Abstract
Saccharomyces cerevisiae plays a primordial role in alcoholic fermentation and has a vast worldwide application in the production of fuel-ethanol, food and beverages. The dominance of S. cerevisiae over other microbial species during alcoholic fermentations has been traditionally ascribed to its higher ethanol tolerance. However, recent studies suggested that other phenomena, such as microbial interactions mediated by killer-like toxins, might play an important role. Here we show that S. cerevisiae secretes antimicrobial peptides (AMPs) during alcoholic fermentation that are active against a wide variety of wine-related yeasts (e.g. Dekkera bruxellensis) and bacteria (e.g. Oenococcus oeni). Mass spectrometry analyses revealed that these AMPs correspond to fragments of the S. cerevisiae glyceraldehyde 3-phosphate dehydrogenase (GAPDH) protein. The involvement of GAPDH-derived peptides in wine microbial interactions was further sustained by results obtained in mixed cultures performed with S. cerevisiae single mutants deleted in each of the GAPDH codifying genes (TDH1-3) and also with a S. cerevisiae mutant deleted in the YCA1 gene, which codifies the apoptosis-involved enzyme metacaspase. These findings are discussed in the context of wine microbial interactions, biopreservation potential and the role of GAPDH in the defence system of S. cerevisiae.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Alcoholic fermentation is the main biotransformation that occurs during winemaking, brewery and fuel-ethanol production. Since these industrial processes are conducted under non-sterile growth conditions, a huge variety of microorganisms is present and can participate in the fermentative process. Although several yeast and bacteria are able to perform alcoholic fermentation, Saccharomyces cerevisiae is the dominant microorganism in all those processes, being usually called the “wine yeast”. During spontaneous wine fermentations, there is a consistent growth pattern in which the non-Saccharomyces species belonging to the natural microflora of grape musts (e.g. Hanseniaspora guilliermondii, Hanseniaspora uvarum, Candida stellata, Kluyveromyces thermotolerans, Kluyveromyces marxianus and Torulaspora delbrueckii) grow during the early stages of fermentation (up to 4–5 % v/v of ethanol) but then begin to die-off giving way to S. cerevisiae strains to complete the fermentation process (Fleet and Heard 1993; Pretorius 2000).
The ability of S. cerevisiae to displace other microbial species during alcoholic fermentation has been always attributed to its higher fermentative power and capacity to withstand the increasingly adverse conditions established in the medium as the fermentation progresses, i.e. high levels of ethanol and organic acids, low pH values, scarce oxygen availability and depletion of certain nutrients (Bisson 1999; Bauer and Pretorius 2000; Hansen et al. 2001). However, the weight of these factors on microbial succession during wine fermentations has been recently questioned and other microbial interactions were proposed by different authors, such as growth arrest mediated by a cell–cell contact mechanism (Nissen and Arneborg 2003; Nissen et al. 2003; Arneborg et al. 2005) and death mediated by killer-like toxins (Comitini et al. 2005; Pérez-Nevado et al. 2006; Osborne and Edwards 2007; Albergaria et al. 2010).
The killer phenomenon has long been recognized among wine yeasts, although the relation between killer activity and the early disappearance of non-Saccharomyces yeasts from wine fermentations was never established because the killer toxins produced by S. cerevisiae that were identified up till now (K1, K2 and K28) are active only against strains of the same species (Pérez et al. 2001). Nevertheless, there are increasingly growing evidences suggesting the involvement of other killer-like toxins in the yeast–yeast and yeast–bacteria interactions in wine fermentations. Indeed, Comitini et al. (2005), as well as Osborne and Edwards (2007) and Nehme et al. (2010), found that certain S. cerevisiae strains produce proteinaceous compounds that are active against malolactic bacteria. Likewise, in a previous work we demonstrated that S. cerevisiae CCMI 885 produces peptides (<10 kDa) that inhibit the growth of H. guilliermondii, T. delbrueckii, K. marxianus and K. thermotolerans (Albergaria et al. 2010). However, the identity of these antimicrobial peptides (AMPs) remained elusive.
In the present work, we purified the previously found AMPs and characterized them regarding their amino acid sequence, encoding genes and antimicrobial spectrum of action. The role of these AMPs in wine microbial interactions was also investigated using S. cerevisiae mutant strains deleted in the AMPs encoding genes.
Materials and methods
Strains and inoculum cultures
The following S. cerevisiae strains were used: CCMI 885 (Culture Collection of Industrial Microorganisms of ex-INETI, Lisbon, Portugal); BY4741 (MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) strain and its isogenic derivative strains Δtdh1 (YJL052w::kanMX4), Δtdh2 (YJR009c::kanMX4) and Δtdh3 (YGR192c::kanMX4) (EUROSCARF, Frankfurt, Germany); BY4742 (MATα his3Δ1 leu2Δ0 lys2Δ0; ura3Δ0) strain and its isogenic derivative strain Δyca1 (EUROSCARF, Frankfurt, Germany). The non-Saccharomyces strains used were: Dekkera bruxellensis ISA 1649, ISA 1700, ISA 1791, ISA 2104, ISA 2116, ISA 2211 (Instituto Superior de Agronomia, Lisbon, Portugal); H. guilliermondii NCYC 2380 (National Collection of Yeast Cultures, Norwich, United Kingdom); K. marxianus PYCC 2671 (Portuguese Yeast Culture Collection, New University of Lisbon, Portugal); K. thermotolerans PYCC 2908 and T. delbrueckii PYCC 4478. Two strains of Oenococcus oeni were used: ISA 4279 and DSM 2529 (German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany). Inoculums of all yeast strains were obtained by transferring one YEPD-agar slant of each strain (pre-grown at 30 °C for 48–72 h) into 50 ml of YEPD medium (10 g/l yeast extract, 20 g/l peptone and 20 g/l glucose) and incubating cultures at 30 °C with 150 rpm of agitation during 16 h (for Dekkera strains incubation took 48–72 h). Inoculums of O. oeni strains were prepared by transferring 1 ml of stock culture into 9 ml of MRS broth and incubating cultures at 25 °C without agitation for 48 h.
Purification of antimicrobial peptides from S. cerevisiae supernatants
AMPs were purified from S. cerevisiae CCMI 885 supernatants of alcoholic fermentations performed in synthetic grape juice (SGJ), prepared as described in Pérez-Nevado et al. (2006), at 25 °C for 7 days. Cell-free supernatants (filtration by 0.22 μm Millipore membranes, Merck Millipore, Algés, Portugal) were first ultrafiltrated through centrifugal filter units (Vivaspin 15R, Sartorius, Göttingen, Germany) equipped with 10 kDa membranes and then concentrated (100-fold) with 2 kDa membranes. This concentrated fraction was first fractionated by gel filtration chromatography, using a Superdex-Peptide column (10/300 GL, GE Healthcare, London, UK) coupled to a High-Performance Liquid Chromatography (HPLC) system (Merck Hitachi, Darmstadt, Germany) equipped with an UV detector (Merck Hitachi, Darmstadt, Germany). One hundred microlitres of fraction was eluted with ammonium acetate 0.1 M at a flow rate of 0.7 ml/min. All fractions (Fig. 1a) were collected into 2 ml Eppendorf, freeze-dried and stored until utilisation. They were all screened for antimicrobial activity and fraction-II was identified as the most active fraction. Fraction-II was then further purified using a strong anion-exchange column (Q-Resource 6 ml, GE Healthcare, London, UK). Peptides were eluted at neutral pH using a gradient of ammonium acetate of 5–500 mM between 10 and 40 min at a flow rate of 1 ml/min. Fractions obtained were collected, vacuum-dried and screened for antimicrobial activity after resuspended in appropriated medium. Three anionic fractions (Fraction II-A, II-B and II-C in Fig. 1b) revealed antimicrobial activity against H. guilliermondii and were analysed by mass spectrometry.
Chromatographic steps used in the purification process of the AMPs. First, the concentrated peptidic (<10 kDa) fraction obtained from S. cerevisiae CCMI 885 supernatants was fractionated by gel filtration chromatography (a) and fractions were collected and checked for antimicrobial activity against Hanseniaspora (c). Since only Fraction-II exhibited strong antimicrobial activity, this fraction was further fractionated using a strong anion-exchange column (b) and the three anionic fractions obtained were tested for antimicrobial activity against Hanseniaspora (d)
Screening of antimicrobial activity and determination of the spectrum of action
Antimicrobial activity of fractions obtained in the different purification steps (gel filtration and anion-exchange chromatography) were tested against the sensitive strain H. guilliermondii using a total protein concentration of 1 mg/ml for gel filtration fractions (Fig. 1c) and 0.5 mg/ml for the anionic fractions (Fig. 1d). The spectrum of action of fraction-II was determined against H. guilliermondii, K. marxianus, K. thermotolerans, T. delbrueckii, D. bruxellensis and O. oeni, following the procedure described below.
All antimicrobial tests were performed in 96-well microplates using YEPD medium for yeast strains and MRS broth for O. oeni strains. Lyophilised fractions were resuspended in the respective growth medium (YEPD or MRS with 30 g/l of ethanol and pH 3.5) to a final protein concentration of 1 mg/ml in total volume of 100 μl. Control assays for each strain were performed using the respective growth medium without addition of the AMPs. The initial cell density in the antimicrobial tests was 105 cells/ml for yeasts and 106 cells/ml for bacteria strains. The microplates were incubated in a thermo-Shaker (Infors HT, Bottmingen, Switzerland) at 30 °C under 700 rpm of agitation for yeasts and at 25 °C without agitation for O. oeni. Growth of the cultures was followed by absorbance measurements at 590 nm using a microplate reader (Dinex Technologies Inc., Chantilly, USA) and also by the enumeration of colonies forming units (CFU) using the classical plating method (as described in the “Cell growth” sub-section), during the time-course of the experiments. All antimicrobial tests were performed in triplicates.
Mass spectrometry (LC-ESI-MS/MS) and sequence analysis
Peptides present in the three anionic fractions were sequenced by liquid chromatography-electrospray ionization-tandem mass spectrometry (LC-ESI-MS/MS). For LC-ESI-MS/MS analysis of peptide mixtures, on-line nano-flow liquid chromatography was performed using the Ultimate 300 RSLC (Dionex, Voisins le Bretonneux, France) with 15 cm nanocapillary columns of an internal diameter of 75 μm (Acclaim Pep Map RSLC, Dionex). The solvent gradient from 4 % to 50 % acetonitrile in 0.5 % formic acid was run at a flow rate of 300 nl/min for 30 min. The eluate was electrosprayed into an LTQ Velos mass spectrometer (Thermo Fisher Scientific, Courtaboeuf, France) through a nanoelectrospray ion source. The LTQ Velos was operated in a CID top 10 mode (i.e. one full scan MS from which 10 major peaks are selected for MS/MS). Raw data files were processed with search engines installed in-house, Mascot (version 2.2, Matrix Science, London, UK) and PEAKS studio (version 5.3, Bioinformatics Solutions Inc., Waterloo, Canada). For peptide identification, the UniProt taxonomy S. cerevisiae (6,650 sequences) protein database was used and the parameters for searching were: none enzyme and possible oxidation of methionine. Peptide mass tolerance and fragment mass tolerance were set to 1.5 and 0.8 Da, respectively. Peptides identification was validated when significant Mascot and PEAKS scores were obtained with false discovery rate <1 %. In addition, a manual validation of MS/MS spectra was performed to be sure of the peptide sequence.
Alcoholic fermentations performed with mixed cultures of H. guilliermondii and S. cerevisiae
Alcoholic fermentations were performed in 150 ml of SGJ (supplemented with 200 mg/l of L-leucine, 120 mg/l of L-histidine, 180 mg/l of L-methionine and 120 mg/l of uracil) using mixed cultures of H. guilliermondii with each of the following S. cerevisiae strains: BY4741 and its isogenic derivatives Δtdh1, Δtdh2, Δtdh3; and BY4742 and its isogenic derivative Δyca1. Fermentations were carried out at 25 °C under slow agitation (80 rpm) with an initial cell density of 105 cells/ml of each species. A single culture fermentation of H. guilliermondii was also performed under the same growth conditions to compare its cell viability under single and mixed culture. All fermentations were performed in duplicates and daily samples were taken to determine cell viability, sugars consumption and ethanol production.
Cell growth
Culturability was determined by the classical plating method both in the antimicrobial tests and alcoholic fermentations. Briefly, samples were plated onto YEPD-agar plates, after appropriate dilution (decimal serial dilution method) and incubated at 25 °C in a Vertical Incubator (Infors, Anjou, Canada) and the number of colony forming units (CFU) enumerated after 2–6 days. In the mixed culture fermentations the CFU counts of H. guilliermondii were obtained on 0.01 % cycloheximide YEPD-agar plates and CFU counts of S. cerevisiae as the difference between total CFU counts on YEPD-agar plates and CFU counts of H. guilliermondii. Enumerations of CFU counts in the antimicrobial tests were determined on YEPD-agar plates both for yeasts and bacteria.
Sugars consumption and ethanol production
Glucose, fructose and ethanol concentrations in alcoholic fermentations were analysed using a High-Performance Liquid Chromatography (HPLC) system (Merck Hitachi, Darmstadt, Germany) equipped with a refractive index detector (L-7490, Merck Hitachi, Darmstadt, Germany). Fermentation samples were first filtrated by 0.45 μm Millipore membranes (Merck Millipore, Algés, Portugal) and then injected on a Sugar-Pak column (Waters Hitachi, Milford, USA) and eluted with a degassed aqueous mobile phase of CaEDTA (50 mg/l) at 90 °C using a flow rate of 0.5 ml/min. All samples were analysed in duplicate.
Results
Purification and identification of AMPs from S. cerevisiae fermentation supernatants
In the previous work (Albergaria et al. 2010), we found that S. cerevisiae CCMI 885 supernatants obtained from alcoholic fermentations performed in SGJ, contained a peptidic fraction (<10 kDa) active against some wine-related yeasts. This peptidic fraction was first fractionated by gel filtration chromatography (Fig. 1a) and all fractions were collected, lyophilised and then screened for antimicrobial activity. Results revealed that fraction-II exhibited strong antimicrobial activity (Fig. 1c) and thus this fraction was further purified by ion-exchange chromatography. Since most AMPs are cationic in nature fraction-II was first pooled into a cation-exchange column (S-Resource) but none of the fractions obtained exhibited antimicrobial activity against H. guilliermondii (data not shown). Thus, fraction-II was pooled into a strong anion-exchange column Fig. 1b and the three anionic (at neutral pH) fractions obtained (fractions II-A, II-B and II-C) were screened for antimicrobial activity against H. guilliermondii (Fig. 1d).
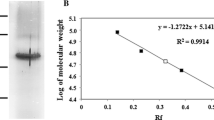
Since all anionic fractions showed antimicrobial activity peptides were sequenced by liquid chromatography-electrospray ionization-tandem mass spectrometry (LC-ESI-MS/MS). Sequence analysis revealed that all peptides present in each anionic fraction correspond to fragments of the S. cerevisiae glyceraldehyde-3-phosphate dehydrogenase (GAPDH) isoenzymes, GAPDH2/3 and GAPDH1 (Table 1), which are encoded by the TDH2, TDH3 and TDH1 genes, respectively. Two main peptides with molecular weights of 1.638 and 1.622 kDa and the following amino acid residues VSWYDNEYGYSTR and ISWYDNEYGYSAR were identified in each fraction (Fig. 2). The theoretical isoelectric point (pI) of these peptides estimated by the ExPASy software (http://www.expasy.ch/tools/peptide-mass.html) is 4.37.
Mass spectrometry spectra (MS/MS) of the two main peptides (double charged) found and validated in each of the anionic fractions obtained by anion-exchange chromatography (fractions II-A, II-B and II-C indicated in Fig. 1b). (a) Peptide VSWYDNEYGYSTR, m/z 820.24, [M + 2H]2+. (b) Peptide ISWYDNEYGYSAR, m/z 812.24, [M + 2H]2+
Spectrum of action of the AMPs
The spectrum of action of the antimicrobial peptides (AMPs) was determined against several wine-related yeasts (H. guilliermondii, K. thermotolerans, T. delbrueckii, K. marxianus and D. bruxellensis) and bacteria (O. oeni). Results showed that under the conditions tested (YEPD or MRS, at pH 3.5) the AMPs inhibited the growth of all these microbial species (Fig. 3), although for some yeast strains (e.g. T. delbrueckii) only a fungistatic effect was observed, while on others a fungicide effect was shown. H. guilliermondii showed to be the most sensitive yeast, with total death occurring after 14 h of incubation with the AMPs, followed by K. marxianus and D. bruxellensis strain ISA 2211 with total death established within 44 h and 96 h, respectively. The present results revealed that these AMPs are active against a wide variety of microorganisms associated with wine fermentations, although the sensitivity of these microbial species towards the AMPs is strain-specific, as shown by the results obtained for different strains of D. bruxellensis and O. oeni (Fig. S1 in the Electronic supplementary material (ESM)). Moreover, these AMPs also showed inhibitory activity when tested in a modified-SGJ (20 g/l of sugars, 30 g/l of ethanol) at pH 3.5 (data not shown).
Viable cells (CFU) and optical density (OD) of Hanseniaspora guilliermondii (a), Kluyveromyces marxianus (b), Torulaspora delbrueckii (c), Kluyveromyces thermotolerans (d), Dekkera bruxellensis strain ISA 2211 (e) and Oenoccocus oeni strain DSM 2529 (f) in the antimicrobial tests (AMT) performed with YEPD medium (at pH 3.5) for yeasts and MRS broth (at pH 3.5) for bacteria, without addition of Fraction-II (control) and with addition of 1 mg/ml of Fraction-II (AMT). Data represented correspond to mean values of triplicate independent assays ± SD (error bars)
Role of GAPDH-derived peptides in S. cerevisiae antagonism
In order to further confirm that GAPDH-derived peptides are involved in the early death of non-Saccharomyces during wine fermentations, we performed alcoholic fermentations with mixed cultures of H. guilliermondii and S. cerevisiae mutant strains deleted in each of the TDH1-3 genes. S. cerevisiae wild-type strain BY4741 was used in mixed culture with H. guilliermondii as positive control and H. guilliermondii in single culture for negative control. Growth profiles of both yeasts (Fig. 4a, b) showed that all strains were able to grow during the first 1–2 days of fermentation, but then H. guilliermondii began to die off (Fig. 4b) while S. cerevisiae kept its cell density at about 107 CFU/ml until the end of fermentation (Fig. 4a). However, death rate of H. guilliermondii significantly varied whether in single or in mixed culture and was faster in the mixed cultures performed with the wild-type strain of S. cerevisiae, particularly after the 3rd day of fermentation. Interestingly, the death rate of H. guilliermondii was much slower in the mixed cultures performed with the ∆tdh1, ∆tdh2 and ∆tdh3 mutants than in the mixed culture performed with the wild-type strain (Fig. 4b). Although ethanol levels had varied among fermentations, when we compare the ethanol profile of the mixed fermentation performed with the wild-type strain and those of ∆tdh1 mutants (Fig. 4c), it can be clearly seen that ethanol cannot explain the differences observed in the death rates of H. guilliermondii on those fermentations (Fig. 4b).
Cell growth of S. cerevisiae (a) and H. guilliermondii (b) and sugar consumption and ethanol production (c) during alcoholic fermentations performed with H. guilliermondii in single and mixed cultures with each of the following S. cerevisiae strains: wild-type BY4741 (Wt), Δtdh1 (TDH1), Δtdh2 (TDH2) and Δtdh3 (TDH3). Data represented correspond to mean values of duplicate independent assays ± SD (error bars)
Recently, Silva et al. (2011) reported that GAPDH is a specific substrate of yeast metacaspase and showed that the in vivo cleavage of GAPDH by metacaspase originated several GAPDH-derived peptide fragments, namely some equal to the ones identified in the present work. In view of this information, we wondered if a S. cerevisiae mutant deleted on the metacaspase YCA1 gene would not prevent the production of GAPDH-derived peptides and thus avoid death of non-Saccharomyces in fermentations with S. cerevisiae. To confirm this hypothesis we performed alcoholic fermentations with H. guilliermondii in co-culture with S. cerevisiae wild-type strain BY4742 and its isogenic mutant ∆yca1. Since cycloheximide is a well-known inhibitor of apoptosis, we also performed fermentation with H. guilliermondii and S. cerevisiae wild-type strain, adding cycloheximide after the first day of fermentation. Fermentation of H. guilliermondii in single culture was used as negative control. Results showed (Fig. 5b) that the use of both cycloheximide and ∆yca1 strain in mixed cultures with H. guilliermondii significantly prevented death of the non-Saccharomyces strain, which was able to keep its culturability at relatively high levels (ca 104 CFU/ml) until the end of fermentation (7 days). In both cases, the culturability of H. guilliermondii after 5 days was four orders of magnitude higher than in the mixed culture fermentation performed with the wild-type strain (without cycloheximide) and comparable to the one observed in the H. guilliermondii single culture fermentation. It is important to notice that ethanol profiles in the mixed cultures performed with the ∆yca1 strain and with the wild-type strain (without cycloheximide) were quite similar (Fig. 5c), which confirmed that minor death of H. guilliermondii in the presence of the S. cerevisiae ∆yca1 strain was not caused by ethanol.
Cell growth of S. cerevisiae (a) and H. guilliermondii (b) and sugar consumption and ethanol production (c) during alcoholic fermentations performed with H. guilliermondii in single and mixed cultures with S. cerevisiae wild-type strain BY4742 (Wt), wild-type strain in the presence of cycloheximide (Wt + CH) and mutant strain Δyca1 (YCA1). Arrow indicates the point at which 0.001 % of cycloheximide was added to the culture. Data represented correspond to mean values of duplicate independent assays ± SD (error bars)
In order to confirm the presence/absence of the previously identified GAPDH-derived anionic AMPs in these mixed cultures, we analysed the chromatographic profiles of S. cerevisiae BY4742 and ∆yca1 supernatants (5 days-old), using the same purification procedure. Results showed (Fig. 6) that the anion-exchange chromatographic profile of S. cerevisiae BY4742 is very similar to the one exhibited by S. cerevisiae CCMI 885 (Fig. 1b) but quite different from that of S. cerevisiae ∆yca1. Moreover, antimicrobial tests performed with these anionic fractions showed that all BY4742 anionic fractions killed H. guilliermondii, while the ∆yca1 anionic fraction only exerted a minor inhibitory effect over the same strain (data not shown). These results confirmed that the bioactive anionic peptides are absent, or at least are present at much lower amounts, in S. cerevisiae ∆yca1 supernatants what explains that H. guilliermondii dies less in co-cultivation with ∆yca1 than with BY4742 or CCMI 885 strains (Fig. 5b).
Discussion
We had previously reported that S. cerevisiae CCMI 885 produces antimicrobial peptides (AMPs < 10 kDa) that inhibit the growth of H. guilliermondii, T. delbrueckii, K. marxianus and K. thermotolerans during alcoholic fermentations performed with mixed cultures (Albergaria et al. 2010). In the present work, we show that these AMPs are derived from the glycolytic enzyme glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Although surprising, this finding is supported by several studies showing that GAPDH, besides its glycolytic role, displays several other activities in different subcellular locations (membrane, cytosol and nucleus), including a primary role in apoptosis and in a variety of critical nuclear pathways (Sirover 2005; Nakajima et al. 2009; Silva et al. 2011). More importantly, in two recent publications, GAPDH-derived AMPs with antifungal activity were isolated, one from the human placental tissue (Wagener et al. 2013) and another from the skin of yellowfin tuna (Seo et al. 2012). Those AMPs correspond to small cationic peptides that match the N-terminal (2-32) amino acid sequence of the human and fish GAPDH protein, respectively, and are active against the pathogenic yeast Candida albicans. Conversely, the AMPs here identified are anionic (at neutral pH), match the C-terminal (309-321) amino acid sequence of the S. cerevisiae GAPDH protein and are active against several wine-related yeasts (e.g. D. bruxellensis) and bacteria (e.g. O. oeni). Regarding the anionic nature of these AMPs it is important to emphasize that at the acidic pH conditions of wine fermentations (pH ranging 3.0–3.5), peptides with pI = 4.37 are not negatively charged. In spite of these differences, the above mentioned findings clearly indicate that GAPDH plays an important role in the defense system of different organisms.
In the yeast S. cerevisiae, three related but not identical GAPDH isoenzymes with different specific activities are encoded by unlinked genes designated TDH1, TDH2 and TDH3 (McAlister and Holland 1985a). McAlister and Holland (1985b) have also shown that none of these TDH genes are individually essential for cell viability, but a functional copy of either TDH2 or TDH3 is required for growth since Δtdh2Δtdh3 cells are not viable. For this reason, in the present work, we have used single mutant strains of S. cerevisiae deleted in each of the TDH1-3 genes in mixed cultures with H. guilliermondii to evaluate their impact on the early death of the non-Saccharomyces yeast. Our results show that deletion of each of these genes in S. cerevisiae reduces its antagonism against H. guilliermondii, thus further demonstrating that GAPDH is involved in wine microbial interactions.
Delgado et al. (2001) found that each of the three GAPDH polypeptides encoded by the TDH1-3 genes is associated with the cell wall of S. cerevisiae. The same authors also demonstrated that GAPDH accumulates in the cell wall of S. cerevisiae in response to starvation and temperature upshift (Delgado et al. 2003). Beyond this stress response, specifically related to the cell-wall-associated GAPDH, a recent work by Silva et al. (2011) identified GAPDH as a specific target of metacaspase in S. cerevisiae, thus proving GAPDH is associated with apoptosis in S. cerevisiae. In a previous work (Albergaria et al. 2010), we showed that S. cerevisiae begins to secrete AMPs to the extracellular medium at the end of the exponential growth phase (1–2 days) in alcoholic fermentations. In addition, our current work also shows that a mutant strain of S. cerevisiae deleted in the metacaspase YCA1 gene significantly prevents death of H. guilliermondii during alcoholic fermentation. Taken together, these findings suggest that the presence of GAPDH-derived peptides in the extracellular media at the end of exponential growth phase might be due to apoptotic cells of S. cerevisiae inducing the cleavage of GAPDH by metacaspases. However, to definitively establish this connection between apoptosis and secretion of AMPs further investigation must be carried out.
Most industrial processes involving alcoholic fermentations with Saccharomyces strains, such as wine, beer or fuel-ethanol production, are carried out under non-sterile growth conditions due to technical and economic reasons, with high risks of microbial contamination. Wine contamination problems can occur at multiple stages of the winemaking process, and can lead to stuck fermentations, low levels of ethanol and the presence of off-flavours in wine. Likewise, contaminations of industrial fuel-ethanol fermentations by yeasts and bacteria have a negative impact on ethanol yield and productivity (Liberal et al. 2007). In both fermentation processes, spoilage microorganisms can include a wide variety of yeasts, namely those of the species D. bruxellensis and Zygosaccharomyces bailii, and bacteria, such as lactic acid and acetic acid bacteria (Loureiro and Malfeito-Ferreira 2003; Liberal et al. 2007). Chemical preservatives such as sulphur dioxide (SO2) are commonly used in winemaking to prevent the development of spoilage microorganisms. However, some wine contaminants such as Pichia spp. and Dekkera spp. can resist to the SO2 levels used on those processes (Barata et al. 2008; Basílio et al. 2008) that cannot be too high in order to allow fermentation by S. cerevisiae strains. In this context, proteins and peptides exhibiting antimicrobial properties might have a remarkable potential for food preservation and control of spoilage microorganisms, although in the winemaking process selectivity is required for not affecting beneficial microorganisms. The use of killer toxins produced by Pichia anomala and Kluyveromyces wickerhamii with fungicidal effects against D. bruxellensis in wine has been reported (Comitini et al. 2004; Comitini and Ciani 2011). In the present work, we found that S. cerevisiae secretes AMPs during alcoholic fermentation that are active at oenological growth conditions against a wide variety of wine-related microbial species, including D. bruxellensis strains. Thus, the possibility of using these AMPs as natural alternative biopreservatives in alcoholic fermentations, wine and/or other food products looks promising and will be further assessed.
References
Albergaria H, Francisco D, Gori K, Arneborg N, Gírio F (2010) Saccharomyces cerevisiae CCMI 885 secretes peptides that inhibit the growth of some non-Saccharomyces wine-related strains. Appl Microbiol Biotechnol 86:965–972. doi:10.1007/s00253-009-2409-6
Arneborg N, Siegumfeldt H, Andersen GH, Nissen P, Daria VR, Rodrigo PJ, Glückstad J (2005) Interactive optical trapping shows that confinement is a determinant of growth in a mixed yeast culture. FEMS Microbiol Lett 245:155–159. doi:10.1016/j.femsle.2005.03.008
Barata A, Caldeira J, Botelheiro R, Pagliara D, Malfeito-Ferreira M, Loureiro V (2008) Survival patterns of Dekkera bruxellensis in wines and inhibitory effect of sulphur dioxide. Int J Food Microbiol 121:201–207. doi:10.1016/j.ijfoodmicro.2007.11.020
Basílio ACM, Araújo PCM, Morais JOF, Silva-Filho EA, Morais MA, Simões DA (2008) Detection and identification of wild yeast contaminants of the industrial fuel ethanol fermentation process. Curr Microbiol 56:322–226. doi:10.1007/s00284-007-9085-5
Bauer FF, Pretorius IS (2000) Yeast stress response and fermentation efficiency: how to survive the making of wine—a review. S Afr J Enol Vitic 21:27–51
Bisson LF (1999) Stuck and sluggish fermentations. Am J Enol Vitic 50:107–119
Comitini F, Ciani M (2011) Kluyveromyces wickerhamii killer toxin: purification and activity towards Brettanomyces/Dekkera yeasts in grape must. FEMS Microbiol Lett 316:77–82. doi:10.1111/j.1574-6968.2004.tb09761.x
Comitini F, De JI, Pepe L, Mannazzu I, Ciani M (2004) Pichia anomala and Kluyveromyces wickerhamii killer toxins as new tools against Dekkera/Brettanomyces spoilage yeasts. FEMS Microbiol Lett 238:235–240. doi:10.1016/j.femsle.2004.07.040
Comitini F, Ferretti R, Clementi F, Mannuzzu I, Ciani M (2005) Interactions between Saccharomyces cerevisiae and malolactic bacteria: preliminary characterization of a yeast proteinaceous compound(s) active against Oenococcus oeni. J Appl Microbiol 99:105–111. doi:10.1111/j.1365-2672.2005.02579.x
Delgado ML, O’Connor JE, Azorin I, Renau-Piqueras J, Gil ML, Gozalbo D (2001) The glyceraldehyde-3-phosphate dehydrogenase polypeptides encoded by the Saccharomyces cerevisiae TDH1, TDH2 and TDH3 genes are also cell wall proteins. Microbiol 147:411–417
Delgado ML, Gil ML, Gozalbo D (2003) Starvation and temperature upshift cause an increase in the enzymaticaly active cell wall-associated glyceraldehyde-3-phosphate dehydrogenase protein in yeast. FEMS Yeast Res 4:297–303. doi:10.1016/S1567-1356(03)00159-4
Fleet GH, Heard GM (1993) Yeast growth during fermentation. In: Fleet GH (ed) Wine microbiology and biotechnology. Harwood Academic Publishers, Basel, Switzerland, pp 27–54
Hansen EH, Nissen P, Sommer P, Nielsen JC, Arneborg N (2001) The effect of oxygen on the survival of non-Saccharomyces yeasts during mixed culture fermentations of grape juice with Saccharomyces cerevisiae. J Appl Microbiol 91:541–547. doi:10.1046/j.1365-2672.2001.01426.x
Liberal AT, Basílio ACM, Resende AM, Brasileiro BTV, Silva-Filho EA, Morais JOF, Simões DA, Morais MA (2007) Identification of Dekkera bruxellensis as a major contaminant yeast in continuous fuel ethanol fermentation. J Appl Microbiol 102:538–547. doi:10.1111/j.1365-2672.2006.03082.x
Loureiro V, Malfeito-Ferreira M (2003) Spoilage yeasts in the wine industry (review). Int J Food Microbiol 86:23–50. doi:10.1016/S0168-1605(03)00246-0
McAlister L, Holland MJ (1985a) Isolation and characterization of yeast strains carrying mutations in the glyceraldehyde-3-phosphate dehydrogenase genes. J Biol Chem 260:15013–15018
McAlister L, Holland MJ (1985b) Differential expression of the three yeast glyceraldehyde-3-phosphate dehydrogenase genes. J Biol Chem 260:15019–15027
Nehme N, Mathieu F, Taillandier P (2010) Impact of the co-culture of Saccharomyces cerevisiae–Oenococcus oeni on malolactic fermentation and partial characterization of a yeast-derived inhibitory peptidic fraction. Food Microbiol 27:150–157. doi:10.1016/j.fm.2009.09.008
Nakajima H, Amano W, Kubo T, Fukuhara A, Ihara H, Azuma YT, Tajima H, Inui T, Sawa A, Takeuchi T (2009) Glyceraldehyde-3-phosphate dehydrogenase aggregate formation participates in oxidative stress-induced cell death. J Cell Biochem 284:34331–34341. doi:10.1074/jbc.M109.027698
Nissen P, Arneborg N (2003) Characterization of early deaths of non-Saccharomyces yeasts in mixed cultures with Saccharomyces cerevisiae. Arch Microbiol 180:257–263. doi:10.1007/s00203-003-0585-9
Nissen P, Nielsen D, Arneborg N (2003) Viable Saccharomyces cerevisiae cells at high concentrations cause early growth arrest of non-Saccharomyces yeasts in mixed cultures by a cell-cell contact-mediated mechanism. Yeast 20:331–341. doi:10.1002/yea.965
Osborne JP, Edwards CG (2007) Inhibition of malolactic fermentation by a peptide produced by Saccharomyces cerevisiae during alcoholic fermentation. Int J Food Microbiol 118:27–34. doi:10.1016/j.ijfoodmicro.2007.05.007
Pérez F, Ramírez M, Regodón JA (2001) Influence of killer strains of Saccharomyces cerevisiae on wine fermentation. Antonie Van Leeuwenhoek 79:393–399. doi:10.1023/A:1012034608908
Pérez-Nevado F, Albergaria H, Hogg T, Gírio F (2006) Cellular death of two non-Saccharomyces wine-related yeasts during mixed fermentations with Saccharomyces cerevisiae. Int J Food Microbiol 108:336–345. doi:10.1016/j.ijfoodmicro.2005.12.012
Pretorius IS (2000) Tailoring wine yeast for the new millennium: novel approaches to the ancient art of winemaking. Yeast 16:675–729. doi:10.1002/1097-0061(20000615)16:8<675::AID-YEA585>3.0.CO;2-B
Seo J-K, Lee MJ, Go H-J, Tae Hyun Park TH, Park NG (2012) Purification and characterization of YFGAP, a GAPDH-related novel antimicrobial peptide, from the skin of yellowfin tuna, Thunnus albacores. Fish & Shellfish Immunol 33:743–752. doi:10.1016/j.fsi.2012.06.023
Silva A, Almeida B, Sampaio-Marques B, Reis MIR, Ohlmeier S, Rodrigues F, Do Vale A, Ludovico P (2011) Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is a specific substrate of yeast metacaspase. Biochem Biophys Acta 1813:2044–2049. doi:10.1016/j.bbamcr.2011.09.010
Sirover MA (2005) New nuclear functions of the glycolytic protein, glyceraldehyde-3-phosphate dehydrogenase, in mammalian cells. J Cell Biochem 95:45–52. doi:10.1002/jcb.20399
Wagener J, Schneider JJ, Baxmann S, Kalbacher H, Borelli C, Nuding S, Küchler R, Wehkamp J, Kaeser MD, Mailänder-Sanchez D, Braunsdorf C, Hube B, Schild L, Forssmann W-G, Korting H-C, Liepke C, Schaller M (2013) A peptide derived from the highly conserved protein GAPDH is involved in tissue protection by different antifungal strategies and epithelial immunomodulation. J Investigat Dermatol 133:144–153. doi:10.1038/jid.2012.254
Acknowledgments
The present work was financed by FEDER funds through POFC-COMPETE and by national funds through Fundação para a Ciência e a Tecnologia (FCT) in the scope of project FCOMP-01-0124-FEDER-014055. M.G.A. and J.C. acknowledge the funding support from FCT (PEst-C/EQB/LA0006/2011). Patrícia Branco is the recipient of a PhD fellowship (SFRH/BD/89673/2012) funded by FCT, Portugal. We want also to thanks to Professors Isabel Sá-Correia (IST/UTL, Lisbon, Portugal), Luísa Marinho (FCUL, Lisbon, Portugal) and Paula Ludovico (ICVS/UM, Braga, Portugal) for kindly providing some mutant strains.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 84 kb)
Rights and permissions
About this article
Cite this article
Branco, P., Francisco, D., Chambon, C. et al. Identification of novel GAPDH-derived antimicrobial peptides secreted by Saccharomyces cerevisiae and involved in wine microbial interactions. Appl Microbiol Biotechnol 98, 843–853 (2014). https://doi.org/10.1007/s00253-013-5411-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-013-5411-y