeast cells, and analyzed by HPLC and GC, respectively. Addition of AAE13 increased resveratrol production by approximately twofold compared to the only expression of 4CL::STS (Fig. 6). However, the total lipid contents were not significantly different between engineered yeast carrying AAE13 and the control in this background (P > 0.05) (Fig. 6).
AAE13 increases resveratrol production and no effect on lipid content. Comparison of the resveratrol production and lipid content between the yeast cells containing co-transformed vectors pAG304GPD-4CL::STS and pAG306GPD-AAE13 (4CLSTS-AAE13) and containing co-transformed vectors pAG304GPD-4CL::STS and pAG306GPD-ccdB (4CLSTS) by adding malonic acid and 4-coumaric acid as a substrate. t tests indicate that all of them are extremely significant difference for resveratrol production (P < 0.01), there is no significant difference for lipid production (P > 0.05)
CoA analysis
LC-MS/MS analysis showed that acetyl-CoA, free CoA, and succinyl-CoA were significantly decreased in yeast overexpressing AAE13, but that malonyl-CoA significantly increased (more than twofold; P < 0.01) in yeast overexpressing AAE13 (Fig. 7). However, in yeast overexpressing 4CL::STS, addition of AAE13 did not produce a significantly higher level of the three acyl-CoAs (Fig. 7).
CoA-esters time course profiles for yeast cells (AAE13 yeast cells harboring pAG305GPD-AAE13, control cells harboring pAG305GPD-ccdB, 4CLSTS-AAE13 cells containing pAG304GPD-4CL::STS and pAG306GPD-AAE13 and 4CLSTS cells containing pAG304GPD-4CL::STS and pAG306GPD-ccdB). Concentrations of intracellular acetyl-CoA (a), intracellular free CoA (b), intracellular succinyl-CoA (c), and intracellular malonyl-CoA (d) between yeast cells AAE13 and control with additional of 0.2 mM malonate. Concentrations of intracellular acetyl-CoA (e), intracellular free CoA (f), and intracellular succinyl-CoA (g) between yeast cells 4CLSTS-AAE13 and 4CLSTS with feeding 4-coumaric acid (12 mg/L) and additional of 0.2 mM malonate. Values are averages of three biological repeats with stand deviations. FCW represents fresh cell weight
Microarray gene expression
To evaluate the effect of AAE13 on yeast growth, whole-genome transcriptional profiles for WAT11 and the AAE13 overexpressor have been studied by microarray. Genes involved in the TCA cycle and amino acid biosynthesis were down-regulated in yeast cells overexpressing AAE13 compared to WAT11 (Fig. 8, Fig. S3, and Table S2 in the Supplementary Material). To confirm microarray results, we chose 13 genes, including the up-regulated YLR142W, YHR137W, YCR034W, and YPR035W and the down-regulated genes YNL037C, YER052C, YGR088W, YDR148C, YJR016C, YGL009C, YHR208W, YLR303W, and YLR304C and the internal control ACT1, for real-time RT-PCR analysis. RNA was extracted from S. cerevisiae overexpressing either AAE13 or containing the empty vector. The results showed that the expression patterns of the 15 selected genes were consistent with the results of the microarray analysis (Fig. S3 in the Supplementary Material). Together, these results demonstrated that AAE13 overexpression inhibits the transcript levels of TCA cycle and some amino acid biosynthesis genes (Fig. 8, Fig. S3, and Table S2 in the Supplementary Material).
Expression profiles of known amino acid biosynthesis and tricarboxylic acid (TCA) cycle-related genes in yeast cells different between overexpressing AAE13 and control. The genes with green squares were down-regulated in yeast cells carrying pAG306GPD-AAE13 and red squares were up-regulated in yeast cells carrying pAG306GPD-ccdB. Each dash indicates one step reaction
Complementation assay
A vector carrying AAE13 was transformed into the temperature-sensitive yeast mutant of AAC1 (aac1-7-1). Two transformants expressing AAE13 as well as aac1 and WAT11 cells carrying empty vectors grew at the permissive temperature of 25 °C on SD or SD-URA medium (Fig. 9, upper panel, top row). At the restrictive temperature of 37 °C, in the aac1 mutant, the four expressing AAE13 could grow although the growth was much slower than the positive control. In contrast, the aac1-negative control harboring the empty vector did not grow (Fig. 9, lower panel, top row). These data showed that AAE13 can partially complement the ACC1 mutant (acc1-7-1) in yeast.
Malonyl-CoA ligase (AAE13) partially complements the temperature-sensitive ACC1 mutant (acc1-7-1). Plates were incubated for 3 days at 25 and 37 °C, respectively. a WAT11 strain carrying empty vector pAG426GPD-ccdB (control); b acc1-7-1 mutant carrying empty vector pAG426GPD-ccdB (control); c acc1-7-1 carrying pAG426GPD-AAE13 (Colony #1); d acc1-7-1 carrying pAG426GPD-AAE13 (Colony #2)
To further confirm that AAE13 is required at the restrictive temperature in the acc1 mutant, the four transformed cell lines, described above, were spread on SD and SD-URA plates in the presence of 0.1 % 5-FOA. The URA3 gene on the pAG426GPD vector backbone encodes orotidine 5′-phosphate decarboxylase, which is required for uracil biosynthesis but also converts 5-FOA to the toxic compound 5-fluorouracil. Transformants were replica-plated onto a SD and SD-URA medium with or without 5-FOA, in order to select for loss of the URA3-containing plasmid. The viable cells were then grown at 37 or 25 °C on the same media. At the permissive temperature of 25 °C, the transformants harboring AAE13 and empty vector grew on SD + 5-FOA plate but did not grow on SD-URA + 5-FOA, indicating loss of their pAG426GPD vector and uracil synthesis capability. At 37 °C, the cells derived from the AAE13 transformants and the negative control did not grow on SD + 5-FOA or SD-URA + 5-FOA, while the positive control could grow on complete SD + 5-FOA. This result confirmed that growth of aac1 yeast was dependent on the presence of AAE13.
Discussion
Malonyl-CoA is an important intermediate in fatty acid and polyketide biosynthesis. Fatty acids are the main targets of engineering for biofuel, especially biodiesel, production. Examples of engineering fatty acid or fatty acid esters from inexpensive renewable materials, such as lignocellulose, have been demonstrated in E. coli, yeast, and algae (Jin et al. 2005; Li et al. 2010; Wang et al. 2013; Work et al. 2010; Yim et al. 2011; Yomano et al. 2009). Modified fatty acids, such as omega-3 and other polyunsaturated fatty acids, are important nutraceutical compounds (Torres-Giner et al. 2010). Many important antibiotics and anti-cancer medicines are polyketides, such as erythromycin A and epothilone B (Wawrik et al. 2005). In plants, polyketides can be found in more than 10,000 phenylpropanoid and flavonoid compounds (Wang et al. 2011a; Wink 2003). Microbial production of these compounds has been reported extensively, with some developed to industrial-scale production (Wang et al. 2010, 2011a, b). Therefore, engineering the availability of malonyl-CoA has been an important target for the production of malonyl-CoA-derived metabolites.
Various approaches have been tested to increase the accumulation of these compounds, the increasing malonyl-CoA concentration by overexpression of ACC is the most tested strategy (Davis et al. 2000; Roesler et al. 1997; Ruenwai et al. 2009). In addition to ACC, plants, animals, as well as some bacteria also generate malonyl-CoA from malonic acid through malonyl-CoA synthetase. The genes encoding malonyl-CoA synthetases have been cloned and characterized from Rhizobium trifolii (An and Kim 1998), Streptomyces coelicolor (Hughes and Keatinge-Clay 2011), Arabidopsis (Chen et al. 2011), and human (Chen et al. 2011; Sloan et al. 2011; Witkowski et al. 2011). Although the physiological function of this biochemical pathway is largely unknown, some of these genes have been biotechnologically exploited to enhance malonyl-CoA-derived metabolites in heterologous systems (Fowler et al. 2009; Xu et al. 2011, 2013). When the bacterial malonyl-CoA synthetase gene MatB was overexpressed in E. coli (Leonard et al. 2008) or S. venezuelae (Park et al. 2011), the accumulation of products from the heterologous flavonoid pathways was significantly increased. Although fatty acid biosynthetic pathways compete with the heterologous flavonoid pathway, the consequence of MatB overexpression on fatty acid biosynthesis was not reported in these papers. Arabidopsis AAE13 is a eukaryotic malonyl-CoA synthetase that differs from the product of the bacterial gene, but the K m and V max of these two enzymes are quite similar (Chen et al. 2011). For example, Arabidopsis AAE13 and R. trifolii MatB have K m values of 0.53 and 0.217 mM and V max values of 24 and 18.98 μmol/min/mg protein, respectively.
When Arabidopsis AAE13 was expressed in S. cerevisiae, the total oil content of the transgenic yeast significantly increased even in the absence of exogenous malonic acid (Figs. 2, 3, and 4), probably due to the utilization of endogenous malonic acid in yeast cells. Kaya and Sano (1996) reported that yeast extract contained malonic acid. Our GC/MS analysis also confirmed that S. cerevisiae WAT11 is a producer of malonic acid (Fig. S4 in the Supplementary Material). Clearly, the internal pool of malonic acid was sufficient to increase the yields of fatty acid and resveratrol.
In comparison, the supplement of exogenous malonic acid at 0.2 or 0.5 mM concentrations caused a small but significant increase in cellular oil content (P < 0.05, Fig. 2). Unfortunately, further increase in the supply of malonic acid caused physiological changes and toxicity. The relatively small beneficial effect of exogenous malonic acid suggests that yeast may not be able to transport this dicarboxylic acid efficiently into the cells. Chen and Tan (2013) recently reported that the overexpression of a dicarboxylic acid plasma membrane transporter from Schizosaccharomyces pombe can promote the uptake of exogenous malonic acid in baker’s yeast. In E. coli, a malonic acid transporter is required for the uptake of malonic acid in MatB overexpressors (Leonard et al. 2008). In contrast, WT Arabidopsis seedlings can uptake and transport exogenous malonic acid efficiently (Chen et al. 2011). The recent finding of Chen and Tan (2013) provides new potential transporters for further improvement of AAE13-overexpressing yeast systems for the production of fatty acids and various malonyl-CoA-derived secondary metabolites.
In addition, several interesting phenotypes were observed, as follows.
CoA limitation
The composition of the CoA pools changes in response to environmental stresses such as the quantity and quality of carbon sources in the medium, growth phase, etc. (Chohnan et al. 1997). Although free CoA and acyl-CoA levels in yeast are much lower than those reported in E. coli, the kinetics of accumulation of these intermediates is quite similar in these two microbes. At first, the occurrence of acetyl-CoA peak is probably due to the rapid consumption of glucose and the subsequent formation of acetyl-CoA from pyruvate. When the cells were starved for carbon sources, acetyl-CoA concentration will be decreased (Chohnan et al. 1997). Acetyl-CoA can be converted to free CoA by the citrate synthase reaction in the mitochondria, then free CoA is given rise to thioester to form succinyl-CoA and propionyl-CoA, etc. (Nakamura et al. 2012). CoA biosynthesis requires various amino acids, with pantothenate (vitamin B5) as an important intermediate (Olzhausen et al. 2009). All genes encoding the CoA biosynthetic enzymes have been identified (Leonardi et al. 2005).
When AAE13 was overexpressed, the CoA-conjugated intermediate compounds showed significant reduction. For example, succinyl-CoA was 17 % of that of control at 48 h, and free CoA was 10 % of that of control at 60 h. The break-up of the balance of various acyl-CoA species due to AAE13 overexpression may cause a detrimental effect on cell metabolism and future engineering should coordinately activate coenzyme A cofactor biosynthesis as well.
AAE13 partially complements an ACC1 mutant
We tested complementation of an ACC1-null mutant in yeast with AAE13. To our surprise, AAE13 could not complement an ACC1-null mutation in yeast by tetrad analysis (data not shown). We speculated two reasons for this observation: firstly, endogenous malonic acid concentration is not high enough or exogenous malonic acid transport into yeast cells is not efficient (Chen and Tan 2013); secondly, the temporal and spatial expression of AAE13 in yeast does not meet the needs of malonyl-CoA production in the ACC1 deletion mutant, although the constitutive GPD promoter was used.
Despite being temperature-sensitive, the complementation itself is very interesting. It is highly unlikely that yeast has a gene that synthesized malonic acid from CoA. In the genome of baker’s yeast, there are nine acyl-CoA synthetase genes (Shockey et al. 2003), among which no malonyl-CoA synthetase homologous genes were found. Yeast FAT2 has the highest hit but its enzyme activity has not been identified (Blobel and Erdmann 1996). We expressed the FAT2 gene in E. coli and purified FAT2 as a His-tagged protein. However, recombinant FAT2 did not show any malonyl-CoA synthetase activity (H. Chen and J. Browse, unpublished data). We also measured the malonyl-CoA synthetase activity in yeast crude protein extracts but did not found any activity. It seems that baker’s yeast lacks this malonyl-CoA-generating pathway and ACC is the sole source for malonyl-CoA. Nevertheless, the overexpression of the AAE13 gene partially complemented a temperature-sensitive mutant of yeast ACC1 (Fig. 9), providing a genetic basis for further investigation.
Over-expression of AAE13 suppressed the TCA cycle in transgenic yeast
When the overall transcription profile was analyzed in yeast strains overproducing resveratrol and lipid, we noticed significant reduction in the expression of TCA cycle-related genes. As shown in Fig. 8, Fig. S3, and Table S2 in the Supplementary Material, the genes for the related enzymes, such as YLR304C (aconitase), YNL037C (isocitrate dehydrogenase), and YDR148C (dihydrolipoyl transsuccinylase), were reduced by at least twofold in transcript abundance. We suspect that this reduction is also related to the reduced level of CoA-conjugated compounds such as succinyl-CoA. Previous reports suggested that succinyl-CoA is a feedback inhibitor of the key enzyme α-ketoglutarate dehydrogenase (Smith et al. 1974). As a result, TCA cycle-related genes showed drastic reduction, probably leading to arrest growth of the transgenic yeast.
This report is the first to demonstrate that overexpression of a malonyl-CoA synthetase leads to significant increase in both lipid and polyketide accumulation. The mechanisms of this increase have been discussed. For engineering purposes, additional modifications of related pathways, such as coenzyme A biosynthesis, may be necessary.
References
An JH, Kim YS (1998) A gene cluster encoding malonyl-CoA decarboxylase (MatA), malonyl-CoA synthetase (MatB) and a putative dicarboxylate carrier protein (MatC) in Rhizobium trifolii—cloning, sequencing, and expression of the enzymes in Escherichia coli. Eur J Biochem 257(2):395–402
Blobel F, Erdmann R (1996) Identification of a yeast peroxisomal member of the family of AMP-binding proteins. Eur J Biochem 240(2):468–476
Broeckling CD, Huhman DV, Farag MA, Smith JT, May GD, Mendes P, Dixon RA, Sumner LW (2005) Metabolic profiling of Medicago truncatula cell cultures reveals the effects of biotic and abiotic elicitors on metabolism. J Exp Bot 56(410):323–336
Chen H, Kim HU, Weng H, Browse J (2011) Malonyl-CoA synthetase, encoded by ACYL ACTIVATING ENZYME13, is essential for growth and development of Arabidopsis. Plant Cell 23(6):2247–2262
Chen WN, Tan KY (2013) Malonate uptake and metabolism in Saccharomyces cerevisiae. Appl Biochem Biotechnol 171(1):44–62
Chohnan S, Furukawa H, Fujio T, Nishihara H, Takamura Y (1997) Changes in the size and composition of intracellular pools of nonesterified coenzyme A and coenzyme A thioesters in aerobic and facultatively anaerobic bacteria. Appl Environ Microbiol 63(2):553–560
Daum G, Paltauf F (1980) Triacylglycerols as fatty acid donors for membrane phospholipid biosynthesis in yeast. Monatsh Chem 111:355–363
Davis MS, Solbiati J, Cronan JE Jr (2000) Overproduction of acetyl-CoA carboxylase activity increases the rate of fatty acid biosynthesis in Escherichia coli. J Biol Chem 275(37):28593–28598
Ekhterae D, Tae HJ, Daniel S, Moller DE, Kim KH (1996) Regulation of acetyl coenzyme-A carboxylase gene in a transgenic animal model. Biochem Biophys Res Commun 227(2):547–552
Folmes CD, Lopaschuk GD (2007) Role of malonyl-CoA in heart disease and the hypothalamic control of obesity. Cardiovasc Res 73(2):278–287
Fowler ZL, Gikandi WW, Koffas MA (2009) Increased malonyl coenzyme A biosynthesis by tuning the Escherichia coli metabolic network and its application to flavanone production. Appl Environ Microbiol 75(18):5831–5839
Halls C, Yu O (2008) Potential for metabolic engineering of resveratrol biosynthesis. Trends Biotechnol 26(2):77–81
Hasslacher M, Ivessa AS, Paltauf F, Kohlwein SD (1993) Acetyl-CoA carboxylase from yeast is an essential enzyme and is regulated by factors that control phospholipid metabolism. J Biol Chem 268(15):10946–10952
Hayashi E, Hasegawa R, Tomita T (1976) Accumulation of neutral lipids in Saccharomyces carlsbergensis by myo-inositol deficiency and its mechanism. Reciprocal regulation of yeast acetyl-CoA carboxylase by fructose bisphosphate and citrate. J Biol Chem 251(18):5759–5769
Hoja U, Marthol S, Hofmann J, Stegner S, Schulz R, Meier S, Greiner E, Schweizer E (2004) HFA1 encoding an organelle-specific acetyl-CoA carboxylase controls mitochondrial fatty acid synthesis in Saccharomyces cerevisiae. J Biol Chem 279(21):21779–21786
Hughes AJ, Keatinge-Clay A (2011) Enzymatic extender unit generation for in vitro polyketide synthase reactions: structural and functional showcasing of Streptomyces coelicolor MatB. Chem Biol 18(2):165–176
Jin YS, Alper H, Yang YT, Stephanopoulos G (2005) Improvement of xylose uptake and ethanol production in recombinant Saccharomyces cerevisiae through an inverse metabolic engineering approach. Appl Environ Microbiol 71(12):8249–8256
Kamisaka Y, Noda N, Sakai T, Kawasaki K (1999) Lipid bodies and lipid body formation in an oleaginous fungus, Mortierella ramanniana var. angulispora. Biochim Biophys Acta 1438(2):185–198
Kamisaka Y, Noda N, Tomita N, Kimura K, Kodaki T, Hosaka K (2006) Identification of genes affecting lipid content using transposon mutagenesis in Saccharomyces cerevisiae. Biosci Biotechnol Biochem 70(3):646–653
Kaya K, Sano T (1996) Algicidal compounds in yeast extract as a component of microbial culture media. Phycologia 35(6S):117–119
Leonard E, Yan Y, Fowler ZL, Li Z, Lim CG, Lim KH, Koffas MA (2008) Strain improvement of recombinant Escherichia coli for efficient production of plant flavonoids. Mol Pharm 5(2):257–265
Leonardi R, Zhang YM, Rock CO, Jackowski S (2005) Coenzyme A: back in action. Prog Lipid Res 44(2–3):125–153
Li Y, Han D, Hu G, Sommerfeld M, Hu Q (2010) Inhibition of starch synthesis results in overproduction of lipids in Chlamydomonas reinhardtii. Biotechnol Bioeng 107(2):258–268
Miyahisa I, Kaneko M, Funa N, Kawasaki H, Kojima H, Ohnishi Y, Horinouchi S (2005) Efficient production of (2S)-flavanones by Escherichia coli containing an artificial biosynthetic gene cluster. Appl Microbiol Biotechnol 68(4):498–504
Nakamura T, Pluskal T, Nakaseko Y, Yanagida M (2012) Impaired coenzyme A synthesis in fission yeast causes defective mitosis, quiescence-exit failure, histone hypoacetylation and fragile DNA. Open Biol 2(9):120117
Olzhausen J, Schübbe S, Schüller HJ (2009) Genetic analysis of coenzyme A biosynthesis in the yeast Saccharomyces cerevisiae: identification of a conditional mutation in the pantothenate kinase gene CAB1. Curr Genet 55(2):163–173
Park SR, Ahn MS, Han AR, Park JW, Yoon YJ (2011) Enhanced flavonoid production in Streptomyces venezuelae via metabolic engineering. J Microbiol Biotechnol 21(11):1143–1146
Perera MA, Choi SY, Wurtele ES, Nikolau BJ (2009) Quantitative analysis of short-chain acyl-coenzyme As in plant tissues by LC-MS-MS electrospray ionization method. J Chromatogr B Anal Technol Biomed Life Sci 877(5–6):482–488
Ralston L, Subramanian S, Matsuno M, Yu O (2005) Partial reconstruction of flavonoid and isoflavonoid biosynthesis in yeast using soybean type I and type II chalcone isomerases. Plant Physiol 137(4):1375–1388
Roesler K, Shintani D, Savage L, Boddupalli S, Ohlrogge J (1997) Targeting of the Arabidopsis homomeric acetyl-coenzyme A carboxylase to plastids of rapeseeds. Plant Physiol 113(1):75–81
Ruenwai R, Cheevadhanarak S, Laoteng K (2009) Overexpression of acetyl-CoA carboxylase gene of Mucor rouxii enhanced fatty acid content in Hansenula polymorpha. Mol Biotechnol 42(3):327–332
Saggerson D (2008) Malonyl-CoA, a key signaling molecule in mammalian cells. Annu Rev Nutr 28:253–272
Schneiter R, Hitomi M, Ivessa AS, Fasch EV, Kohlwein SD, Tartakoff AM (1996) A yeast acetyl coenzyme A carboxylase mutant links very-long-chain fatty acid synthesis to the structure and function of the nuclear membrane-pore complex. Mol Cell Biol 16(12):7161–7172
Schneiter R, Guerra CE, Lampl M, Tatzer V, Zellnig G, Klein HL, Kohlwein SD (2000) A novel cold-sensitive allele of the rate-limiting enzyme of fatty acid synthesis, acetyl coenzyme A carboxylase, affects the morphology of the yeast vacuole through acylation of Vac8p. Mol Cell Biol 20(9):2984–2995
Shin GH, Veen M, Stahl U, Lang C (2012) Overexpression of genes of the fatty acid biosynthetic pathway leads to accumulation of sterols in Saccharomyces cerevisiae. Yeast 29(9):371–383
Shintani D, Roesler K, Shorrosh B, Savage L, Ohlrogge J (1997) Antisense expression and overexpression of biotin carboxylase in tobacco leaves. Plant Physiol 114(3):881–886
Shockey JM, Fulda MS, Browse J (2003) Arabidopsis contains a large superfamily of acyl-activating enzymes. Phylogenetic and biochemical analysis reveals a new class of acyl-coenzyme a synthetases. Plant Physiol 132(2):1065–1076
Sloan JL, Johnston JJ, Manoli I, Chandler RJ, Krause C, Carrillo-Carrasco N, Chandrasekaran SD, Sysol JR, O'Brien K, Hauser NS, Sapp JC, Dorward HM, Huizing M, Barshop BA, Berry SA, James PM, Champaigne NL, de Lonlay P, Valayannopoulos V, Geschwind MD, Gavrilov DK, Nyhan WL, Biesecker LG, Venditti CP (2011) Exome sequencing identifies ACSF3 as a cause of combined malonic and methylmalonic aciduria. Nat Genet 43(9):883–886
Smith CM, Bryla J, Williamson JR (1974) Regulation of mitochondrial α-ketoglutarate metabolism by product inhibition at α-ketoglutarate dehydrogenase. J Biol Chem 249(5):1497–505
Tehlivets O, Scheuringer K, Kohlwein SD (2007) Fatty acid synthesis and elongation in yeast. Biochim Biophys Acta 1771(3):255–270
Tong L (2005) Acetyl-coenzyme A carboxylase: crucial metabolic enzyme and attractive target for drug discovery. Cell Mol Life Sci 62(16):1784–1803
Torres-Giner S, Martinez-Abad A, Ocio MJ, Lagaron JM (2010) Stabilization of a nutraceutical omega-3 fatty acid by encapsulation in ultrathin electrosprayed zein prolamine. J Food Sci 75(6):N69–79
Urban P, Mignotte C, Kazmaier M, Delorme F, Pompon D (1997) Cloning, yeast expression, and characterization of the coupling of two distantly related Arabidopsis thaliana NADPH-cytochrome P450 reductases with P450 CYP73A5. J Biol Chem 272(31):19176–19186
Wang X, Yomano LP, Lee JY, York SW, Zheng H, Mullinnix MT, Shanmugam KT, Ingram LO (2013) Engineering furfural tolerance in Escherichia coli improves the fermentation of lignocellulosic sugars into renewable chemicals. Proc Natl Acad Sci U S A 110(10):4021–4026
Wang Y, Chen H, Yu O (2010) Metabolic engineering of resveratrol and other longevity boosting compounds. Biofactors 36(5):394–400
Wang Y, Chen S, Yu O (2011a) Metabolic engineering of flavonoids in plants and microorganisms. Appl Microbiol Biotechnol 91(4):949–956
Wang Y, Halls C, Zhang J, Matsuno M, Zhang Y, Yu O (2011b) Stepwise increase of resveratrol biosynthesis in yeast Saccharomyces cerevisiae by metabolic engineering. Metab Eng 13(5):455–463
Wawrik B, Kerkhof L, Zylstra GJ, Kukor JJ (2005) Identification of unique type II polyketide synthase genes in soil. Appl Environ Microbiol 71(5):2232–2238
Wink M (2003) Evolution of secondary metabolites from an ecological and molecular phylogenetic perspective. Phytochemistry 64(1):3–19
Witkowski A, Thweatt J, Smith S (2011) Mammalian ACSF3 protein is a malonyl-CoA synthetase that supplies the chain extender units for mitochondrial fatty acid synthesis. J Biol Chem 286(39):33729–33736
Work VH, Radakovits R, Jinkerson RE, Meuser JE, Elliott LG, Vinyard DJ, Laurens LM, Dismukes GC, Posewitz MC (2010) Increased lipid accumulation in the Chlamydomonas reinhardtii sta7-10 starchless isoamylase mutant and increased carbohydrate synthesis in complemented strains. Eukaryot Cell 9(8):1251–1261
Xu P, Ranganathan S, Fowler ZL, Maranas CD, Koffas MA (2011) Genome-scale metabolic network modeling results in minimal interventions that cooperatively force carbon flux towards malonyl-CoA. Metab Eng 13(5):578–587
Xu P, Gu Q, Wang W, Wong L, Bower AG, Collins CH, Koffas MA (2013) Modular optimization of multi-gene pathways for fatty acids production in E. coli. Nat Commun 4:1409
Yim H, Haselbeck R, Niu W, Pujol-Baxley C, Burgard A, Boldt J, Khandurina J, Trawick JD, Osterhout RE, Stephen R, Estadilla J, Teisan S, Schreyer HB, Andrae S, Yang TH, Lee SY, Burk MJ, Van Dien S (2011) Metabolic engineering of Escherichia coli for direct production of 1,4-butanediol. Nat Chem Biol 7(7):445–452
Yomano LP, York SW, Shanmugam KT, Ingram LO (2009) Deletion of methylglyoxal synthase gene (mgsA) increased sugar co-metabolism in ethanol-producing Escherichia coli. Biotechnol Lett 31(9):1389–1398
Zha W, Rubin-Pitel SB, Shao Z, Zhao H (2009) Improving cellular malonyl-CoA level in Escherichia coli via metabolic engineering. Metab Eng 11(3):192–198
Acknowledgements
We thank Dr. John Browse of Washington State University for the gift of AAE13 gene in pENTR/D-TOPO prior to its publication. This work is support in part by three federal grants: one is from US Department of Energy (DESC0001295), one from National Science Foundation (MCB-0923779), and one from US Department of Agriculture (2010-65116-20514), and National 863 projects (2013AA102801-03).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 329 kb)
Rights and permissions
About this article
Cite this article
Wang, Y., Chen, H. & Yu, O. A plant malonyl-CoA synthetase enhances lipid content and polyketide yield in yeast cells. Appl Microbiol Biotechnol 98, 5435–5447 (2014). https://doi.org/10.1007/s00253-014-5612-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-014-5612-z