Abstract
Thraustochytrids are ubiquitous marine osmo-heterotrophic fungi-like microorganisms with only about 40 identified species till now. In this study, a total of 60 thraustochytrid strains were isolated from marine coastal habitats. Analysis of 18S rRNA gene sequences revealed that they belonged to three genera, i.e., Schizochytrium, Aurantiochytrium, and Thraustochytrium. All of the isolates were found to show considerable cellulolytic and lipolytic activities. Strains of Aurantiochytrium sp. and Thraustochytrium sp. were found to produce the highest levels of extracellular polysaccharides (EPS), which reached 345 μg ml−1 in the growth media. Fourier transform infrared (FTIR) spectra of the EPS samples derived from two thraustochytrids (PKU#Sed1 and #SW1) displayed peaks for carbohydrates, proteins, lipids, uronic acids, and nucleic acids. Fatty acid profiles of four thraustochytrids comprised of palmitic acid (C16:0) and docosahexaenoic acid (DHA) as their major constituents. Schizochytrium sp. demonstrated the highest DHA production at 44 % of total fatty acids (TFA) with biomass and DHA yield of 7.1 and 1.6 g l−1, respectively, on the fourth day of growth. All the four isolates exhibited considerable production of palmitic acid (16:0) in their fatty acid profiles ranging from 35 to 50 % TFA. This is the first report on extracellular enzymes, EPS, and DHA production from thraustochytrids isolated from the coastal habitats of China.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Microbial activities contribute significant role towards mineralization and recycling of nutrients in the biogeochemical cycles of marine ecosystems. Bacteria have been studied extensively from diverse ecosystems due to their significant ecological functions (Benner et al. 1986; Dighton 2003). Recently, thraustochytrids, the osmo-heterotrophic fungi-like protists, have attracted researchers' attention due to their high abundance reported from various oceanic habitats (Raghukumar and Damare 2011). They have also been demonstrated to be actively involved in the breakdown, scavenging, and mineralization of highly refractory organic matter by producing a range of extracellular enzymes (Bongiorni et al. 2005a). Thus, thraustochytrids may be an important component of the microbial food webs of marine ecosystems (Raghukumar et al. 1994; Raghukumar and Raghukumar 1999).
At this point, only about 40 thraustochytrid species are cultivated and identified. They belong to the genera, Althornia, Diplophrys, Japonochytrium, Thraustochytrium, Aurantiochytrium (formerly known as Schizochytrium), and Ulkenia (Honda et al. 1999; Bongiorni et al. 2005b; Raghukumar 2002). Some of these thraustochtrids have been reported to be rich source for hydrolytic enzymes. Still, isolation and identification of thraustochytrids has only been in limited number of geographic locations to date (Bongiorni et al. 2005b; Yang et al. 2010; Wilkens and Maas 2012). Thraustochytrid strains isolated from coastal seawaters of Japan and Fiji, which were identified based on 18S rRNA genes and PUFA profiles, exhibited to form monophyletic cluster (Huang et al. 2003). The phylogeny of Labyrinthulomycetes based on multigene analysis suggested the evolutionary loss of chloroplasts and gain of ectoplasmic gliding (Tsui et al. 2009). However, the true diversity of thraustochytrids remained to be explored from diverse marine habitats.
Apart from extracellular enzymes, thraustochytrids have also been shown to produce considerable quantities of extracellular polysaccharides (EPSs) (Jain et al. 2005). EPSs are significant component of biogeochemical processes and serve a major role towards formation of microbial biofilms and aggregates. It also functions as food source for marine animals by sequestering dissolved and particulate nutrients from the water phase facilitating their utilization as nutrient and energy sources (Decho 1990; Bhosle et al. 1995). Being composed of polysaccharides, proteins, lipids, and nucleic acids, the microbial EPS have been suggested to have several biotechnological applications as antitumor agents and anticoagulants and in cosmetic and food industries (Sutherland 1998). Physiologically, they prevent the thraustochytrid cells from desiccation (Jain et al. 2005). However, compared with bacteria and fungi, very few studies have emphasized the EPS production of thraustochytrids and require more attention in order to understand their ecological significance (Jain et al. 2005).
Production of ω-3 polyunsaturated fatty acids (PUFAs) is another interesting characteristic of thraustochytrids. The ω-3 PUFAs, especially docosahexaenoic acid (DHA), assist towards reduction of coronary heart disease, stroke, and rheumatoid arthritis (Kinsella 1987). DHA has been reported as an essential component for normal development of neural tissue of eyes and brain in infants and various other disorders (Simopoulos 1989; Takahata et al. 1998). Currently, main industrial sources of DHA are from fish oils and microalgal species (Volkman et al. 1989). However, thraustochytrids have emerged recently as an easier and less expensive means of producing high biomass and oils rich in DHA (Lewis et al. 1999). Several studies have attempted the optimization of media components and growth conditions for enhanced DHA production from thraustochytrids (Bajpai et al. 1991; Singh et al. 1996; Yaguchi et al. 1997; Arafiles et al. 2011). Moreover, thraustochytrids have been demonstrated to play a major role as food source for marine planktonic animals due to their high PUFA content, contributing significantly towards microbial food web in the marine ecosystems (Raghukumar and Damare 2011).
The objective of the present study was to isolate thraustochytrids from the various coastal marine habitats of Shenzhen coastal waters, including mangrove, seawater, and sediment samples. The resulting strains were identified based on the full-length 18S rRNA gene sequence. These thraustochytrid strains were screened for their potential of producing three different extracellular enzymes, i.e., protease, lipase, and cellulase, EPS, and PUFAs.
Materials and methods
Sample collection and thraustochytrid isolation
Mangrove leave, sediment, and seawater samples were collected from coastal marine habitats of Pearl River Delta region of China (Table 1). These samples were carried in sterile plastic bags and bottles immediately back to the laboratory for isolation. Direct plating method was used for mangrove and sediment samples as follows: (a) For mangrove leaves samples, discs of 5 mm diameter were cut and placed directly on modified Vishniac's (MV) medium (Damare and Raghukumar 2006), after surface sterilization with a 0.5 % sodium hypochlorite solution (Newell and Fell 1982). Sediment samples were suspended in sterile seawater and plated directly on MV media plates. The MV media contained 0.075 g streptomycin and 0.05 g of ampicillin per 100 ml. After incubation for 1 week at room temperature, pure isolates were obtained by repeated subculturing at an interval of 20–25 days. (b) The pine pollen method was used for isolation of thraustochytrids from seawater samples (Gaertner 1968; Porter 1990). Two milliliters of seawater sample was incubated with 3 ml of sterile seawater in sterile Petri dishes. A small quantity of pine pollen, sterilized in an oven at 100 °C for 48 h, was added as “bait.” The plates were incubated at room temperature for 3–7 days with regular monitoring for thraustochytrid growth. The resulting thraustochytrid colonies were subcultured in seawater/pine pollen containing antibiotics (0.075 % streptomycin and 0.05 % ampicillin) for the axenic cultures. The pine pollen containing axenic cultures was subcultured on MV media plates amended with antibiotics and incubated at room temperature. Microscopic observation of the isolates was done to determine bacterial contamination. A slight modification of the pine pollen isolation method was applied for the seawater samples collected in July 2012 by addition of different combinations of 0.05 % antibiotics (penicillin, ampicillin, rifampicin, and tetracycline) and antifungal agents (Nystatin, Amphotericin B, and Itraconizol) to the MV plates to check the effect on the diversity of isolated thraustochytrids. All of the thraustochytrid isolates were maintained at 5 °C on MV Medium. Four promising strains (Schizochytrium sp. PKU#Mn4, Aurantiochytrium sp. PKU#Sed1, Thraustochytrium sp. PKU#SW1, and Thraustochytrium sp. PKU#SW2) have been deposited in China General Microbiological Culture Collection Center (CGMCC no. 8091-8094).
Isolation and sequencing of 18S rRNA gene
Thraustochytrid isolates were identified by amplification and sequence analysis of complete 18S region of SSU rRNA gene. Total genomic DNA of thraustochytrid cells was extracted using Ezup Soil DNA extraction kit (Sangon Biotech, China), following the manufacturer's guideline. The small subunit 18S rRNA gene was amplified by polymerase chain reaction (PCR) in the DNA T100TM Thermal cycler (Bio-Rad, USA), using the 18S rRNA gene specific primers, 18S001 (5′-AACCTGGTTGATCCTGCCAGTA-3′) and 18S13 (5′-CCTTGTTACGACTTCACCTTCCTCT-3′) (Honda et al. 1999). Approximately 25 ng of genomic DNA was taken in 50 μl reaction volume containing 25 μl of Taq PCR mix (Generay, China) and 5 pmol of each primer. The PCR program was run for initial denaturation step at 95 °C for 5 min, followed by 35 cycles of 1 min at 94 °C, 1 min at 50 °C and 1 min at 72 °C, and a final extension at 72 °C for 20 min. The PCR products were purified using Gel DNA extraction kit (NewTopBio, China) and ligated into pGEM®-T Easy Vector (Promega, USA). The ligated product was transformed into Escherichia coli Top-10 competent cells (TianGen, China) by heat shock method. Clones containing positive insert were sent to BGI (Shenzhen, China) for sequencing analysis using M13 universal primers.
Phylogenetic analyses
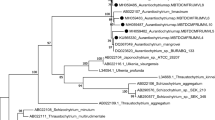
Forward and reverse sequences were edited and assembled using Chromas Pro version 1.34 (Technelysium Pty Ltd, Tewantia, Queensland, Australia). The final sequences were compared to the nucleotide sequences of reference organisms available in the GenBank database (Altschul et al. 1990). The 18S rRNA gene sequences (>1,500 bp) were aligned with their closest match using the program, ClustalW (Thompson et al. 1994). Gaps and ambiguously aligned sequences were removed manually from further analyses. Phylogenetic analyses were carried out using distance setting (Maximum parsimony) in MEGA 4 software (Tamura et al. 2007) with 1,000 bootstrap replicates. In phylogenetic analysis, the two sequences belonging to Aplanochytrium and Labyrinthula sp. were selected as out-groups for the rooted tree. Out of the 60 thraustochytrids, 10 strains isolated during different seasons and belonging to 3 distinct genera were selected as representatives (Table 1). The identification details of 10 representative thraustochytrid strains are shown in Table 2. The 18S rRNA gene sequences of these 10 strains were submitted to GenBank under the access number of JX847360 and JX847360-847378.
Qualitative assay for extracellular enzymes
The enzymatic activity of all the 60 thraustochytrid isolates was analyzed in this study. The strains were screened initially using qualitative plate assay for three different enzymes (protease, cellulase, and lipase) by streaking them on the media plates, supplemented with specific substrates. Protease activity was detected using MV plates supplemented with 1 % skimmed milk powder (Damare et al. 2006). The thraustochytrid isolates were grown on these media plates at room temperature for 5 days. Clear zones produced around the thraustochytrid colonies on plates indicated protease activity.
Cellulase (CMCase) activity was detected using carboxymethylcellulose (CMC)-MV plates (Carder 1986). The CMC-MV plates comprised 0.5 % carboxymethylcellulose–sodium salt (CMC-Na), 1.0 % glucose, 0.15 % peptone, 0.01 % yeast extract, 100 % seawater, and 2.0 % agar. After growing the thraustochytrid isolates for 5 days at room temperature, these plates were stained with 0.1 % Congo red solution for 20 min at room temperature. The resulting plates were washed twice with 1.0 M NaCl and were kept overnight at 4 °C. Clear zones around the colonies indicated the CMCase activities (Nagano et al. 2011; Carder 1986). Lipase activity was detected using MV plates supplemented with 0.01 % phenol red, 1 % olive oil, and 10 mM CaCl2. The pH was adjusted to 7.3–7.4 with 1 M NaOH (Singh et al. 2006). After incubation, a change in color from pink to yellow indicated the lipase activity.
Quantitative analysis of enzymatic activities
On the basis of qualitative screening, four thraustochytrid strains, namely, Schizochytrium sp. (PKU#Mn4), Aurantiochytrium sp. (PKU#Sed1), and Thraustochytrium spp. (PKU#SW1 and PKU#SW2) sp. were selected for quantitative studies. Inoculums were prepared by growing isolates in M4 medium (2 % glucose, 0.025 % KH2PO4, 0.15 % peptone, 0.1 % yeast extract, pH 7) (Jain et al. 2005) at 30 °C on a shaker at 150 rpm for 2 days. These inoculums were further added at a final concentration of 5 % to fresh M4 medium containing individual enzyme-specific substrates and incubated on a shaker at 30 °C, 150 rpm for 4 days. All these experiments were performed in triplicates. Individual enzymes were quantitated in the supernatants of the isolates.
The protease activity was assayed using casein and azocasein as substrates. Two hundred microliters of crude culture filtrate was incubated with equal volumes of 0.6 % casein and 2 % azocasein solution prepared in 0.05 M Tris–HCl buffer, pH 9. After incubating for 30 min at 37 °C, the reaction was stopped by adding 1 ml of 10 % trichloroacetic acid solution. The resulting solutions were centrifuged at 8,000 rpm for 10 min. The casein and azocasein hydrolytic activity was measured immediately following the methods described by Than et al. (2004) and Damare et al. (2006), respectively. One casein digestion unit (CU)/azocasein digestion unit (ACU) was defined as the increase in absorbance at 275/440 nm by 0.001/min under the above condition (Bramono et al. 2006; Than et al. 2004).
Cellulase activity was assayed following the method described by Raghukumar et al. (1994). A reaction mixture containing 200 μl culture filtrate and 200 μl of 0.5 % CMC in 0.05 M sodium phosphate buffer, pH 7 was incubated at 37 °C for 30 min. To stop the reaction, 1 ml of dinitrosalicylic acid reagent was added to above reaction mixture and then was boiled for 5 min. CMCase activity was measured at 575 nm. One unit of CMCase activity was defined as the amount of enzyme liberating 1 μmol of reducing sugar per minute under the above assay conditions.
For lipase assay, 6 mg para-nitrophenyl palmitate (pNPP), dissolved in 2 ml isopropanol and 18 ml 50 mM sodium phosphate buffer, pH 8, was used as the substrate. One milliliter each of culture filtrate and sodium phosphate buffer, pH 8, was added to 1 ml of the pNPP substrate solution. After incubating at 37 °C for 30 min, the released product, para-nitrophenol (pNP), was measured at 410 nm. One unit of lipase activity was defined as the amount of enzyme liberating 1 μmol of pNP per minute under these assay conditions (Gupta et al. 2002; Pinsirodom and Parkin 2001). In addition, lipase activity was analyzed by growing one of the best lipase producing isolate, PKU#Sed1, in the presence of different surfactants in the growth media. The isolate was inoculated (5 % inoculum) in M4 broth amended with 1 % concentration of five different substrates (Tween 80, Tween 20, Triton X-100, Nonidet P-40, and sodium deoxycholate) and incubated on a shaker at 150 rpm, 30 °C for 4 days. Lipase activity was assayed as described above.
Growth and EPS production
Growth and EPS production was monitored in M4 medium for the four isolates, Mn4, Sed1, SW1, and SW2. Cultures were incubated at 30 °C on a shaker, at 150 rpm. Growth was analyzed by measuring the absorbance (turbidity) at 660 nm at regular intervals. The EPS samples were prepared by centrifuging the cell suspension at 10,000 rpm for 15 min and filtering the supernatant serially through a Whatman GF/F glass-fiber filter and 0.45-μm membrane filters. The filtrate was collected and dialyzed using a 3,500 Da cut-off membrane against distilled water for 48 h. The EPS was estimated in the dialyzed filtrate by analyzing total carbohydrates by phenol-sulfuric acid method (Dubois et al. 1956). The experiment was carried out until the culture reached the stationary phase. One of the isolates, Sed1, showing highest level of EPS production was grown in the presence of different initial concentrations of glucose (0, 1, 3, 5, and 7 %) in the growth media for 5 days. EPS was isolated from all these samples and estimated as described above in order to analyze the effect of glucose concentration on EPS production.
Microscopy and FTIR spectrum of EPS
Thraustochytrid isolates grown in M4 broth were analyzed by phase contrast microscopy for the presence of EPS following the Alcian Blue staining method described by Decho (1993). The grown cultures were stained with 1 % Alcian Blue in 3 % acetic acid, pH 2.5, to test for acidic polysaccharides (Decho 1993). Two of the isolates (Sed1 and SW1) producing high EPS were subjected for Fourier transform infrared (FTIR) spectral analysis. FTIR spectra were recorded for EPS samples between 4,000 and 500 cm−1 with an FTIR spectrometer (IRPrestige-21). The absorption spectra of the two strains (Sed1 and SW1) were recorded in the FTIR technique as dried films on Thallium Bromo-Iodide KRS-5 crystal. The filtrate containing EPS was prepared in a similar way as described in the previous section. To this filtrate, three volumes of cold absolute ethanol was added and the mixture was left overnight. The precipitated EPS was centrifuged at 10,000 rpm for 30 min, lyophilized and stored at −20 °C for further analyses. This lyophilized EPS was spread on a KRS-5 crystal, and spectrum was recorded with the spectrometer. The FTIR spectrum of the EPS samples was obtained after subtracting the background spectrum. The EPS sample was transferred to a quartz mortar and mixed with potassium bromide crystal. This mixture was compressed on to a tablet and subjected to FTIR measurement.
Analysis of fatty acid production during different growth stages
All the four thraustochytrid isolates were grown in M4 medium. Every day, an aliquot of the grown culture was removed, and the cells were centrifuged at 5,000 rpm for 10 min. After washing twice with autoclaved seawater, the cells were stained with 0.01 % w/v Nile Red in acetone. Nile Red (9-diethylamino-5H-benzo[α]-phenoxazine-5-one) is a phenoxazone dye that fluoresces golden yellow after binding with intracellular lipid droplets under an epifluorescence microscope (Greenspan et al. 1985). The Nile Red stained cells may be examined by the fluorescence emitted at wavelengths of ≤570 nm, due to interaction of stain with extremely hydrophobic environments, i.e., neutral lipid droplets. The thraustochytrid cells of four isolates were analyzed for the intracellular lipid production periodically for up to 4 days using an Olympus BX53 epifluorescence microscope. The cultures grown for 4 days were harvested by centrifugation at 5,000 rpm, rinsed with distilled water, and centrifuged again. The cell pellets were freeze-dried, weighed, and stored at −80 °C for lipid extraction and fatty acid analysis.
Lipid extraction and fatty acid analysis
Lipid extraction was done by the direct transesterification method described by Kamlangdee and Fan (2003). Two milliliters of 4 % sulfuric acid in methanol (transesterification reaction mix) was added directly to freeze-dried cells (50–100 mg), vortexed for 20 s, and incubated at 80 °C for 1 h. After transesterification, the mixtures were allowed to cool to room temperature. After cooling, 1 ml each of water and hexane were added to all the mixtures. The fatty acid methyl esters (FAMEs) in the hexane layer were mixed, centrifuged, and collected. The FAME samples (total fatty acids) were dried using a nitrogen stream and weighed. These samples were redissolved in 1 ml of hexane before injecting into gas–liquid chromatograph for analysis with a 7890-5975 GC-MS (Agilent, USA) at School of Environment and Energy, Peking University. It was equipped with a DB-5ms capillary column (60 m × 250 μm × 0.25 μm). Helium was used as the carrier gas, and the flow rate was kept at 3 ml/min. A volume of 1 μl was injected under splitless injection mode. After injection, the column temperature was maintained at 70 °C for 2 min followed by programming at 10° min−1 to 230 °C, then at 3° min−1 to 290 °C, which was held for 5 min. The injection port temperature was held at 280 °C. Quantification of fatty acid contents was done by comparing their peak areas with that of the standard mixtures (Sigma-Aldrich, USA).
Results
Culturable diversity of thraustochytrids
A total of 60 thraustochytrid cultures were isolated from mangrove, sediment, and seawater samples collected during different seasons (Table 1). They belonged to three different genera of thraustochytrids i.e., Aurantiochytrium sp., Schizochytrium sp., and Thraustochytrium sp., on the basis of NCBI blast results of 18S rRNA gene sequences (Fig. 1 and Tables 1 and 2). Application of two isolation techniques, pine pollen baiting (Fig. S1a-b) and direct plating, in this study resulted in the identification of 60 thraustochytrids, belonging to three genera of thraustochytrids (Fig. S1c–e). Only one isolate of Aurantiochytrium sp. recovered from sediment samples, indicating low abundance of thraustochytrids in sediments. A modification of pine pollen method by incorporating different types of antibiotic and antifungal agents in the media plates led to isolation of only two genera of thraustochytrids, i.e., Aurantiochytrium and Thraustochytrium from seawater samples.
Maximum parismony (MP) phylogenetic tree based on 18S rRNA genes from ten representative thraustochytrid isolates with sequences from Aplanochytrium and Labyrinthula sp. as an outgroup. Topology was built using Mega 4 from a ClustalW 1.83 alignment for ∼1,600 bp long sequences. Numbers above or below branches indicate bootstrap values (>50 %) from 1,000 replicates
Interestingly, the abundance and kind of isolates varied with different seasons and nature of samples for above three genera of thraustochytrids (Fig. 2, Table 1). Schizochytrium sp. was predominantly isolated only from decaying mangrove samples (Table 2). Other two genera (Aurantiochytrium and Thraustochytrium) were isolated from all types of marine samples in the present study and found to be more or less equally distributed among different periods of time (Fig. 2). A majority of the cultures were obtained from decaying mangrove leaves during March and June 2012 (Table 1). In addition, no other groups of Labyrinthulomycetes (i.e., Aplanochytrium and Labyrinthulids) except thraustochytrids could be recovered in culture in this study. All the other isolates matched with their respective closest relatives at a 99 % identity (Table 2).
Protease activity
None of the 60 thraustochytrid isolates showed protease activity on media plates supplemented with skimmed milk powder. The results of qualitative plate assay for the 10 representative thraustochytrids isolates is summarized in Table S1. However, a negligible protease activity was detected when assayed quantitatively (data not shown). The extent of quantitative protease enzyme production varied for different thraustochytrid isolates (the maximum value was 2.611 U for the isolate, PKU#SW1, when assayed with casein as the substrate).
Cellulase activity
All of the thraustochytrid isolates showed positive cellulase activity on CMC-MV plate (Table S1). A prominent clearance zone was observed on CMC-MV plate both when these strains grown directly as well as with their cell free culture supernatant (Fig. S2a, b). Four of these isolates showing good CMCase activity on plate were selected for quantitative estimation studies. Two of the isolates, PKU#SW1 and #SW2, showed high activity of CMCase on second day of growth, with a value reaching up to 0.6 μmol ml−1 min−1 for the isolate #SW1 (Fig. 3). The isolates, PKU#Sed1 and #Mn4 showed a maximum CMCase activity of 0.068 and 0.2 μmol ml−1 min−1, respectively, on the first day of growth. The cellulolytic activities of the thraustochytrids strains in this study were found to maximize on first or second day of growth decreasing significantly on third day (Fig. 3).
Lipase activity
All the thraustochytrid isolates displayed excellent lipase activity on media plates (Fig. S2c, d, Table S1). Both the isolates and their cell-free culture supernatant turned MV plates containing olive oil and phenol red from pink to yellow within a few hours of incubation, indicating positive reaction for lipase production (Fig. S2c, d). Four of the thraustochytrid isolates, PKU#Sed1 (Aurantiochytrium sp.), #Mn4 (Schizochytrium sp.), #SW1 (Thraustochytrium sp.), and #SW2 (Thraustochytrium sp.), which displayed strong lipase activity on plate, were selected for further quantitative analysis. The lipase-producing abilities of the abovementioned four isolates were tested both in the presence and absence of olive oil in the growth media (Table S1). The growth patterns and lipase enzyme activities varied among different isolates. All of these four isolates showed increased growth (OD660) in the presence of olive oil in the media. However, three of these isolates (PKU#Mn4, #SW1, and #SW2) displayed higher lipase activity in the absence of olive oil with the exception of PKU#Sed1, whose extracellular lipase enzyme production was enhanced up to 11.87 U ml−1 only in the presence of olive oil (Table S1). Lipase production by the isolate PKU#Sed1 was further investigated in the presence of different surfactants in the growth media, with Tween 80 to be the best for lipase activity (∼25 U ml−1) (Fig. 4).
EPS production
All the four thraustochytrid isolates demonstrated EPS production in shake flask cultures. Phase contrast microscopy showed positive staining of EPS with Alcian Blue (Fig. S3). The stained cells were enclosed within EPS sheath. The EPS production rate was further confirmed quantitatively (Fig. 5) with enhanced EPS production, which increased with the age of culture, reaching the maximum in their stationary phase. The EPS concentration ranged from 100 to 200 μg ml−1 for these isolates. Two of the isolates, Sed1 and SW1, showed higher EPS production of 197 and 196 μg ml−1 on the fifth day of growth and were selected further for FTIR analysis. Interestingly, the growth pattern was found to be distinct for Sed1 and SW1. The isolate, SW1, demonstrated equally high concentration of EPS production despite of lesser growth (absorbance at 660 nm) than Sed1 (Fig. 5). The EPS production was increased linearly with glucose concentration in the growth media for the isolate Sed1. The maximum concentration of EPS was observed as 345.3 μg ml−1 on the 5th day of growth (stationary phase) in the presence of 7 % initial glucose concentration (Table S2). The EPS production yield ranged from 100 to 200 μg/ml and was found to increase with the age of cultures, attaining highest values in stationary phase (Fig. 5). An increased glucose concentration of 7 % in the growth media resulted in elevated level of EPS (345 μg ml−1) from the isolate Sed1 (Table S2).
FTIR spectra of EPS samples
FTIR spectral analysis for the isolates, Sed1 and SW1, revealed appearance of peaks pertaining to functional groups of proteins, carbohydrates, phospholipids, and nucleic acids (Fig. S4). The peaks around 3,300 cm−1 (−OH and/or –NH stretching) and 1,047 cm−1 confirmed the presence of carbohydrates in EPS of both the isolates. The –COC– group vibrations in the area across 1,100 cm−1 are indicative of uronic acid in the EPS. The peaks in the area 1540–1647 cm−1 representing vibrations of the –CONH– group of amide I (−CO–) and amide II (−NH) showed the presence of proteins in the EPS (Sheng et al. 2006). The peaks at 2,855 and 2,925 cm−1 in the EPS sample of Sed1 corresponded to the respective symmetrical and asymmetrical –CH– vibrations of lipids (Fig. S4). In the 1,246 cm−1 region, a phosphate group absorption band was observed indicating the presence of nucleic acids (Fig. S4) (Sheng et al. 2006). However, the absence of peaks around 800 cm−1, indicated the EPS samples devoid of sulfated polysaccharides in this study (Fig. S4).
Fatty acid production
Epifluorescence microscopy showed positive Nile Red staining for intracellular lipids for all the four thraustochytrid isolates. The intensity of fluorescence for stained lipids increased with the age of cultures, reaching maximum on the fourth day of growth (Fig. 6). Since the isolates exhibited highest fluorescence on the fourth day of growth, the subsequent analysis for fatty acid was done on the same day. The biomass, TFA, and DHA produced by thraustochytrid isolates grown in the liquid media have been represented in Table S1. The productivity of TFA and DHA varied distinctly between different thraustochytrid genera, being the highest for the isolate, Mn4 (Schizochytrium sp.). The maximum TFA and DHA production values were 51.5 and 44.3 %, respectively, by this isolate eventually resulting in a DHA yield of 1.6 g l−1 (Table S1). Other isolates exhibited DHA yield in the range of 27–41 % TFA (Table S1). Highest biomass (7.5 g l−1) was produced by the isolate Sed1.
The fatty acid profile of four thraustochytrid isolates comprised of saturated, monounsaturated, and polyunsaturated constituents. The predominant PUFA was DHA for all the isolates (Fig. 7 and Table S1). The PUFA profile contained eicosapentaenoic acid (EPA)/DHA and arachidonic acid (AA)/EPA/DHA for different thraustochytrids. However, the percentage of AA (∼0.03–0.1 %) or EPA (∼0.3–0.7 %) was comparatively very less than DHA (∼27–44 %). AA was present only in the PUFA profile of the isolates belonging to Thraustochytrium sp. Maximum EPA production was showed by Sed1 (Aurantiochytrium sp.) at 0.7 % of TFA (Table S3). Among saturated fatty acids, palmitic acid (16:0) was the major one produced at a higher percentage (ranged from 35.2 to 51.9 %) by all the isolates. Isolates belonging to Thraustochytrium sp. (SW1 and SW2) also demonstrated considerable production of myristic acid (14:0) and pentadecyclic acid (15:0) (Table S3). All the four isolates showed a poor representation of monounsaturated fatty acids (MUFAs) such as palmitoleic acid (16:1), oleic acid (18:1) and stearic acid (18:0) in their fatty acid profiles.
Discussion
Culturable diversity
The small subunit ribosomal RNA (18S rRNA) has been commonly used for the identification of thraustochytrids (Honda et al. 1999). The low evolutionary rate of this gene facilitates the accurate identification at specific level as evident from previous studies (Cavalier-Smith et al. 1994; Van de Peer et al. 2000). A study based on 18S rRNA gene sequences of 14 thraustochytrid strains emphasized the improved molecular phylogenetic analyses as much reliable alternative for their identification (Honda et al. 1999). Therefore, the identification method used in the present study may provide more reliable diversity measurements of thraustochytrids. These isolation results suggest that the two genera of thraustochytrids i.e., Aurantiochytrium and Thraustochytrium as the abundant planktonic thraustochytrid communities in seawater ecosystem of Pearl River Delta.
Several studies have reported isolation of Schizochytrium sp. from mangrove habitats previously (Raghukumar 2002). The isolation of this species only from mangroves and absence in the seawater and sediment samples in the present study may be attributed to its substrate specific inhabitation. This hypothesis is further supported by findings of previous studies where the zoospores of this species have been demonstrated to be attracted chemotactically to fallen leaves in mangrove waters (Fan et al. 2002). Other examples of substrate-specific associations include the inhabitation of algae and seagrasses by Aplanochytrium sp. (Raghukumar 2002; Leander et al. 2004). The number of thraustochytrids associated with algal, mangrove and sea-grass detritus were found to increase with their advanced decomposition stage (Raghukumar 2002). This may be pertained to leaching out of dissolved organic matter, including phenolics and tannins in decomposed state, which may inhibit growth of thraustochytrids during the early stages of detritus (Sathe-Pathak et al. 1993; Raghukumar et al. 1995).
The observations in the present study are in close agreement with the previous reports where seasonal and spatial scale fluctuations affected thraustochytrid densities in the marine habitats of Mediterranean sandy shores (Bongiorni et al. 2004). The closest relatives of thraustochytrids isolates found in the present study have been reported from several other marine habitats in the previous studies, suggesting their abundance in mangrove and coastal seawater ecosystems (Table 2). One of the isolates, PKU#Mn16 closely (98 %) related to Thraustochytriidae sp., which has been reported as the good source of PUFAs such as DHA (Jain et al. 2007). One of the thraustochytrid strain, PKU#Mn11 belonged to Aurantiochytrium sp. which has been demonstrated earlier to produce various hydrolytic extracellular enzymes (Taoka et al. 2009).
Enzymatic activities
Most of the previous studies have demonstrated thraustochytrids as an efficient producer of protease activities (Sharma et al. 1994; Bongiorni et al. 2005a; Taoka et al. 2009). However, a certain selection was observed in the pattern of peptidase enzyme production by diverse thraustochytrid isolates (Bongiorni et al. 2005a). Fourteen strains of Aplanochytrium were isolated from zooplankton samples and exhibited only protease activity suggesting their involvement in degradation of complex proteinaceous component of zooplankton cadavers (Damare and Raghukumar 2006).
The role of thraustochytrids in the degradation and mineralization of leaf litter was supported by the production of cellulase from the isolate Schizochytrium mangrovei derived from mangrove leaves (Raghukumar et al. 1994). The thraustochytrid strain Schizochytrium aggregatum was also shown to produce the extracellular cellulase enzyme, β-1,4-glucanase (Bremer and Talbot 1995). However, no cellulase activity was detected among six thraustochytrid strains belonging to Schizochytrium, Thraustochytrium and Aurantiochytrium sp., ascribing this result to the use of different cellulose substrate than CMC for plate assay (Taoka et al. 2009). Recently, qualitative analysis of 14 thraustochytrid strains belonging to the genera Aplanochytrium, Botryochytrium, Oblongichytrium, Parietichytrium, Schizochytrium, Sicyoidochytrium, Thraustochytrium and Ulkenia displayed cellulolytic activity (Nagano et al. 2011). The only strain showing negative results for cellulolytic enzyme activity belonged to the genus Aurantiochytrium (Nagano et al. 2011). In contrast, the Aurantiochytrium strain (PKU#Sed1) used in the present study demonstrated celluolytic activity qualitatively and quantitatively. The variation in enzymatic activities observed in these results underlines the importance of particular thraustochytrid strains towards their role in association with different geographical habitats.
The cellulolytic activities of the thraustochytrids strains in this study were found to maximize on first or second day of growth decreasing significantly on third day suggesting the catabolic repression of cellulase production due to accumulation of end product, glucose. Similar findings have been reported for cellulase synthesis in some fungi being controlled by end or intermediate product repression (Eriksson and Hamp 1978; Sukumaran et al. 2005). Interestingly, the Thraustochytrium sp., which was isolated from the seawater in this study, demonstrated higher cellulolytic activity than the mangrove isolates. An elaborated study on inducible mechanism driving higher cellulase production in the presence of different substrates may reveal functional role of this species in seawater habitats of Pearl River Delta.
The principle behind the color change during lipase production is that the phenol red dye has an end point at pH 7.3-7.4 where it is pink. When the organism producing lipase is grown on plates containing phenol red, a decrease in pH from 7.3 to 7.0 results due to the release of free fatty acids from olive oil which in turn changes the plate color yellow (Singh et al. 2006). This method has been applied in earlier studies for successful demonstration of qualitative lipase activity by various organisms (Kanchana et al. 2011; Rajan et al. 2011). The effect of different oils on production of lipases has not been completely understood (Lima et al. 2003). The induction of enzyme may be influenced both by carbon chain lengths and degree of unsaturations of fatty acids in these oils. Different oil substrates are composed mostly of oleic and linoleic acids, the former being more influential towards enhanced lipase production (Jaeger and Reetz 1998). Olive oil, a rich source of oleic acid, was found to be the best carbon source, inducing lipase production in the Thraustochytrium sp. (Kanchana et al. 2011). However, the higher lipase production without olive oil in the growth media by three thraustochytrid isolates in the present study suggests its constitutive nature rather than inducible (Table S1).
Tween 80 belongs to the class of surface-active materials which may enhance the lipase production either by affecting the permeability of the cell or by a surfactant effect on cell-bound lipase. It is evident from earlier studies also that Tween 80 stimulates the microbial lipase production, when supplied in the growth media (Boekema et al. 2007). Recently, a study emphasized the role of Tween 80 as a carbon source, enhancing biomass and total lipid yield from an isolate, Thraustochytrium aureum ATCC 34304, with a little mention on effect of Tween 80 on its lipase production (Taoka et al. 2011). The lipase activity was assayed spectro-photometrically in the present study and found to be comparatively 100-fold higher than that reported by Bongiorni et al. (2005a)
EPS production
EPS plays significant role in various ecological systems predominantly by constituting the components of reduced-carbon reservoir, microbial aggregates and nutrient sources and its production have been emphasized in bacteria and fungi by several researchers (Whitfield 1988; Gutierrez et al. 1996; Wingender et al. 2001; Flemming and Wingender 2010). All the four thraustochytrid strains in this study showed positive staining for Alcian Blue suggesting the presence of acidic polysaccharides in their EPS. This result is in agreement with earlier report where acidic polysaccharides were observed in the EPS of thraustochytrids (Jain et al. 2005). The acidic character may be attributed to the phenolic and polyaromatic compounds (containing peptide and carbohydrate moieties with carboxylic substituents) present in the EPS (Flemming and Wingender 2010). The acidic polysaccharides have been reported from a range of marine organisms such as bacteria, cyanobacteria and diatoms. These acidic polysaccharides may assist towards ion exchange and detoxification mechanisms by accumulation of toxic metal ions and are considered to influence the architectural integrity of a biofilm in a particular environment (Flemming and Wingender 2010).
The EPS production yield was found to increase with the age of cultures, attaining highest values in stationary phase (Fig. 5). Similar observations have been mentioned in previous studies towards increased production of EPS at the end of growth phase of thraustochytrids, bacteria and phytoplankton (Myklestad and Haug 1972; Williams and Wimpenny 1977, 1978; Jain et al. 2005). Moreover, the composition and quantity of the microbial EPS have been observed to vary with kind of microorganisms, their surrounding environmental and nutrient factors (Mayer et al. 1999). The effect of nutrient surfaces on EPS production of thraustochytrids have been demonstrated in earlier reports (Bremer 1976; Raghukumar et al. 2000). Among nutrients, carbohydrates have been reported to particularly induce production of enhanced amounts of EPS in microorganisms (Sutherland 1985).
FTIR spectra
The presence of carbohydrates, proteins, lipids and nucleic acid in the EPS sample of thraustochytrid isolates were revealed by IR spectra in the present study. Likewise, EPS from the thraustochytrid isolates SC1 and CW1 were demonstrated to be composed of sugars, sulfated polysaccharides, proteins and lipids in earlier study (Jain et al. 2005). Sulfated polysaccharides are well known to have an important role towards providing protection from desiccation by retention of moisture during adverse conditions (Decho 1990). Among proteins, the non-enzymatic constituent such as cell surface-associated and extracellular carbohydrate-binding proteins of the EPS mainly supports the mechanical stability and complexity of microbial biofilms. A fraction of these proteins may be indicative of extracellular enzymes of microbes that degrade EPS components thereby rendering the matrix an external digestive system that breaks down range of complex biopolymers such as cellulose, chitin and lipids (Flemming and Wingender 2010). Interestingly, the presence of nucleic acid in the EPS sample of thraustochytrid in the present study suggests them to be a reservoir facilitating horizontal gene transfer between biofilm cells (Flemming and Wingender 2010). Uronic acid confers overall negative charge to the EPS samples, enabling them capable for complexing of metals in the environmental biofilms (Flemming and Wingender 2010).
Fatty acid production
The fatty acid profiles of the four thraustochytrid isolates used in this study were found to comprise DHA as the major PUFA. Strains belonging to Aurantiochytrium and Schizochytrium sp. were best producers for biomass, TFA and DHA (Table S1). Previous studies have also demonstrated Schizochytrium and Aurantiochytrium sp. as the best candidates for DHA production (Yaguchi et al. 1997; Yokochi et al. 1998; Jakobsen et al. 2008). The DHA production ranged from 27-44 % TFA for the four thraustochytrids in the present study which is comparable with the DHA yield from these organisms reported in earlier studies (Yang et al. 2010). An Aurantiochytrium strain, BL10 was found to produce DHA up to 16 g l-1 under optimized media conditions (Yang et al. 2010). The fatty acid profiles of this strain contained palmitic acid (C16:0; ∼60 % TFA) and DHA (∼39 % TFA) as the major constituent (Yang et al. 2010). These results are in close agreement with the present study where thraustochytrid isolate, Sed1 (Aurantiochytrium sp.) exhibited palmitic acid (C16:0; ∼52 %) and DHA (∼41 %) as the predominating fatty acids in its profile (Table S3). Among PUFAs, DHA was the major fatty acid produced by all the four thraustochytrids. EPA and AA were found at a very low percentage among other PUFAs. However, the fatty acid composition of thraustochytrids has been demonstrated to vary with the cultivation conditions, i.e., carbon/nitrogen ratio in the growth media (Yang et al. 2010). Aurantiochytrium sp. has been proposed to be an efficient model organism for osmoregulation studies due to its higher growth rate and tolerance towards greater concentrations of glucose (Yang et al. 2010). Present study also revealed the highest growth rate for the thraustochytrid isolate, Sed1 (Aurantiochytrium sp.) (Table S1) suggesting it to be a potential culture for future scale up processes. Also, the highest DHA production was 1.6 g l-1 for the strain Mn4 (Schizochytrium sp.) without optimization of growth media components (different carbon and nitrogen sources) in this study. Therefore, it is likely that under the optimized media conditions, the isolates used in the present study may display greater DHA producing abilities.
All the four isolates exhibited a high production of palmitic acid (35-51 % TFA) (Table S3), suggesting them as the potential source for utilization towards biodiesel production. The high levels of saturated fatty acids and MUFAs such as palmitic acid (16:0) and oleic acid (18:1) are crucially important for the biofuel industry (Fisher et al. 2008). The saturated fatty acids and MUFAs improve the biodiesel quality by increasing its oxidative as well as the thermal stability (Monyem et al. 2000; Knothe 2007). A thraustochytrid isolate, Thraustochytrium striatum ATCC 24473, was demonstrated to produce 33.92 % (w/w of TFA) of palmitic acid (16:0) and 48.21 % of oleic acid (18:1) (Li and Ward. 1994). Similarly, two of the isolates belonging to Thraustochytrium sp. showed high production of palmitic acid (35 and 36 % TFA) in this study. However, the concentration of oleic acid (18:1) was comparatively low (maximum 1.9 % TFA). Among other low carbon fatty acids (LCFAs), a considerable amount of myristic acid (14:0) and pentadecylic acid (15:0) was produced only by Thraustochytrium sp. (Table S3). Recently, a few other studies have also reported thraustochytrids as the great reservoir of LCFAs and MUFAs indicating the possibility of their use for biofuel production (Lee Chang et al. 2012).
In conclusion, culture-based approaches identified three major genera, i.e., Aurantiochytrium, Thraustochytrium, and Schizochytrium from coastal habitats of Pearl River Delta. The extracellular enzymes, EPS, and DHA production exhibited by these thraustochytrid isolates indicate their potential biotechnological applications and ecological role in this ecosystem. The considerable quantitative production of cellulase and lipase enzymes from three genera of thraustochytrids suggests them as promising source for industrial applications. Best cellulase and lipase enzyme producing strains were Thraustochytrium (PKU#SW1) and Aurantiochytrium sp. (PKU#Sed1), respectively. The high amount of EPS produced by these isolates suggests their possible ecological role towards biofilm and microbial aggregate formation. The simpler PUFA profiles suggest the thraustochytrid isolated from coastal habitats of Pearl River Delta, China, an excellent source for DHA purification, scale up, and commercialization processes in the future. Moreover, the high percentage of low carbon fatty acids produced by these thraustochytrids proposes their application in biofuel industries.
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215(3):403–410
Arafiles K, Alcantara J, Batoon J, Galura F, Cordero P, Leaño E, Dedeles G (2011) Cultural optimization of thraustochytrids for biomass and fatty acid production. Mycosphere 2:521–531
Bajpai P, Bajpai PK, Ward OP (1991) Production of docosahexaenoic acid by Thraustochytrium aureum. Appl Microbiol Biotechnol 35:706–710
Benner R, Moran MA, Hodson RE (1986) Biogeochemical cycling of lignocellulosic carbon in marine and freshwater ecosystems: relative contributions of prokaryotes and eukaryotes. Limnol Oceanogr 31:89–100
Bhosle NB, Garg A, Sawant SS, Wagh A (1995) Isolation and partial chemical analysis of exopolysaccharides from the marine fouling diatom Navicula subinflata. Bot Mar 38:103–110
BoekeMa B, Beselin A, Breuer M, Hauer B, Koster M, Rosenau F, Jaeger KE, Tommassen J (2007) Hexadecane and Tween 80 stimulate lipase production in Burkholdetia glumae by different mechanisms. Appl Environ Microbiol 73(12):3838–3844
Bongiorni L, Pignataro L, Santangelo G (2004) Thraustochytrids (fungoid protists): an unexplored component of marine sediment microbiota. Sci Mar 68:43–48
Bongiorni L, Jain R, Raghukumar S, Aggarwal RK (2005a) Thraustochytrium gaertnerium sp. nov.: a new thraustochytrid stramenopilan protist from mangroves of Goa, India. Protist 156(3):303–315
Bongiorni L, Pusceddu A, Danovaro R (2005b) Enzymatic activities of epiphytic and benthic thraustochytrids involved in organic matter degradation. Aquat Microb Ecol 41:299–305
Bramono K, Yarnazaki M, Tsuboi R, Ogawa H (2006) Comparison of proteinase, lipase and alpha-glucosidase activities from the clinical isolates of Candida species. Jpn J Infect Dis 59(2):73–76
Bremer GB (1976) The ecology of marine lower fungi. In: Jones EBG (ed) Recent advances in aquatic mycology. Elek Science, London, pp 313–333
Bremer GB, Talbot G (1995) Cellulolytic enzyme-activity in the marine protist Schizochytrium aggregatum. Bot Mar 38(1):37–41
Carder JH (1986) Detection and quantitation of cellulase by congo red staining of substrates in a cup-plate diffusion assay. Anal Biochem 153(1):75–79
Cavalier-Smith T, Allsopp M, Chao EE (1994) Thraustochytrids are chromists, not fungi: 18S rRNA signatures of Heterokonta. Philos Trans R Soc Lond Ser B Biol Sci 346(1318):387–397
Damare V, Raghukumar S (2006) Morphology and physiology of the marine straminipilan fungi, the aplanochytrids isolated from the equatorial Indian Ocean. Indian J Mar Sci 35(4):326–340
Damare S, Raghukumar C, Muraleedharan UD, Raghukumar S (2006) Deep-sea fungi as a source of alkaline and cold-tolerant proteases. Enzyme Microb Technol 39(2):172–181
Decho AW (1990) Microbial exopolymer secretions in ocean environment: their role(s) in food webs and marine processes. Oceanogr Mar Biol Annu Rev 28:73–153
Decho AW (1993) Methods for the observation and use in feeding experiments of microbial exopolymer. In: Kemp PF, Sherr BF, Sherr EB, Cole JJ (eds) Handbook of methods in aquatic microbial ecology. Lewis, Chelsea, pp 685–694
Dighton J (2003) The role of fungi in ecosystem processes. Marcel Dekker, New York
Dubois M, Gilles KA, Hamilton JK, Rebers PA, Smith F (1956) Colorimetric method for the determination of sugars and related substances. Anal Chem 28:350–356
Eriksson KE, Hamp SG (1978) Regulation of Endo-1,4-β-glucanase production in Sporotrichum pulverulentum. Eur J Biochem 90(1):183–190
Fan KW, Vrijmoed LLP, Jones EBG (2002) Physiological studies of subtropical mangrove thraustochytrids. Bot Mar 45(1):50–57
Fisher L, Nicholls D, Sanderson K (2008) Production of biodiesel. World Intellect Prop Organ 067605
Flemming H, Wingender J (2010) The biofilm matrix. Nat Rev Microbiol 8:623–633
Gaertner A (1968) Eine Methode des quantitativen Nachweises niederer mit Pollen koederbarer Pilze im Meerwasser und im Sediment. Verӧff Inst Meeresforsch Bremerh 3:75–92
Greenspan P, Mayer EP, Fowler SD (1985) Nile Red: a selective fluorescent stain for intracellular lipid droplets. J. Cell Biol 100:965–973
Gupta N, Rathi P, Gupta R (2002) Simplified para-nitrophenyl palmitate assay for lipases and esterases. Anal Biochem 311(1):98–99
Gutierrez A, Martinez AT, Prieto A (1996) Structural characterization of extracellular polysaccharides produced by fungi from the genus Pleurotus. Carbohydr Res 281:143–154
Honda D, Yokochi T, Nakahara T, Raghukumar S, Nakagiri A, Schaumann K, Higashihara T (1999) Molecular phylogeny of labyrinthulids and thraustochytrids based on the sequencing of 18S ribosomal RNA gene. J Eukaryot Microbiol 46(6):637–647
Huang JZ, Aki T, Yokochi T, Nakahara T, Honda D, Kawamoto S, Shigeta S, Ono K, Suzuki O (2003) Grouping newly isolated docosahexaenoic acid-producing thraustochytrids based on their polyunsaturated fatty acid profiles and comparative analysis of 18S rRNA genes. Mar Biotechnol 5(5):450–457
Jaeger KE, Reetz MT (1998) Microbial lipases form versatile tools for biotechnology. Trends Biotechnol 16(9):396–403
Jain R, Raghukumar S, Tharanathan R, Bhosle NB (2005) Extracellular polysaccharide production by thraustochytrid protists. Mar Biotechnol 7:184–192
Jain R, Raghukumar S, Sambaiah K, Kumon Y, Nakahara T (2007) Docosahexaenoic acid accumulation in thraustochytrids: search for the rationale. Mar Biol 151(5):1657–1664
Jakobsen AN, Aasen IM, Josefsen KD, Strøm AR (2008) Accumulation of docosahexaenoic acid-rich lipid in thraustochytrid Aurantiochytrium sp. strain T66: effects of N and P starvation and O2 limitation. Appl Microbiol Biotechnol 80:297–306
Kamlangdee N, Fan KW (2003) Polyunsaturated fatty acids production by Schizochytrium sp. isolated from mangrove. J Sci Technol 25:643–650
Kanchana R, Muraleedharan UD, Raghukumar S (2011) Alkaline lipase activity from the marine protists, thraustochytrids. World J Microb Biotechnol 27(9):2125–2131
Kinsella JE (1987) Seafoods and fish oils in human health and disease. Marcel Dekker, New York
Knothe G (2007) Some aspects of biodiesel oxidative stability. Fuel Process Technol 88:669–677
Leander CA, Porter D, Leander BS (2004) Comparative morphology and molecular phylogeny of aplanochytrids (Labyrinthulomycota). Eur J Protistol 40(4):317–328
Lee Chang KJ, Dunstan GA, Abell GC, Clementson LA, Blackburn SI, Nichols PD, Koutoulis A (2012) Biodiscovery of new Australian thraustochytrids for production of biodiesel and long-chain omega-3 oils. Appl Microbiol Biotechnol 93:2215–2231
Lewis TE, Nichols PD, McMeekin TA (1999) The biotechnological potential of thraustochytrids. Mar Biotechnol 1:580–587
Li ZY, Ward OP (1994) Production of docosahexaenoic acid by Thraustochytrium roseum. J Ind Microbiol 13:238–241
Lima VMG, Krieger N, Sarquis MIM, Mitchell DA, Ramos LP, Fontana JD (2003) Effect of nitrogen and carbon sources on lipase production by Penicillium aurantiogriseum. Food Technol Biotechnol 41(2):105–110
Mayer C, Moritz R, Kirschner C, Borchard W, Maibaum R, Wingender J, Flemming HC (1999) The role of intermolecular interactions: studies on model systems for bacterial biofilms. Int J Biol Macromol 26:3–16
Monyem A, Canakci M, Van Gerpen JH (2000) Investigation of biodiesel thermal stability under simulated in-use conditions. Appl Eng Agric 16:373–378
Myklestad SM, Haug A (1972) Production of carbohydrates by the marine diatom Chaetoceros affinis var. willei (Gran) Hustedt. II. Preliminary investigation of the extracellular polysaccharide. J Exp Mar Biol Ecol 9:137–144
Nagano N, Matsui S, Kuramura T, Taoka Y, Honda D, Hayashi M (2011) The distribution of extracellular cellulase activity in marine eukaryotes, thraustochytrids. Mar Biotechnol 13(2):133–136
Newell SY, Fell JW (1982) Mycoflora of turtlegrass (Thalassia testudinum Konig) as recorded after seawater incubation. Bot Mar 23:265–275
Pinsirodom P, Parkin KL (2001) Lipase assays. Current protocols in food analytical chemistry. Wiley, New York
Porter D (1990) Labyrinthulomycota. In: Margulis L, Corliss JO, Melkonian M, Chapman D (eds) Handbook of Protoctista. Jones and Bartlett, Boston, pp 388–398
Raghukumar S (2002) Ecology of the marine protists, the Labyrinthulomycetes (Thraustochytrids and Labyrinthulids). Eur J Protistol 38(2):127–145
Raghukumar S, Damare VS (2011) Increasing evidence for the important role of Labyrinthulomycetes in marine ecosystems. Bot Mar 54:3–11
Raghukumar S, Raghukumar C (1999) Thraustochytrid fungoid protists in faecal pellets of the tunicate Pegea confoederata, their tolerance to deep-sea conditions and implication in degradation processes. Mar Ecol Prog Ser 190:133–140
Raghukumar S, Sharma S, Raghukumar C, Sathe-Pathak V (1994) Thraustochytrid and fungal component of marine detritus. IV. Laboratory studies on decomposition of the leaves of the mangrove Rhizophora apiculata Blume. J Exp Mar Biol Ecol 183:113–131
Raghukumar S, Sathepathak V, Sharma S, Raghukumar C (1995) Thraustochytrid and fungal component of marine detritus. III. Field studies on decomposition of leaves of the mangrove Rhizophora apiculata. Aquat Microb Ecol 9(2):117–125
Raghukumar S, Anil AC, Khandeparkar L, Patil JS (2000) Thraustochytrid protists as a component of marine microbial films. Mar Biol 136:603–609
Rajan A, Kumar DRS, Nair AJ (2011) Isolation of a novel alkaline lipase producing fungus Aspergillus fumigatus MTCC 9657 from aged and crude rice bran oil and quantification by HPTLC. Int J Biol Chem 5:116–126
Sathe-pathak V, Raghukumar S, Raghukumar C, Sharma S (1993) Thraustochytrid and fungal component of marine detritus. I. Field studies on decomposition of the brown alga Sargassum cinereum. J Agric Ind J Mar Sci 22(3):159–167
Sharma S, Raghukumar C, Raghukumar S, Sathe-pathak V, Chandramohan D (1994) Thraustochytrid and fungal component of marine detritus II. Laboratory studies on decomposition of the brown alga Sargassum cinereum. J Agric J Exp Mar Biol Ecol 175(2):227–242
Sheng G, Yu H, Wang C (2006) FTIR-spectctral analysis of two photosynthetic H2-producing strains and their extracellular polymeric substances. Appl Microbiol Biotechnol 73:204–210
Simopoulos AP (1989) Summary of the NATO advanced research workshop on dietary omega 3 and omega 6 fatty acids: biological effects and nutritional essentiality. J Nutr 119:521–528
Singh A, Wilson S, Ward OP (1996) Docosahexaenoic acid (DHA) production by Thraustochytrium sp. ATCC 20892. World J Microbiol Biotechnol 12:76–81
Singh R, Gupta N, Goswami VK, Gupta R (2006) A simple activity staining protocol for lipases and esterases. Appl Microbiol Biotechnol 70(6):679–682
Sukumaran RK, Singhania RR, Pandey A (2005) Microbial cellulases—production, applications and challenges. J Sci Ind Res 64(11):832–844
Sutherland IW (1985) Biosynthesis and composition of gram negative bacterial extracellular and wall polysaccharides. Annu Rev Microbiol 39:243–270
Sutherland IW (1998) Novel and established applications of microbial polysaccharides. Trends Biotechnol 16:41–46
Takahata K, Monobe KI, Tada M, Weber PC (1998) The benefits and risks of n-3 polyunsaturated fatty acids. Biosci Biotechnol Biochem 62:2079–2085
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24(8):1596–1599
Taoka Y, Nagano N, Okita Y, Izumida H, Sugimoto S, Hayashi M (2009) Extracellular enzymes produced by marine eukaryotes, thraustochytrids. Biosci Biotechnol Biochem 73(1):180–182
Taoka Y, Nagano N, Okita Y, Izumida H, Sugimoto S, Hayashi M (2011) Effect of Tween 80 on the growth, lipid accumulation and fatty acid composition of Thraustochytrium aureum ATCC 34304. J Biosci Bioeng 111:420–424
Than PP, Del Castillo CS, Yoshikawa T, Sakata T (2004) Extracellular protease production of bacteriolytic bacteria isolated from marine environments. Fish Sci 70(4):659–666
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22(22):4673–4680
Tsui CKM, Marshall W, Yokoyama R, Honda D, Lippmeier JC, Craven KD, Peterson PD, Berbee ML (2009) Labyrinthulomycetes phylogeny and its implications for the evolutionary loss of chloroplasts and gain of ectoplasmic gliding. Mol Phylogenet Evol 50(1):129–140
Van de Peer Y, Baldaufrid SL, Doolittle WF, Meyerid A (2000) An updated and comprehensive rRNA phylogeny of (crown) eukaryotes based on rate-calibrated evolutionary distances. J Mol Evol 51(6):565–576
Volkman JK, Jeffrey SW, Nichols PD, Rogers GI, Garland CD (1989) Fatty acid and lipid composition of 10 species of microalgae used in mariculture. J Exp Mar Biol Ecol 128:219–240
Whitfield C (1988) Bacterial extracellular polysaccharides. Can J Microbiol 34:415–420
Wilkens SL, Maas EW (2012) Development of a novel technique for axenic isolation and culture of thraustochytrids from New Zealand marine environments. J Appl Microbiol 112(2):346–352
Williams AG, Wimpenny JWT (1977) Exopolysaccharide production by Pseudomonas NCIB 11264 grown in batch culture. J Gen Microbiol 102:13–21
Williams AG, Wimpenny JWT (1978) Exopolysaccharide production by Pseudomonas NCIB 11264 grown in continuous culture. J Gen Microbiol 104:47–57
Wingender J, Strathmann M, Rode A, Leis A, Flemming HC (2001) Isolation and biochemical characterization of extracellular polymeric substances from Pseudomonas aeruginosa. Methods Enzymol 336:302–314
Yaguchi T, Tanaka S, Yokochi T, Nakahara T, Higashihara T (1997) Production of high yields of docosahexaenoic acid by Schizochytrium sp. strain SR21. J Am Oil Chem Soc 74:1431–1434
Yang HL, Lu CK, Chen SF, Chen YM (2010) Isolation and characterization of Taiwanese heterotrophic microalgae: screening of strains for docosahexaenoic acid (DHA) production. Mar Biotechnol 12(2):173–185
Yokochi T, Honda D, Higashihara T, Nakahara T (1998) Optimization of docosahexaenoic acid production by Schizochytrium limacinum SR21. Appl Microbiol Biotechnol 49:72–76
Acknowledgments
This work was partially funded by National Science Foundation of China grant 31170109 (GYW) and Shenzhen Development and Reform Commission grant 835 (GYW). Members of Shenzhen Key Lab of Nano-Micro Material Research, Peking University, are acknowledged for their technical support during FTIR analysis of EPS samples. All the authors are thankful to the Shenzhen Marine environment and resource monitoring center for the facilities extended during sampling.
Author information
Authors and Affiliations
Corresponding author
Additional information
Y. Liu and P. Singh contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 233 kb)
Rights and permissions
About this article
Cite this article
Liu, Y., Singh, P., Sun, Y. et al. Culturable diversity and biochemical features of thraustochytrids from coastal waters of Southern China. Appl Microbiol Biotechnol 98, 3241–3255 (2014). https://doi.org/10.1007/s00253-013-5391-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-013-5391-y