Abstract
Coenzyme A (CoA) is a ubiquitous cofactor required for numerous enzymatic carbon group transfer reactions. CoA biosynthesis requires contributions from various amino acids with pantothenate as an important intermediate which can be imported from the medium or synthesized de novo. Investigating function and expression of structural genes involved in CoA biosynthesis of the yeast Saccharomyces cerevisiae, we show that deletion of ECM31 and PAN6 results in mutants requiring pantothenate while loss of PAN5 (related to panE from E. coli) still allows prototrophic growth. A temperature-sensitive mutant defective for fatty acid synthase activity could be functionally complemented by a gene significantly similar to eukaryotic pantothenate kinases (YDR531W). Enzymatic studies and heterologous complementation of this mutation by bacterial and mammalian genes showed that YDR531W encodes a genuine pantothenate kinase (new gene designation: CAB1, “coenzyme A biosynthesis”). A G351S missense mutation within CAB1 was identified to cause the conditional phenotype of the mutant initially studied. Similar to CAB1, genes YIL083C, YKL088W, YGR277C and YDR106C responsible for late CoA biosynthesis turned out as essential. Null mutants could be complemented by their bacterial counterparts coaBC, coaD and coaE, respectively. Comparative expression analyses showed that some CoA biosynthetic genes are weakly de-repressed with ethanol as a carbon source compared with glucose.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
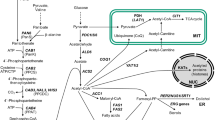
Coenzyme A (CoA) is a ubiquitous co-substrate for a large number of enzymes involved in the transfer of acyl groups. Acetyl-CoA as the most important thioester derivative at the sulfhydryl group of CoA plays a central role for the oxidative degradation of sugars and fatty acids via the tricarboxylic acid cycle but is also indispensable for anabolic pathways such as fatty acid biosynthesis, sterol biosynthesis, formation of ketone bodies in mammals and gluconeogenesis from C2 substrates in many microorganisms. For biosynthesis of CoA, pantothenate (vitamin B5) is a general intermediate that may be formed de novo from several amino acids or can be taken up from outside the cell, using a specific permease. Once inside the cell, five reactions are required to convert pantothenate into CoA (reviewed by Leonardi et al. 2005).
The yeast Saccharomyces cerevisiae is able to synthesize pantothenate de novo (White et al. 2001) although growth of some laboratory strains requires pantothenate supplementation (Stolz and Sauer 1999). Although biosynthesis of pantoate in S. cerevisiae presumably follows the pathway established in bacteria (structural genes panB and panE), a homolog of aspartate decarboxylase (panD in E. coli) providing β-alanine is absent from yeast. Instead, β-alanine is obtained by oxidative conversion of polyamines (White et al. 2001, 2003; summarized in Fig. 1). The use of external pantothenate requires the high-affinity transporter of the plasma membrane, Fen2 (Stolz and Sauer 1999).
To finally synthesize CoA, pantothenate must react with cysteine and ATP. As an initial step, pantothenate is phosphorylated by the ATP-dependent pantothenate kinase (panK; encoded by coaA in E. coli) which is considered as the rate-limiting enzyme for the entire pathway. Indeed, CoA could competitively inhibit panK activity in E. coli presumably by its interference with ATP binding (Vallari et al. 1987) while acylated CoA thioesters were less efficient. In contrast to bacteria, mammalian panK enzymes are strongly inhibited by acetyl-CoA (and malonyl-CoA, although less effective) while CoA, surprisingly, is a mild activator (Rock et al. 2000). In the following reaction, 4′-phosphopantothenate forms an amide bond with cysteine which is subsequently decarboxylated to give 4′-phosphopantetheine. The nucleotide moiety of CoA is then provided by ATP and the resulting dephospho-CoA finally needs to be phosphorylated. Interestingly, an alternative pathway catalyzed by the CoA-synthesizing protein complex CoA-SPC of 400 kDa has been postulated for yeast (Bucovaz et al. 1980, 1997), proposing an early transfer of the ADP moiety directly to pantothenate. While this pathway would bypass the need of 4′-phosphopantothenate, genomic data from yeasts and fungi support the existence of genuine panK enzymes (Calder et al. 1999).
Completion of the mammalian pathway of CoA biosynthesis by comparitive genomics (Daugherty et al. 2002) allowed the identification of genes encoding 4′-phosphopantothenoylcysteine synthetase (PPCS), 4′-phosphopantothenoylcysteine decarboxylase (PPCDC), 4′-phosphopantetheine adenylyltransferase (PPAT) and dephospho-CoA kinase (DPCK) for which putative homolog exist in S. cerevisiae. These findings suggest that biosynthesis of CoA follows a uniform pathway in all living systems. Not only CoA as a mobile cofactor is essential for metabolism but also its phosphopantethein component which becomes covalently linked to eukaryotic fatty acid synthases (Fichtlscherer et al. 2000).
In this work, we analyzed biosynthesis of CoA in the yeast S. cerevisiae by genetical and biochemical methods. Focusing on yeast panK, we characterized a temperature-sensitive mutant which has been previously isolated in a screen for fatty acid-requiring mutants. We could identify a missense mutation at a completely conserved residue within panK causing this defect.
Materials and methods
Strains of S. cerevisiae and E. coli, media and growth conditions
All strains of S. cerevisiae used in this work (compiled in Table 1) are isogenic to strain JS91.15-23. Synthetic complete (SC) media used for selective growth of transformants have been described (Schüller et al. 1992). To obtain selective medium lacking pantothenate (SCD-Pan), a mixture of pure substances composed identical to yeast nitrogen base (Invitrogen) was used. Strains were incubated at 30 or 37°C as indicated.
For bacterial expression of the GST-CAB1 fusion gene, strain BL21-CodonPlus (Stratagene/Agilent) grown in YT-G and supplemented with ampicillin and chloramphenicol was used.
Plasmid constructions and site-directed mutagenesis
Plasmids constructed and used for this work are listed in Table 2. The murine panK3 cDNA clone was purchased from OriGene via AMS Biotechnologie (Wiesbaden, Germany). To overexpress and epitope-tag CoA biosynthetic genes, MET25-containing vector p426-MET25HA (2 μm URA3 MET25 prom-HA3; Mumberg et al. 1994) was used. Bacterial genes coaA, coaBC, coaD and coaE were amplified using gene-specific primers and subsequently cloned into p426-MET25HA. To achieve a regular gene dosage, plasmid YCplac33 (Gietz and Sugino 1988) was used. Reporter gene fusions for genes of pantothenate and CoA biosynthesis were constructed using YEp356 and related vectors (Myers et al. 1986). For molecular characterization of the cab1 ts mutant allele, primers CAB1-Bam and CAB1-Hind were used to amplify the reading frame of YDR531W/CAB1. The PCR product obtained was cloned into pUC19 to give pJO19. Plasmids from two independent amplifications were subsequently used for DNA sequencing. The ydr531w(G351S) missense mutation was introduced into the coding region of wild-type YDR531W using the QuikChange site-directed mutagenesis kit (Stratagene/Agilent). The desired mutant allele in the resulting plasmid pJO62 was confirmed by DNA sequencing.
For bacterial expression of GST fusion genes by IPTG induction, derivatives of pGEX-2TK (GE Healthcare) were used.
Enzyme assays
Assay of pantothenate kinase followed the procedure previously described by Vallari et al. (1987). In brief, D-[1-14C] pantothenate (14C-pan; supplied by Biotrend, Cologne, Germany) was converted into phosphopantothenate which was subsequently bound to ion-exchange filter and analyzed by liquid scintillation counting. 5500 Bq of 14C-pan (2.75 nmol) were incubated in buffer (100 mM Tris/HCl, 2.5 mM MgCl2, 2.5 mM ATP, pH 7.4) with 75 μg of total protein in a volume of 40 μl. After incubation at 30 or 37°C for 10 min, the mixture was transferred to a DE-81 ion-exchange filter disk and washed three times with 1% acetic acid in technical ethanol. Dried filter disks were transferred into scintillation solution (Beckman Ready-Solv MP) and analyzed with a Perkin Elmer Packard Tri-Carb 2900TR scintillation counter. Assay of β-galactosidase activities has been previously described (Schwank et al. 1995).
Miscellaneous procedures
Transformation of S. cerevisiae and PCR amplification have been previously described (Schwank et al. 1995). DNA sequencing was performed by Agowa (Berlin, Germany).
Results
Genes of S. cerevisiae involved in pantothenate biosynthesis
De novo biosynthesis of pantothenate utilizes derivatives of amino acid metabolism, 2-oxoisovalerate and β-alanine (cf. Fig. 1). Three enzymes are specifically required for the conversion of these molecules into pantothenate, 2-oxoisovalerate hydroxymethyltransferase, dehydropantoate reductase and pantoate-β-alanine ligase which are encoded by genes panB, panE and panC in E. coli, respectively. Since ECM31 (=YBR176W) of S. cerevisiae shows significant similarity to E. coli panB (36.4% identity, 55% similarity), we constructed a deletion mutant (strain JWH3, Δecm31::HIS3) which indeed failed to grow on a selective medium lacking pantothenate (Fig. 2; also shown by White et al. 2001). The same result was obtained with a null mutant defective for PAN6 (=YIL145C; strain JWH2, Δpan6::HIS3) which is similar to panC from E. coli (40% identity, 52% similarity).
In contrast, a null mutant lacking PAN5 (=YHR063C; strain KLY4-17, Δpan5::HIS3) which is similar to E. coli dehydropantoate reductase gene panE (22.8% identity, 40% similarity) was still able to grow in the absence of pantothenate, although slightly less efficient than the wild-type (cf. Fig. 2). We thus reasoned that a second gene encoding an isoenzyme may exist in S. cerevisiae. Indeed, the gene product of YDL144C is also similar to E. coli panE over its entire length (21.4% identity, 33.3% similarity). However, even a strain with a double deletion (KLY6, Δpan5::HIS3 Δydl144c::kanMX) was not auxotrophic for pantothenate (Fig. 2). Identical results were obtained with double deletion mutations introduced into other strain backgrounds (not shown). We conclude that the remaining dehydropantoate reductase is encoded by a S. cerevisiae gene unrelated in sequence to bacterial enzymes.
Mutations pan6 and fen2 show synthetical lethality
Owing to the existence of the high-affinity pantothenate transporter Fen2 (Stolz and Sauer 1999), biosynthetic mutants of S. cerevisiae can utilize external pantothenate. We thus wished to investigate whether a pan6 fen2 double mutant is still viable. After mating of haploid single mutants JWH2 (Δpan6::HIS3) and JS07.1-6 (Δfen2::LEU2) and subsequent sporulation of the resulting diploid strain no viable progeny with a His+ Leu+ phenotype could be observed (18 tetrads). Media for sporulation, spore germination, and phenotypic characterization of ascospores were supplemented with an excess of pantothenate (50 μM). We, thus, conclude that pan6 and fen2 are synthetically lethal. Our data also argue against the existence of a low-affinity pantothenate transporter and suggest that passive diffusion of pantothenate across the membrane does not occur.
Characterization of a temperature-sensitive mutant defective for pantothenate kinase activity
A previous screen for mutants defective for fatty acid synthase (FAS) activity also led to the identification of strains containing full-length FAS subunits α and β but were devoid of pantetheine (Schweizer et al. 1973). Strains carrying the cab1 ts mutation (for explanation of the gene designation see below) show regular growth at 30°C but fail to proliferate at 37°C. The temperature-sensitive mutant JS91.14-24 was used to isolate the wild-type gene which could restore growth at 37°C. Plasmids obtained by functional complementation all contained the APA2-YDR531W gene pair. Since YDR531W encodes a protein with significant similarity to the pantothenate kinase of A. nidulans (43.8% identity, 55% similarity; not shown), failure to synthesize CoA may cause the mutant phenotype. Indeed, plasmid pSBS5 containing the coding region of YDR531W under control of the MET25 promoter could also complement the temperature-sensitive phenotype of strain JS91.14-24 (Fig. 3a). However, this finding could be also the result of dosage-dependent suppression of a mutation in a distinct gene.
Complementation of conditional phenotypes of pantothenate kinase mutants. a Strain JS91.14-24 (cab1 ts) was transformed with plasmids pJO57 and pJO62 encoding wild-type YDR531W (CAB1) and a G351S variant activated by the natural control region (YCp vector). b Strain JS91.14-24 (cab1 ts) was transformed with plasmids pSBS5, pFK1, pJO73 and pJO72 encoding homologous and heterologous panK genes (coaA: E. coli, panK3: M. musculus) and the coaA-related gene YGR205W (S. cerevisiae). Genes were activated by the MET25 promoter (2 μm vector). c Test for allelism of ydr531w and cab1 ts. Serial dilutions of strains JOY2 and JOY2D were grown on rich medium (YEPD) at 30 and 37°C, respectively. d Carbon source-dependent complementation of ydr531w null mutant (strain KLY5) by GAL1-activated YDR531W (plasmid pKL7)
We thus constructed a null mutant allele (Δydr531w::HIS3; in plasmid pSBS7) which was subsequently introduced into the diploid strain JS01.3 (his3/his3 YDR531W/YDR531W). Following sporulation of the resulting transformant SSH1 (containing a single mutant allele Δydr531w::HIS3), no viable ascospores with a His+ phenotype were obtained. Since the same result was obtained with a medium containing fatty acids, YDR531W is an essential gene, at least under conditions tested. This finding agrees with the data of the yeast systematic gene deletion project (Winzeler et al. 1999; Giaever et al. 2002) and is further supported by the finding that a haploid Δydr531w mutant transformed with a GAL1-YDR531W fusion can grow in galactose-containing medium but not in the presence of glucose (Fig. 3d). We thus repeated the disruption experiment of YDR531W with a diploid obtained by mating of JS91.14-24 with a wild-type strain (JOY2; his3/his3 YDR531W/cab1 ts). Considering cab1 ts as a mutant allele of YDR531W, introduction of the ydr531w null allele into JOY2 should result in transformants with a temperature-sensitive phenotype. Indeed, the predicted phenotype was observed for strain JOY2D (Δydr531w::HIS3/cab1 ts; Fig. 3c), confirming that cab1 ts is a mutant allele of YDR531W.
For a precise mapping of the mutation responsible for the temperature-sensitive phenotype, we amplified the coding region of YDR531W using DNA from strain JS91.14-24 as a template. Comparative DNA sequencing revealed the existence of five missense mutations (D208E, N255H, S261P, A327T and G351S). Phylogenetic comparison of pantothenate kinase (panK) sequences from fungal and higher eukaryotic species showed that among these variants G351 is the only residue which is entirely conserved (cf. Fig. 4). We, thus, introduced the G351S mutation into a functional YDR531W gene by site-directed mutagenesis. In contrast to the wild-type sequence, the expression of ydr531w(G351S) driven by its natural promoter failed to complement the temperature-sensitivity of strain JS91.14-24 (Fig. 3a). We conclude that alteration of the highly conserved residue G351 is indeed responsible for the conditional phenotype caused by the mutant allele studied initially.
Sequence comparison of eukaryotic pantothenate kinases. Entirely conserved residues are emphasized by gray shading. Missense mutations identified in the cab1 ts allele are indicated by #. The G351S mutation (depicted inverse) is sufficient to confer the temperature-sensitive phenotype. Residues involved in binding of the competitive inhibitor acetyl-CoA are indicated by vertical line (hydrogen bond) or * (hydrophobic interaction). An Aspergillus nidulans, Hs Homo sapiens; Mm Mus musculus, Sc Saccharomyces cerevisiae
Although the coaA gene encoding panK in E. coli fails to display any significant similarity to the gene product of YDR531W, we expressed the bacterial gene in strain JS91.14-24. Interestingly, introduction of a MET25-coaA fusion (plasmid pFK1) could substantially restore growth at 37°C (Fig. 3b), supporting YDR531W as the genuine panK gene of S. cerevisiae. Yeast expression of the murine panK3 gene (its product is 31.6% identical and 47.7% similar to YDR531W) with plasmid pJO73 could functionally complement temperature-sensitivity of strain JS91.14-24 as well. Interestingly, S. cerevisiae gene YGR205W encodes a protein which is distantly related to bacterial coaA (15.2% identity, 29.1% similarity) but resembles prokaryotic pantothenate kinases at the structural level (de La Sierra-Gallay et al. 2004). In contrast to coaA of E. coli, overexpression of YGR205W using multi-copy plasmid pJO72 did not confer growth of strain JS91.14-24 at the non-permissive temperature (Fig. 3b).
As a final proof of function, we assayed panK activity in crude extracts from strain JS91.14-24 and multicopy transformants overexpressing YDR531W. As is shown in Table 3, enzyme activity in strain JS91.14-24 was reduced to about 50% of the wild-type level at 30°C and was undetectable at 37°C. The use of the heterologous MET25 promoter on a multicopy plasmid increased enzyme activity by a factor of more than 50. A substantial panK activity was also assayed in yeast transformants expressing bacterial coaA, explaining the result of the heterologous complementation experiment described above. Conversely, the expression of YDR531W as a GST fusion in E. coli similarly increased enzyme activity in protein extracts. From these results, we conclude that YDR531W indeed encodes the initial enzyme of coenzyme A biosynthesis from pantothenate and suggest CAB1 (coenzyme A biosynthesis) as its new adequate gene designation.
Genetic analysis of genes presumably involved in late CoA biosynthesis
Because of sequence similarity, the four remaining steps to finally synthesize coenzyme A from phosphopantothenate may be catalyzed by the products of S. cerevisiae genes YIL083C (phosphopantothenoylcysteine synthetase, PPCS), YKL088W (phosphopantothenoylcysteine decarboxylase, PPCDC), YGR277C (phosphopantetheine adenylyltransferase, PPAT) and YDR196C (dephospho-CoA kinase, DPCK). Similar to what has been described for YDR531W/CAB1, null mutant alleles for these four genes were individually introduced into a diploid strain and the heterozygous situation at all four loci could be shown. Following sporulation, no haploid progeny carrying the selection marker used for disruption of the respective locus could be identified, again confirming the results of the systematic gene deletion project (Winzeler et al. 1999; Giaever et al. 2002). Haploid strains with a chromosomal deletion of one the four genes were obtained only in the presence of a plasmid containing the respective wild-type gene.
It should be mentioned that two genes (VHS3 and SIS2/HAL3) exhibit significant sequence similarity to YKL088W. Vhs3 and Sis2 are highly related to each other over their entire length; both proteins are able to bind to and inhibit the Ser/Thr-specific phosphatase Ppz1 involved in regulation of halotolerance and ion homeostasis. Null mutation vhs3 and sis2 are synthetically lethal (Ruiz et al. 2004). Similarity of Ykl088w with Vhs3 and Sis2 is most apparent at the C-terminus of the respective proteins (Ykl088w and Vhs3: 22.2% identity, 37.6% similarity; Ykl088w and Sis2: 20.8% identity, 34.5% similarity over entire length). To assay for dosage-dependent suppression, we introduced multi-copy plasmids containing MET25 fusions of VHS3 and SIS2, respectively, into a diploid with a heterozygous Δykl088w null mutation. Following sporulation, we were unable to obtain haploid progeny with a chromosomal Δykl088w mutation (only 2 viable spores per tetrad). In contrast, introduction of a MET25-YKL088W fusion plasmid allowed growth of haploid Δykl088w spores. We conclude that even at an elevated level of gene expression, VHS3 and SIS2 are unable to replace YKL088W and may not be considered as genuine genes of coenzyme A biosynthesis.
To obtain further evidence for a function of YIL083C, YKL088W, YGR277C and YDR196C in coenzyme A biosynthesis, we assayed for heterologous complementation of mutants by the corresponding bacterial genes. E. coli genes coaBC (encoding bifunctional PPCS and PPCDC), coaD (encoding PPAT) and coaE (encoding DPCK) were fused with the MET25 promoter and subsequently introduced into heterozygous diploids carrying deletion mutations Δyil083c, Δykl088w (MET25-coaBC), Δygr277c (MET25-coaD) and Δydr196c (MET25-coaE), respectively. Similar to authentic S. cerevisiae genes, plasmid-dependent expression of coaBC allowed growth of haploid null mutants Δyil083c and Δykl088w (not shown). The same result could be obtained for coaD in the Δygr277c mutant and coaE in the Δydr196c mutant. We conclude that bacterial genes of CoA biosynthesis are able to fulfill their function even in S. cerevisiae, at least when adequately overexpressed. This finding supports the hypothesis that yeast null mutants used for heterologous complementation are indeed defective for CoA biosynthesis.
Expression analysis of coenzyme A biosynthetic genes
Since coenzyme A and its derivatives influence several metabolic pathways, we wished to investigate whether the expression of the biosynthetic genes is affected by the carbon source and by availability of amino acids and pantothenate, respectively. We thus constructed lacZ fusion genes for ECM31, PAN6, CAB1, YIL083C, YKL088W, YGR277C and YDR196C and transformed the resulted plasmids into regulatory wild-type strain JS42. Transformants were grown under selective conditions in SCD (2% glucose, supplementation with amino acids), SCE (3% ethanol as the sole carbon source), SM (no amino acids added) and SCD-Pan (2% glucose, no pantothenate), respectively. As shown in Table 4, for some fusions a modest increase of reporter gene expression was found when transformants were cultivated in the presence of the non-fermentable carbon source ethanol, compared with glucose as a substrate. However, a de-repression factor of 2.5 (for ECM31) or below does not justify to use the designation “glucose repression”. Similarly, the complete absence of amino acids (in SM) did not significantly alter gene expression when compared with the supplementation in SCD medium. Surprisingly (at least for pantothenate biosynthetic genes ECM31 and PAN6), variation of pantothenate supply failed to influence reporter gene expression, as well. We thus consider the seven genes for which expression was assayed as typical “house-keeping” genes which are transcribed at a constitutive low level.
Discussion
The yeast S. cerevisiae is able to synthesize pantothenate de novo from intermediates of amino acid metabolism. Comparative in silico analyses indicated that yeast genes ECM31, PAN5 and PAN6 (similar to E. coli panB, panE and panC, respectively) may be involved in pantothenate biosynthesis. In agreement with a previous study (White et al. 2001), null mutants ecm31 and pan6 were indeed auxotrophic for pantothenate, in contrast to the pan5 mutant phenotype. Yeast gene YDL144C encodes a second protein related to panE but even a double mutant pan5 ydl144c could still grow in the absence of pantothenate. This indicates that activity of dehydropantoate reductase may be provided by a distinct protein which is not similar to the bacterial enzyme.
It has been reported that S. cerevisiae strains auxotrophic for pantothenate show a defect of biosynthesis of β-alanine (Stolz and Sauer 1999) which in yeast is derived from polyamines via a complex oxidative pathway (White et al. 2001, 2003) instead of using decarboxylation of aspartate (panD gene product in E. coli; Cronan et al. 1982). In the absense of de novo biosynthesis of pantothenate, growth is rescued by external supplementation requiring a functional pantothenate transporter (Fen2; Stolz and Sauer 1999). From our failure to obtain a pan6 fen2 double mutant, we conclude that viability of yeast requires either a functional pantothenate transporter (Fen2) or intact biosynthetic genes (ECM31, PAN5 and PAN6). In contrast to what was found for the fission yeast Schizosaccharomyces pombe (viability of a pan6 liz1 [=fen2] double null mutant at 10 μM pantothenate; Stolz et al. 2004), pantothenate uptake via passive diffusion may not be effective in S. cerevisiae. Synthetic lethality of mutations pan6 and fen2 also argues against the existence of a low-affinity pantothenate transporter. This finding parallels the situation in E. coli where panC panF double mutants could not be constructed (panF encoding pantothenate permease; Vallari and Rock 1985).
While previous work on the enzymology of yeast CoA biosynthesis from pantothenate proposed the existence of an alternative pathway (Bucovaz et al. 1980, 1997) comparative genomic analyses taking advantage of the characterization of the human enzymes (Daugherty et al. 2002) supported a uniform order of events. Owing to the absence of a CoA uptake system, biosynthetic genes must be considered as indispensable for cellular viability. Even with fatty acid supplementation, we were unable to obtain viable haploid mutants for the five candidate genes (YDR531W, YIL083C, YKL088W, YGR277C and YDR196C), confirming the results of genome-wide deletion studies (Winzeler et al. 1999; Giaever et al. 2002). In this work, we focused on the essential YDR531W gene which encodes the yeast pantothenate kinase (panK) according to the following criteria: (1) similarity of its gene product to the human enzyme; (2) characterization of a temperature-sensitive mutant (cab1 ts) lacking panK activity under non-permissive conditions; (3) functional complementation of this mutant by YDR531W as well as by the bacterial panK gene coaA and mammalian panK3; (4) lack of complementation of the ydr531w deletion mutation and cab1 ts mutation; (5) identification of missense mutations within the ydr531w allele of the ts mutant strain; (6) strongly increased panK activity after overexpression of YDR531W in a yeast wild-type strain. Taken together, we consider YDR531W as the genuine yeast panK gene and consequently suggest CAB1 (coenzyme A biosynthesis) as its adequate designation. Our finding of heterologous complementation of cab1 ts by mammalian panK3 offers the opportunity of functional in vivo studies with gene variants from other eukaryotes.
Among the five missense mutations found within the cab1 ts mutant, A327T and G351S appeared as most promising to cause the temperature-sensitive phenotype. G351 is the only residue which is entirely conserved among eukaryotic panK enzymes. Superimposition of the Cab1 sequence to the recently solved crystal structure of human panK (panK3; Hong et al. 2007) suggests that G351 should be located in the interior of the enzyme and may not be involved in ATP binding or dimerization. Introduction of the G351S mutation into a functional CAB1 gene led to a variant which could no longer complement cab1 ts. However, the remaining four missense mutations were not assayed individually for their influence on enzyme function. The strong conservation of residues responsible for acetyl-CoA binding among panK3 and the CAB1 gene product (7 residues involved in hydrogen bonds, 13 residues mediating hydrophobic interactions; cf. Fig. 4; Hong et al. 2007) indicates that the yeast panK may also show competitive inhibition by this nucleotide.
Similar to what was found for E. coli coaA and yeast cab1 ts, heterologous complementation studies showed that bacterial genes could also replace yeast genes of late CoA biosynthesis (complementation of Δyil083c and Δykl088w by bifunctional coaBC, Δygr277c by coaD and Δydr196c by coaE). This result provides evidence that the corresponding yeast wild-type genes are indeed involved in coenzyme A biosynthesis. Interestingly, the coaBC gene product shows significant similarity to the carboxy-terminal half of Ykl088w. In contrast, no similarity of protein CoaBC and Yil083c could be detected. Sequences of CoaD and Ygr277c are also unrelated while CoaE and Ydr196c show similarities almost over their entire length (28.2% identity, 40.2% similarity). To introduce a systematic gene nomenclature providing functional information, we thus suggest to rename YIL083C (new designation: CAB2), YKL088W (CAB3), YGR277C (CAB4) and YDR196C (CAB5).
Beyond its central metabolic function, CoA biosynthesis may exhibit a (direct or indirect) function for ribosomal biogenesis. Using a genome-wide strategy (“diploid shuffle”), temperature-sensitive alleles for all essential genes of S. cerevisiae have been constructed recently (Ben-Aroya et al. 2008). Interestingly, the functional characterization of the mutant alleles obtained indicated that YGR277C and YDR196C influence processing of rRNA precursors. Based on the incomplete cleavage of the 35S rRNA precursor the authors conclude that a CoA derivative may be important for processing of the primary rRNA transcript.
Expression analysis of pantothenate and CoA biosynthetic genes showed that these genes are transcribed at a low level which is hardly affected by external stimuli. Although the existence of a specific transporter for external pantothenate may suggest a regulatory feedback on its de novo biosynthesis, transformants containing lacZ fusions of ECM31 and PAN6 did not provide evidence for pantothenate repression of these biosynthetic genes. This is in contrast to several other anabolic pathways which are transcriptionally controlled by key metabolites such as inositol (Chen et al. 2007) and thiamin (Nosaka et al. 2005). Similarly, variation of amino acid availability (minimal medium vs. synthetic complete medium) did not significantly alter expression of seven lacZ fusions assayed. Variation of the carbon source (glucose vs. ethanol) resulted in a 2-2.5-fold increase of reporter gene expression for four of the seven tested fusion genes. These findings essentially agree with the previous microarray data analyzing gene expressions patterns in the course of the diauxic shift (DeRisi et al. 1997; Gasch et al. 2000). De-repression of CoA biosynthetic genes makes some sense, considering the requirement of acetyl-CoA for gluconeogenesis in the presence of a C2 substrate. The moderate increase of gene expression for ECM31, PAN6, CAB1, and YIL083C with ethanol instead of glucose as the sole carbon source was no longer observed in mutants snf1 and cat8 sip4 (not shown) lacking important regulators of the gluconeogenic pathway (Schüller 2003). Since neither promoter contains a sequence reminiscent of a Cat8 and Sip4 binding sites (Roth et al. 2004), the increase of gene expression observed may be indirectly influenced by these transcription factors. Instead, in silico inspection of the promoter sequences of CoA biosynthetic genes gave evidence for binding sites of pleiotropic transcription factors such as Abf1 (ATCTTACATGACG; −180/−168 of CAB1), Reb1 (TGACCCG; −196/−190 of YIL083C) and Rap1 (GTCCATACGC; −185/−176 of YKL088W) possibly mediating their almost constitutive expression at a low level.
References
Ben-Aroya S, Coombes C, Kwok T, O’Donnell KA, Boeke JD, Hieter P (2008) Toward a comprehensive temperature-sensitive mutant repository of the essential genes of Saccharomyces cerevisiae. Mol Cell 30:248–258
Bucovaz ET, Tarnowski SJ, Morrison WC, Macleod RM, Morrison JC, Sobhy CM, Rhoades JL, Fryer JE, Wakim JM, Whybrew WD (1980) Coenzyme A-synthesizing protein complex of Saccharomyces cerevisiae. Mol Cell Biochem 30:7–26
Bucovaz ET, Macleod RM, Morrison JC, Whybrew WD (1997) The coenzyme A-synthesizing protein complex and its proposed role in CoA biosynthesis in bakers’ yeast. Biochimie 79:787–798
Calder RB, Williams RS, Ramaswamy G, Rock CO, Campbell E, Unkles SE, Kinghorn JR, Jackowski S (1999) Cloning and characterization of a eukaryotic pantothenate kinase gene (panK) from Aspergillus nidulans. J Biol Chem 274:2014–2020
Chen M, Hancock LC, Lopes JM (2007) Transcriptional regulation of yeast phospholipid biosynthetic genes. Biochim Biophys Acta 1771:310–321
Cronan JE Jr, Littel KJ, Jackowski S (1982) Genetic and biochemical analyses of pantothenate biosynthesis in Escherichia coli and Salmonella typhimurium. J Bacteriol 149:916–922
Daugherty M, Polanuyer B, Farrell M, Scholle M, Lykidis A, de Crécy-Lagard V, Osterman A (2002) Complete reconstitution of the human coenzyme A biosynthetic pathway via comparative genomics. J Biol Chem 277:21431–21439
de La Sierra-Gallay IL, Collinet B, Graille M, Quevillon-Cheruel S, Liger D, Minard P, Blondeau K, Henckes G, Aufrere R, Leulliot N, Zhou CZ, Sorel I, Ferrer JL, Poupon A, Janin J, van Tilbeurgh H (2004) Crystal structure of the YGR205w protein from Saccharomyces cerevisiae: close structural resemblance to E. coli pantothenate kinase. Proteins 54:776–783
DeRisi JL, Iyer VR, Brown PO (1997) Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278:680–686
Fichtlscherer F, Wellein C, Mittag M, Schweizer E (2000) A novel function of yeast fatty acid synthase. Subunit α is capable of self-pantetheinylation. Eur J Biochem 267:2666–2671
Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO (2000) Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell 11:4241–4257
Giaever G, Chu AM, Ni L, Connelly C, Riles L, Véronneau S, Dow S, Lucau-Danila A, Anderson K, André B, Arkin AP, Astromoff A, El-Bakkoury M, Bangham R, Benito R, Brachat S, Campanaro S, Curtiss M, Davis K, Deutschbauer A, Entian KD, Flaherty P, Foury F, Garfinkel DJ, Gerstein M, Gotte D, Güldener U, Hegemann JH, Hempel S, Herman Z, Jaramillo DF, Kelly DE, Kelly SL, Kötter P, LaBonte D, Lamb DC, Lan N, Liang H, Liao H, Liu L, Luo C, Lussier M, Mao R, Menard P, Ooi SL, Revuelta JL, Roberts CJ, Rose M, Ross-Macdonald P, Scherens B, Schimmack G, Shafer B, Shoemaker DD, Sookhai-Mahadeo S, Storms RK, Strathern JN, Valle G, Voet M, Volckaert G, Wang CY, Ward TR, Wilhelmy J, Winzeler EA, Yang Y, Yen G, Youngman E, Yu K, Bussey H, Boeke JD, Snyder M, Philippsen P, Davis RW, Johnston M (2002) Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387–391
Gietz RD, Sugino A (1988) New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74:527–534
Hong BS, Senisterra G, Rabeh WM, Vedadi M, Leonardi R, Zhang YM, Rock CO, Jackowski S, Park HW (2007) Crystal structures of human pantothenate kinases Insights into allosteric regulation and mutations linked to a neurodegeneration disorder. J Biol Chem 282:27984–27993
Leonardi R, Zhang YM, Rock CO, Jackowski S (2005) Coenzyme A: back in action. Prog Lipid Res 44:125–153
Mumberg D, Müller R, Funk M (1994) Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res 22:5767–5768
Myers AM, Tzagoloff A, Kinney DM, Lusty CJ (1986) Yeast shuttle and integrative vectors with multiple cloning sites suitable for construction of lacZ fusions. Gene 45:299–310
Nosaka K, Onozuka M, Konno H, Kawasaki Y, Nishimura H, Sano M, Akaji K (2005) Genetic regulation mediated by thiamin pyrophosphate-binding motif in Saccharomyces cerevisiae. Mol Microbiol 58:467–479
Rock CO, Calder RB, Karim MA, Jackowski S (2000) Pantothenate kinase regulation of the intracellular concentration of coenzyme A. J Biol Chem 275:1377–1383
Roth S, Kumme J, Schüller HJ (2004) Transcriptional activators Cat8 and Sip4 discriminate between sequence variants of the carbon source-responsive promoter element in the yeast Saccharomyces cerevisiae. Curr Genet 45:121–128
Ruiz A, Muñoz I, Serrano R, González A, Simón E, Ariño J (2004) Functional characterization of the Saccharomyces cerevisiae VHS3 gene: a regulatory subunit of the Ppz1 protein phosphatase with novel, phosphatase-unrelated functions. J Biol Chem 279:34421–34430
Schüller HJ (2003) Transcriptional control of nonfermentative metabolism in the yeast Saccharomyces cerevisiae. Curr Genet 43:139–160
Schüller HJ, Hahn A, Tröster F, Schütz A, Schweizer E (1992) Coordinate genetic control of yeast fatty acid synthetase genes FAS1 and FAS2 by an upstream activation site common to genes involved in the membrane lipid biosynthesis. EMBO J 11:107–114
Schwank S, Ebbert R, Rautenstrauss K, Schweizer E, Schüller HJ (1995) Yeast transcriptional activator INO2 interacts as an Ino2p/Ino4p basic helix-loop-helix heteromeric complex with the inositol/choline-responsive element necessary for expression of phospholipid biosynthetic genes in Saccharomyces cerevisiae. Nucleic Acids Res 23:230–237
Schweizer E, Kniep B, Castorph H, Holzner U (1973) Pantetheine-free mutants of the yeast fatty-acid-synthetase complex. Eur J Biochem 39:353–362
Stolz J, Sauer N (1999) The fenpropimorph resistance gene FEN2 from Saccharomyces cerevisiae encodes a plasma membrane H+-pantothenate symporter. J Biol Chem 274:18747–18752
Stolz J, Caspari T, Carr AM, Sauer N (2004) Cell division defects of Schizosaccharomyces pombe liz1 - mutants are caused by defects in pantothenate uptake. Eukaryot Cell 3:406–412
Vallari DS, Rock CO (1985) Isolation and characterization of Escherichia coli pantothenate permease (panF) mutants. J Bacteriol 164:136–142
Vallari DS, Jackowski S, Rock CO (1987) Regulation of pantothenate kinase by coenzyme A and its thioesters. J Biol Chem 262:2468–2471
White WH, Gunyuzlu PL, Toyn JH (2001) Saccharomyces cerevisiae is capable of de novo pantothenic acid biosynthesis involving a novel pathway of β-alanine production from spermine. J Biol Chem 276:10794–10800
White WH, Skatrud PL, Xue Z, Toyn JH (2003) Specialization of function among aldehyde dehydrogenases: the ALD2 and ALD3 genes are required for β-alanine biosynthesis in Saccharomyces cerevisiae. Genetics 163:69–77
Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, Chu AM, Connelly C, Davis K, Dietrich F, Dow SW, El Bakkoury M, Foury F, Friend SH, Gentalen E, Giaever G, Hegemann JH, Jones T, Laub M, Liao H, Liebundguth N, Lockhart DJ, Lucau-Danila A, Lussier M, M’Rabet N, Menard P, Mittmann M, Pai C, Rebischung C, Revuelta JL, Riles L, Roberts CJ, Ross-MacDonald P, Scherens B, Snyder M, Sookhai-Mahadeo S, Storms RK, Véronneau S, Voet M, Volckaert G, Ward TR, Wysocki R, Yen GS, Yu K, Zimmermann K, Philippsen P, Johnston M, Davis RW (1999) Functional characterization of the S cerevisiae genome by gene deletion and parallel analysis. Science 285:901–906
Acknowledgments
We thank Prof. Eckhart Schweizer (Erlangen) for kindly providing the cab1 ts mutant strain (initially designated ts6629) and Sonja Burghardt, Karola Hahn, Felix Kliewe, Kati Landsberg and Jan Witthöft for support.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by K. Breunig.
Rights and permissions
About this article
Cite this article
Olzhausen, J., Schübbe, S. & Schüller, HJ. Genetic analysis of coenzyme A biosynthesis in the yeast Saccharomyces cerevisiae: identification of a conditional mutation in the pantothenate kinase gene CAB1 . Curr Genet 55, 163–173 (2009). https://doi.org/10.1007/s00294-009-0234-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00294-009-0234-1