Abstract
This study aimed to develop an economically viable enzyme for the optimal production of steviol (S) from stevioside (ST). Of 9 commercially available glycosidases tested, S-producing β-glucosidase (SPGase) was selected and purified 74-fold from Penicillium decumbens naringinase by a three-step column chromatography procedure. The 121-kDa protein was stable at pH 2.3–6.0 and at 40–60 °C. Hydrolysis of ST by SPGase produced rubusoside (R), steviolbioside (SteB), steviol mono-glucoside (SMG), and S, as determined by HPLC, HPLC-MS, and 1H- and 13C-nuclear magnetic resonance. SPGase showed higher activity toward steviol mono-glucosyl ester, ST, R, and SMG than other β-linked glucobioses. The optimal conditions for S production (30 mM, 64 % yield) were 47 mM ST and 43 μl of SPGase at pH 4.0 and 55 °C. This is the first report detailing the production of S from ST hydrolysis by a novel β-glucosidase, which may be useful for the pharmaceutical and agricultural areas.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Steviol glucosides from the leaves of Stevia rebaudiana Bertoni (Compositae) or Stevia suavissimus (Rosaceae) have the same backbone (steviol, S) and differ in their carbohydrate residues content; that is, mono-, di-, and trisaccharides containing glucose and/or rhamnose at positions C-13 and C-19 (Fig. 1). These structural differences impart the glucosides’s relative sweetness and quality of taste. The major glycosides are stevioside (ST), rubusoside (R), steviol mono-glucoside (SMG), steviol mono-glucosyl ester (SME), steviolbioside (SteB), rebaudiosides A–F (RebA–F), and dulcoside A, which are mainly used as low calorie sweeteners. ST, the major component in S. rebaudiana Bertoni, is a natural sweetener that is about 143-fold sweeter than sucrose at a concentration of 0.025 % but has a slightly bitter taste and aftertaste (Darise et al. 1984). Since the start of the industrial production of ST in 1996, it has been used primarily as a sweetener for seasonings, pickles, and salted foods. More recently, amphipathic steviol glucosides have been applied as superior natural solubilizers that enhance the solubility of various pharmaceutically important compounds with very low water-solubility, such as paclitaxel, curcumin, capsaicin, cyclosporine, nystatin, and erythromycin (Zhang et al. 2011; 2012). S is the important aglycone of steviol glucosides and is used as a pharmaceutical compound for the improvement of cognitive functions such as learning, memory, alertness, and psychotic stability (Fowler et al. 2009), as well as being a plant growth factor (Oliveira et al. 2008). To date, the economical production of S from steviol glucosides is not yet established.
With regard to the industrialization process, some researchers have tried to produce S from steviol glucosides using chemical treatments. After acid hydrolysis of ST under extremely acidic conditions, the produced S rearranges into isosteviol automatically (Kohoda et al. 1976). An alternative S-producing method from ST involves the treatment with sodium periodate and sodium hydroxide (Ogawa et al. 1980). However, this process calls for a highly diluted system and a large excess of the expensive sodium periodate to achieve useful yields. Thus, this process is not economical for the preparation of large amounts of S notwithstanding the problems of it using excess toxic compounds.
Therefore, enzyme-based approaches are useful methods for the mass production of S from steviol glucosides. β-Glucosidase (β-d-glucoside glucohydrolase, E.C. 3.2.1.21) catalyzes the hydrolysis of β-glucosidic linkages of various oligosaccharides and aryl-glycosides. Many β-glucosidases have been isolated from microorganisms, plants, and animals, and as such their substrate specificities have been characterized in detail. Among the reported β-glucosidases, only four enzymes that hydrolyze the glucose moiety in ST have been described in detail. β-Glucosidases from Clavibacter michiganense (Nakano et al. 1998) and Flavobacterium johnsoniae (Okamoto et al. 2000) hydrolyze the β-glucosidic linkage of the 19-carboxyl group of steviol glucosides; however, they do not degrade β-glucosidic linkages of the 13-hydroxyl group of RebA or ST. Recently, 2 R-hyper producing enzymes have been isolated; one is a β-glucosidase from Aspergillus aculeatus (Ko et al. 2012) and the other a β-galactosidase from Aspergillus sp. (Wan et al. 2012). To the best of our knowledge, there is no detailed description about a multi-functional β-glucosidic enzyme capable of hydrolyzing the sophorosyl residue at the 13-hydroxyl group and one glucosyl residue at the 19-carboxyl group of ST. Although some reports have described a S-producing procedure using commercial pectinases (Pectinol 59L and Cytolase PCL5), “Pectinol 59L” is no longer commercially available (Ruddat et al. 1965) and “Cytolase PCL5” did not show any biochemical properties and mechanism for S-production (Wehrli 2011). In our studies of commercially available β-linkage-hydrolyzing enzymes, we identified an enzyme capable of producing S from ST via SteB, R, SME, and SMG. We also optimized the production process of S by using a combination of factorial design and response surface methodology (RSM). RSM is an effective statistical tool that is widely used in the optimization of fermentation processes. It has several advantages, including lower experimental numbers, suitability for experiments with multiple factors, the ability to search for relationship between factors, and the ability to find the most suitable condition. These benefits allow the user to forecast responses with minimal errors with respect to parameter effects. RSM contains many reported designs such as the Box–Behnken design, the Graeco–Latin design, and the central composite design (CCD; Ashipala and He 2008).
In this study, a novel S-producing β-glucosidase, SPGase, was selected to produce S from ST via SteB, R, SME, and SMG. This report details the purification, substrate specificities, and kinetic parameters of the enzyme. Moreover, we describe the parameters required for the maximum production of S from ST by RSM.
Materials and methods
Materials and enzymes
Compounds SMG, SME, and SteB were prepared in our laboratory following the method described by Nakano et al. (1998). Compound R was prepared as described by Ko et al. (2012). Compounds S, ST, and RebA; Aspergillus niger hemicellulase, hesperidinase, and β-glucanase; Aspergillus aculeatus Viscozyme L; Trichoderma longibrachiatum β-glucanase; Penicillium decumbens naringinase; Trichoderma reesei ATCC 26921 β-glucanase; Clostridium thermocellum thermostable β-glucanase; almond β-glucosidase; and all other chemicals were purchased from Sigma-Aldrich (St. Louis, MO, USA).
Enzyme purification
Purification of S-producing β-glucosidase from P. decumbens naringinase was carried out using a DEAE–Sepharose fast flow column (GE Healthcare, Piscataway, NJ, USA) and Superose 12 10/300 GL gel filtration (GE Healthcare) column chromatography according to the manufacturer’s protocol. The enzyme solution (32 mg/ml) was loaded onto a 1st DEAE–Sepharose fast flow column (1.6 × 2.5 cm) equilibrated with 40 mM sodium acetate buffer (NAB, pH 6.0). The absorbed proteins were eluted with a 0–1.0 M sodium chloride linear gradient at a flow rate of 1 ml/min. The active fractions were then concentrated and loaded onto a Superose 12 column (2.6 × 30 cm) equilibrated with 40 mM NAB (pH 6.0) in 50 mM sodium chloride. The enzyme solution was loaded onto a 2nd DEAE–Sepharose fast flow column (1.6 × 2.5 cm) equilibrated with 40 mM NAB (pH 5.0). The absorbed proteins were eluted with a 0–0.4 M sodium chloride linear gradient at a flow rate of 1 ml/min. Finally, the active fractions were dialyzed against 40 mM NAB (pH 5.0) and then the final purified protein was concentrated using an Amicon Ultra 10,000 MWCO (Millipore, Billerica, MA, USA).
Enzyme assay
The β-glucosidase activity was determined by measuring the release of glucose from para-nitrophenyl β-glucoside (pNPG-β). When pNPG-β, 0.80–5 mM) was used as a substrate, the reaction was stopped by the addition of a sodium carbonate solution (final concentration, 0.67 M) to the enzyme reaction mixture. The amount of 4-nitrophenol liberated from pNPG-β was measured as the absorption at 400 nm in a cuvette (length, 1 cm) with a molar extinction coefficient of 55,560 M−1 cm−1. The activity of the S-producing β-glucosidase was determined by the release of glucose from ST, R, SME, SMG, SteB, RebA, and glucobioses. One unit of enzyme activity was defined as the amount of enzyme hydrolyzing 1 μmol of glucose per minute. The reaction mixture consisted of the substrate (30 mM), the enzyme solution, and 40 mM NAB (pH 5.0). The liberated glucose was analyzed by the Tris–glucose oxidase–peroxidase assay with our modification by using the Glucose AR-II Test (Wako Pure Chemical Industries, Osaka, Japan). The protein concentration was measured by the Bradford method using bovine serum albumin as the standard (Bradford 1976).
Effects of pH and temperature on enzyme activity
The enzyme (33.2 μg/ml) was incubated at 55 °C in 32 mM Britton–Robinson buffer (pH 2–12) with 30 mM ST to determine the optimum pH. In addition, the enzyme was kept at 4 °C for 20 h in 32 mM Britton–Robinson buffer (pH 2–12), and the residual enzyme activity was examined to determine the pH stability. The optimal temperature was determined by incubating the enzyme at 30–80 °C for 5 h in 40 mM NAB (pH 5.0). The thermal stability was ascertained by incubating the enzyme at 30–80 °C for 12 h in 40 mM NAB (pH 5.0), and the residual enzyme activity was examined using 50 mM ST at 55 °C.
Synthesis, purification, and product analysis
The hydrolysis products of ST by various glycosidases were analyzed by thin-layer chromatography (TLC) and high-performance liquid chromatography (HPLC). Nine commercially available glycosidases (3.7–4 U) from A. niger, T. longibrachiatum, A. aculeatus, T. reesei, C. thermocellum, and almond were incubated for 24 h with 30 mM ST at the pH and temperature described in Fig. 2. The reaction mixture was placed in a water bath for 5 min to halt the enzyme activity. At the designated time intervals, aliquots (10 μl) were removed and the reaction products were analyzed by TLC using pre-coated silica gel 60 F254 plates (Merck, Darmstadt, Germany) developed in a solvent system consisting of acetonitrile:water [85:15 (v/v)], with steviol glucosides as the standard compounds. The plates were dipped into a solution containing 0.03 g of N-(1-naphthyl) ethylenediamine and 5 ml of concentrated sulfuric acid prepared in 95 ml of methanol, heated at 100 °C (Su and Robyt 1993), and then visualized. Phosphomolybdic acid staining solution gave highly sensitive spot of S in the TLC plate (Burstein 1953).
HPLC profiles of the stevioside hydrolysis products and HPLC-MS analysis of three isolated products. a HPLC profiles of stevioside hydrolysis products by commercially available glycosidases. a Standard steviol glucosides in order of retention time: stevioside (1), rubusoside (2), steviol mono-glucoside (3), and steviol (4), respectively. b Standard steviol glucosides in order of retention time rebaudioside A (1), steviolbioside (2), and steviol mono-glucoside ester (3), respectively. c T. reesei β-glucanase (pH 5.0, 35 °C). d A. niger β-glucanase (pH 5.0, 35 °C). e A. niger hesperidinase (pH 4.0, 40 °C). f almond β-glucosidase (pH 5.0, 35 °C). g A. niger hemicellulase (pH 4.5, 40 °C). h T. longibrachiatum β-glucanase (pH 5.0, 35 °C). i C. thermocellum β-glucanase (pH 5.8, 70 °C). j P. decumbens naringinase (pH 5.0, 37 °C). k A. aculeatus Viscozyme L (pH 4.5, 60 °C) in a. Each enzyme was reacted with 30 mM stevioside in Britton–Robinson buffer (pH 4.0–5.8) for 24 h, and then the reaction mixture was analyzed by HPLC as described in the “Materials and methods” section. Methods. HPLC-MS data of isolated P1 (b), P2 (c), and P3 (d) as unknown compounds from reaction mixtures (j, P. decumbens naringinase)
Chromatographic separation for quantitative analysis was achieved using a 1200 series quaternary HPLC system (Agilent Technologies, Palo Alto, CA, USA) consisting of a G1311A quaternary pump with a G1322A vacuum degasser, a G1329A thermostatted autosampler, a G1316A column oven set at 30 °C, a Eclipse XDB-phenyl column (5 μm, 4.6 × 150 mm), and a G1314B VW detector set at 210 nm. After desalting with Amberlite MB-3 (Organo, Tokyo, Japan), the resultant digests were separated on a C18 column with a step wise methanol gradient (0–100 %). The fractions containing the reaction products were collected and desalted again with Amberlite MB-3, followed by lyophilization. Finally, three products (P1, P2, and P3 in Fig. 2) were isolated by preparative HPLC using an LC-6AD instrument (Shimadzu, Kyoto, Japan) under the following conditions: YMC-Pack Pro C18 RS column (250 × 20 mm ID, 5 μm; YMC Co., Ltd. Kyoto, Japan); mobile phase, 30 % (v/v) methanol; flow rate, 1 ml/min; room temperature; detection, SPD-20A UV/VIS detector (Shimadzu). High performance liquid chromatography–mass spectrometry (HPLC-MS)-based analysis for the isolated compounds was performed using an HCT ultra PTM Discovery Ion Trap system (Bruker Daltonik, GmbH, Bremen, Germany) in the negative electrospray ionization mode.
For structural elucidation of the hydrolysis products, approximately 50 mg of the isolated R, SMG, and S were dissolved in 550 μl of deuterated water (D2O) and placed into 3-mm nuclear magnetic resonance (NMR) tubes. NMR spectra were acquired on a Unity INOVA 500 spectrometer (Varian, Palo Alto, CA, USA) operating at 600 MHz for 1H-NMR and 150 MHz for 13C-NMR at 25 °C. Linkages between the S and glucose were evaluated using the spectra obtained via homonuclear correlation spectroscopy, heteronuclear single quantum coherence, and heteronuclear multiple bond correlation.
Optimization procedure and experimental design
A 5-level CCD with four factors was applied to produce the S optimization procedure using the Design Expert 8.01 software (SAS Institute Inc., Cary, NC, USA), including the six replicates at the central point, which were utilized in the fitting of a second-order response surface. The ST concentration (x 1), enzyme (1.53 U/ml) amount (x 2), reaction pH (x 3), and reaction temperature (x 4) were utilized to prepare each of the 27 cultivation conditions summarized in Table 3. Optimization was conducted using a desirability function to determine the effects of x 1, x 2, x 3, and x 4 on S production. A total of 27 experiments composed of 16 factorial points, 8 axial points, and 6 center points were conducted to determine the 14 coefficients of the model as follows:
where y is the predicted response; β0 is the intercept, β1, β2, β3, and β4 are linear coefficients, β11, β22, β33, and β44 are squared coefficients, and β12, β13, β14, β23, β24, and β34 are interaction coefficients. Once an appropriate model was obtained, it was used to determine the predicted optimum conditions for the process.
Results
Screening for a steviol-producing β-glucosidase from commercially available glycosidases
We screened 9 commercially available glycosidases from A. niger, T. longibrachiatum, A. aculeatus, T. reesei, C. thermocellum, and almonds to identify an enzyme capable of converting ST into S. Each enzyme was incubated with 30 mM ST at the optimal pH for 1 day, and the hydrolysis products were analyzed by TLC or HPLC. Most enzymes did not hydrolyze ST, or exhibited very low hydrolysis activity (<1 %) toward it. Two enzymes, A. aculeatus Viscozyme L and P. decumbens naringinase, hydrolyzed the glucosidic linkage at the sophoroside at the 13-hydroxyl group or the glucose at the 19-carboxyl group of ST. As shown in Fig. 2, P. decumbens naringinase produced 3 products from ST, as demonstrated by peak 1 (P1; retention time, 22.7 min), peak 2 (P2; retention time, 24 min), and peak 3 (P3, retention time, 29.8 min). The molecular masses of the constituents identified in P1, P2, and P3 (Fig. 2) were determined to be 642, 478, and 318 Da, respectively, by HPLC-MS analysis, and their respective structures were elucidated as R, SME, and S by 1H- and 13C-NMR spectroscopies. P. decumbens naringinase was further purified, characterized, and finally used to produce S from ST via R, SME, and SMG.
Purification of steviol-producing β-glucosidase from P. decumbens and its biochemical characterization
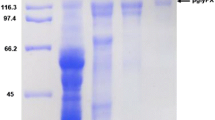
To characterize the biochemical properties, we purified the S-producing β-glucosidase from P. decumbens followed by a three-step column chromatography utilizing DEAE–Sepharose and Superose 12 gel filtrations (Table 1). The final protein (19.30 U/mg) was purified approximately 74.2-fold from the crude enzyme, resulting in a high recovery of activity (41.58 %). The purified protein showed a single band on a 12 % sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel with a molecular mass of 121 kDa (Fig. 3). The enzyme showed the highest activity at pH 4.0 and maintained more than 95 % of its maximal activity in the pH range of 2.3–6.0 (Fig. 4). The enzyme showed the highest activity at 60 °C in a 30 min-reaction, and was stable at up to 40–60 °C for 5 h (Fig. 4). In addition, the effect of metal ions on the enzyme was studied. Metal ions such as Ca2+, Mg2+, Li+, Mn2+, and EDTA at 1 mM had no significant effect on the enzyme activity. However, the enzyme was inhibited by 47 % by 1 mM Zn2+ (data not shown).
Effects of pH (a) and temperature (b) on the activity and stability of the purified enzyme. a Circle activity profile at various pH values using 33.2 μg/ml enzyme, 30 mM stevioside in Britton–Robinson buffer at 37 °C for 5 h; filled circle pH stability profile using 99.6 ng enzyme in 40 mM Britton–Robinson buffer at 4 °C for 20 h. b Filled circle activity profile at various temperatures using 99.6 ng enzyme in 40 mM NAB (pH 5.0) for 5 h at various temperatures. Circle thermal stability using 99.6 ng enzyme in 40 mM NAB (pH 4.5) at 30−80 °C after a 12-h incubation. After sampling the reaction mixture at designated times, the remaining enzyme activity was assayed by a standard method
Substrate specificities of the steviol-producing β-glucosidase from P. decumbens
The substrate specificities of the S-producing β-glucosidase and almond β-glucosidase were investigated with different β-linked glucobioses and steviol glucosides, as summarized in Table 2. The enzymes had similar activities with respect to pNPG-β, and the S-producing β-glucosidase exhibited higher specificities toward ST, R, SMG, and SME than the β-glucobioses. However, it rarely hydrolyzed RebA, which has a β-glucosyl (1 → 3) derivative of the C-13-hydroxyl group of ST (Fig. 5c). In addition, the enzyme did not show hydrolysis activity towards laminaribiose. These results are thought to be due to steric hindrance, with respect to enzyme catalysis of the complex glucosidic linkage of RebA. The enzyme kinetic parameters demonstrated that SMG was the best substrate, and this enzyme specifically hydrolyzes catalysis between the glucosidic moiety and S, such as in R, SMG, SME, and ST (Table 2). The catalytic efficiency of the enzyme toward the steviol glucosides followed the order SMG > R > SME > ST. According to the reaction time of the S-producing β–glucosidase from P. decumbens, ST was converted to R or SteB was converted to S via SME, R was converted to mainly SME or SMG, and SMG or SME were converted to S, indicating that the transformation pathway of ST is as follows: major pathway, ST → R → SMG → S; minor pathway 1, ST → SteB → SMG → S; minor pathway 2, ST → R → SME → S (Fig. 6). These results indicate that the enzyme hydrolyzes only the glucose moiety in ST. Finally, we named the enzyme S-producing β-glucosidase (SPGase) from ST.
HPLC profiles of the hydrolysis product of the steviol-producing β-glucosidase with various steviol glucosides. The reaction mixture containing the steviol-producing β-glucosidase (33.2 μg/ml) and 30 mM of each substrate was reacted in 40 mM NAB (pH 5.0) at 50 °C for 24 h. a Standard steviol glucosides as stevioside, rubusoside, steviol mono-glucoside, and steviol, respectively. b Standard steviol glucosides as rebaudioside A, steviolbioside, and steviol mono-glucosyl ester, respectively. c Reaction mixture with rebaudioside A. d Reaction mixture with stevioside. e Reaction mixture with rubusoside. f Reaction mixture with steviolbioside. g Reaction mixture with steviol mono-glucoside. h Reaction mixture with steviol mono-glucosyl ester
Time–course analysis of stevioside hydrolysis by the steviol-producing β-glucosidase. The reaction mixture containing the steviol-producing β-glucosidase (33.2 μg/ml) and 20 mM each substrate was reacted in 40 mM NAB (pH 5.0) at 50 °C for 10–70 h. a—a Standard steviol glucosides as stevioside, rubusoside, steviol mono-glucoside, and steviol, respectively; b standard steviol glucosides as rebaudioside A, steviolbioside, and steviol mono-glucosyl ester, respectively; the reaction mixture incubated for c 10, d 20, e 30, f 40, g 50, h 60, and i 70 h, respectively. b The proposed stevioside hydrolysis pathway of the steviol-producing β-glucosidase from reaction mixture a. The solid-lined arrow indicates strong modification, and the dash-lined arrow indicates a weak enzyme reaction
Determination of the polynomial equation coefficients for optimal production of steviol
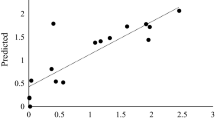
RSM was used to study the interaction of these variables in relation to S production. From the preliminary experiments, 4 important factors were selected to optimize the S production: enzyme amount (20–60 μl), ST concentration (5–80 mM), temperature (30 °C–80 °C) and reaction pH (2–6). The CCD of S production is listed in Table 3. As ascertained from the central points of the corresponding contour plots (Fig. 7), the optimal values for the 4 variables for S production were an ST concentration of 47 mM and a SPGase amount of 43 μl reacted at pH 4.0 and 55 °C. S production was measured to be 30 mM (64 % yield) under these conditions. The results indicate that S production can be expressed in terms of the following regression equation:
Response surface and contour plots of steviol production. Shown are the mutual interactions between stevioside concentration and enzyme amount (a), stevioside concentration and pH (b), stevioside concentration and temperature (c), enzyme amount and pH (d), enzyme amount and temperature (e), and pH and temperature (f). Other parameters, except for the two parameters in each figure, are zero level in coded units
\( y=29.47-0.39{x_1}-0.71{x_2}+1.57{x_3}+2.67{x_4}+0.86{x_1}{x_2}-0.56{x_1}{x_3}-0.73{x_1}{x_4}+0.76{x_2}{x_3}-0.063{x_2}{x_4}+5.85{x_3}{x_4}-15.04x_1^2-16.10x_2^2-11.07x_3^2-10.55x_4^2 \) where x 1, x 2, x 3, and x 4 are coded values of the ST concentration (mM), enzyme amount (μl), reaction pH, and reaction temperature (°C), respectively. The regression equation gave a high R 2 value (0.877) using an ANOVA test. This is an estimate of the fraction of overall variation in the data accounted for by the model, and thus, the model is capable of explaining 87 % of the variation in response. Based on the model, the predicted response for S production was 30 mM, and the observed experimental value was 30 mM at 43 μl of SPGase with 47 mM ST at 55 °C and pH 4.0, demonstrating almost identical results for the predicted and observed S productions.
Discussion
In the present study, a novel S-producing β-glucosidase, SPGase, isolated from P. decumbens naringinase was screened and purified, and its biochemical properties were characterized. In addition, the conditions for production of S were optimized by RSM using 4 important factors; namely, ST concentration, enzyme activity, temperature, and pH.
β-Glucosidases are widely found in molds, yeasts, bacteria, and higher plants, and many have been purified and characterized. Thus far, only 3 enzymes, namely, F. johnsoniae β-glucosidase (FJGase, Okamoto et al. 2000), C. michiganense β-glucosidase (CMGase), and A. aculeatus β-glucosidase (SSGase), have been reported to be capable of hydrolyzing the β-glucosyl linkages of steviol glucosides. However, the presently discovered SPGase was very different from these 3 enzymes in its properties and extremely broad substrate specificities. For example, the novel enzyme has a larger molecular mass (121 kDa) than FJGase (72 kDa), CMGase (65 kDa), and SSGase (79 kDa). The optimal pH and pH stability (pH 4.0 and pH 2.0–6.0, respectively) of SPGase were similar to those of SSGase (pH 5.0 and pH 3.0–7.0, respectively), but more acidic than the values for FJGase (pH 7.0 and pH 3.0–9.0, respectively) and CMGase (pH 7.5 and pH 6.0–10, respectively). In addition, a large difference was found with respect to specificities toward steviol glucosides. FJGase and CMGase exhibited high specificities toward the β-glucosidic linkage at the 19-carboxyl group of RebA, whereas SPGase and SSGase could not hydrolyze the RebA. Of the four enyzmes, only SSGase could not hydrolyze the β-glucosidic linkage at the 19-carboxyl group of ST, R, and SME, respectively. To put it concretely, SSGase is a specific β-glucosidase for producing R from ST. FJGase degraded R to form S via SMG and SME, whereas it could not hydrolyze ST to R. FJGase could not degrade RebB and SteB with the di-glucosidic unit at the 13-hydroxyl group and it preferred smaller glucosidic linkages at the 19-carboxyl group such as R, SMG, and SME. CMGase could not hydrolyze the β-glucosidic linkage at the 13-hydroxyl group of the steviol glucosides, preferring instead to degrade glucose at the 19-carboxyl group of RebA, which has bulky side chains at the 13-hydroxyl group. On the other hand, SSGase, FJGase, and CMGase could not produce S by a one-enzyme system, indicating that they are useful enzymes only for the partial hydrolysis of steviol glucosides. Only SPGase produced S from ST via R, SteB, SME, and SMG, being able to hydrolyze both sides of the β-glucosidic linkages at the 13-hydroxyl group and 19-carboxyl group of ST. Finally, we suggest that SPGase displayed multi-functional activities, and this activity is essential for S production from ST. The 4 enzymes displayed similar low activities toward the β-glucobioses. This is consistent with the fact that the other three enzymes have much higher specificities toward steviol glucosides than toward β-glucooligosaccharides. The natural substrates for the enzymes seem to be steviol glucosides and not the oligosaccharides derived from the naturally abundant glucose polymers such as cellulose and laminaran. To the best of our knowledge, this is the first study characterizing a β-glucosidase capable of producing S from ST.
In this study, we screened S-producing β-glucosidases among the commercially available enzymes. SPGase produced S via R and SMG as a main pathway and its production condition was optimized with 4 important factors. This optimized condition may be applicable for the economical production of S from the relatively cheap ST for the pharmaceutical and agricultural areas. This study preludes the beginning of detailed investigations into SPGase, and future studies should entail cloning the full-length gene as well as protein engineering experiments. The present results demonstrate the potential of this novel β-glucosidase from P. decumbens naringinase for use in the industrial production of S.
References
Ashipala OK, He Q (2008) Optimization of fibrinolytic enzyme production by Bacillus subtilis DC-2 in aqueous two-phase system (poly-ethylene glycol 4000 and sodium sulfate). Bioresour Technol 99:4112–4119
Bradford MM (1976) Rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Burstein S (1953) Reduction of phosphomolybdic acid by compounds processing conjugated double bonds. Anal Chem 25:422–424
Darise M, Mizutani K, Kasai R, Tanaka O, Kitahata S, Okada S, Ogawa S, Murakami F, Chen FH (1984) Enzymic transglucosylation of rubusoside and the structure-sweetness relationship of steviol-bisglycosides. Agric Biol Chem 48:2483–2488
Fowler A, Goralczyk R, Kilpert C, Mechan AO, Mohajeri MH, Mussler B, Wyss A (2009) Novel neutraceutical compositions containing Stevia extract or Stevia extract constituents and uses thereof. Patent Cooperation Treaty Publication No.WO2009/071277
Ko JA, Kim YM, Ryu YB, Jeong HJ, Park TS, Park SJ, Wee YJ, Kim JS, Kim D, Lee WS (2012) Mass production of rubusoside using a novel stevioside-specific β-glucosidase from Aspergillus aculeatus. J Agric Food Chem 60:6210–6216
Kohoda H, Kasai R, Yamasaki K, Murakami K, Tananka O (1976) New sweet diterpene glucosides from Stevia rebaudiana. Phytochemistry 15:981–983
Nakano H, Okamoto K, Yatake T, Kiso T, Kitahata S (1998) Purification and characterization of a novel β-glucosidase from Clavibacter michiganense that hydrolyzes glucosyl ester linkage in steviol glucosides. J Biosci Bioeng 85:162–168
Ogawa T, Nozaki M, Matsui M (1980) Total synthesis of stevioside. Tetrahedron 36:2642–2648
Okamoto K, Nakano H, Yatake T, Kiso T, Kitahata S (2000) Purification and some properties of a β-glucosidase from Flavobacterium johnsonae. Biosci Biotechnol Biochem 64:333–340
Oliveira BH, Stiirmer JC, Filho JDS, Ayub RA (2008) Plant growth regulation activity of steviol and derivatives. Phytochemisty 69:1528–1533
Ruddat M, Heftmann E, Lang A (1965) Biosynthesis of steviol. Arch Biochem Biophys 110:496–499
Su D, Robyt JF (1993) Control of the synthesis of dextran and acceptor-products by Leuconostoc mesenteroides B-512FM dextransucrase. Carbohydr Res 248:471–476
Wan HD, Tao GJ, Kim D, Xia YM (2012) Enzymatic preparation of a natural sweetener rubusoside from specific hydrolysis of stevioside with β-galactosidase from Aspergillus sp. J Mol Catal B: Enzym 82:12–17
Wehrli C (2011) Process for the enzymatic preparation of steviol from stevioside. Patent Cooperation Treaty Publication No. WO 2011/089031
Zhang F, Koh GY, Jeansonne DP, Hollingsworth J, Russo PS, Vicente G, Stout RW, Liu Z (2011) A novel solubility-enhanced curcumin formulation showing stability and maintenance of anticancer activity. J Pharm Sci 100:2778–2789
Zhang F, Koh GY, Hollingsworth J, Russo RS, Stout RW, Liu Z (2012) Reformulation of etoposide with solubility-enhancing rubusoside. Int J Pharm 434:453–459
Acknowledgments
This research was supported by a National Research Foundation grant funded by the Korean government (Ministry of Education, Science, and Technology No. 2009-0081749), Bio-industry Technology Development Program (No. 112147-3), Ministry for Food, Agriculture, Forestry and Fisheries and the KRIBB Research Initiative Program, Republic of Korea.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Jin-A Ko and Young Bae Ryu contributed equally to this work.
Rights and permissions
About this article
Cite this article
Ko, JA., Ryu, Y.B., Kwon, HJ. et al. Characterization of a novel steviol-producing β-glucosidase from Penicillium decumbens and optimal production of the steviol. Appl Microbiol Biotechnol 97, 8151–8161 (2013). https://doi.org/10.1007/s00253-013-4883-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-013-4883-0