Abstract
Caffeic acid is a valuable aromatic compound that possesses many important pharmacological activities. In structure, caffeic acid belongs to the hydroxycinnamic acid family and can be biosynthesized from the aromatic amino acid tyrosine. In the present paper, the caffeic acid biosynthesis pathway was reconstituted in engineered Escherichia coli to produce caffeic acid from simple biomass sugar glucose and xylose. Different engineering approaches were utilized to optimize the production. Specifically, two parallel biosynthesis routes leading from tyrosine to caffeic acid were studied. The copy number of the intermediate biosynthesis genes was varied to find appropriate gene doses for caffeic acid biosynthesis. Three different media, including a MOPS medium, a synthetic medium, and a rich medium, were also examined to improve the production. The highest specific caffeic acid production achieved was 38 mg/L/OD. Lastly, cultivation of engineered E. coli in a bioreactor resulted in a production of 106 mg/L caffeic acid after 4 days.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
There is presently a rising interest in microbial production of valuable aromatic compounds from inexpensive simple carbon sources or renewable biomass feedstocks (Gosset 2009). Notably, several industrially important products, such as styrene, p-hydroxystyrene, and p-hydroxybenzoate, have been successfully produced in engineered bacteria (Qi et al. 2007; McKenna and Nielsen 2011; Verhoef et al. 2007). Driving the progress is a recent development of metabolic engineering on microbes’ aromatic amino acid biosynthesis pathway (or shikimate pathway) in which l-phenylalanine, l-tryptophan, or l-tyrosine can be used as precursors for production of desired products. However, the reported titers and yields of these products are still low, largely due to the poor availability of the substrate aromatic amino acids. Efforts have therefore been made to increase the endogenous pool size of these amino acids. For example, a number of Escherichia coli strains have been successfully constructed by different engineering approaches to overproduce tyrosine with high yields (Lutke-Eversloh and Stephanopoulos 2008; Santos and Stephanopoulos 2008; Juminaga et al. 2012). Based on this accomplishment, microbial biosynthesis of several tyrosine-derived compounds, including p-coumaric acid, melanin, flavonoids, tyrosol, and alkaloids, has been achieved (Santos and Stephanopoulos 2008; Santos et al. 2011; Satoh et al. 2012; Nakagawa et al. 2011).
Caffeic acid (3,4-dihydroxy cinnamic acid) is an important hydroxycinnamic acid that can be derived from tyrosine. It is an intriguing compound because it possesses various pharmacological activities, including antioxidant (Maurya and Devasagayam 2010; Mori and Iwahashi 2009), antitumor (Rajendra-Prasad et al. 2011; Hudson et al. 2000; Morton et al. 2000), antiviral (Ikeda et al. 2011), antidepressive (Takeda et al. 2002), and antidiabetic (Jung et al. 2006; Cheng et al. 2003) activities. In addition, caffeic acid phenethyl ester has been well documented to have anticarcinogenic and antidiabetic activities (Chuu et al. 2012; Natarajan et al. 1996; Grunberger et al. 1988; Sud’ina et al. 1993; Celik et al. 2009). As a key precursor for lignin formation, caffeic acid is widely found in plants, but despite its reported pharmaceutical values, there are relatively few studies for extracting caffeic acid from natural plant sources, mainly due to the complexity and inefficiency of the separation and purification process (Ye et al. 2010; Wang et al. 2009; Leonardis et al. 2005). Therefore, engineering a microbe to make a high level of caffeic acid is of great research interest.
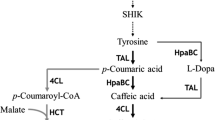
Caffeic acid can be biosynthesized from tyrosine through a two-step pathway, in which tyrosine is converted to p-coumaric acid and then to caffeic acid by tyrosine ammonia lyase (TAL) and 4-coumarate 3-hydroxylase (Coum3H), respectively (Rosler et al. 1997; Berner et al. 2006). Alternatively, 4-coumarate:CoA ligase (4CL) can convert p-coumaric acid to coumaroyl-CoA which becomes caffeoyl-CoA by 4-coumarate 3-hydroxylase (Gross and Zenk 1974; Kneusel et al. 1989). Notably, it has been reported that certain CoA thioesterases can hydrolyze caffeoyl-CoA to make caffeic acid (Ramirez-AhumadaMdel et al. 2006). E. coli native CoA thioesterases such as hydroxyphenylacetyl-CoA thioesterase have also been found to have promiscuous activity against a variety of substrates and thus could act on caffeoyl-CoA to generate caffeic acid (Song et al. 2006; Zhuang et al. 2008). Therefore, another potential pathway for caffeic acid biosynthesis is to convert tyrosine to coumaric acid, coumaroyl-CoA, caffeoyl-CoA, and finally, caffeic acid by four enzymatic reactions (Fig. 1).
Very recently, Lin et al. reported the production of caffeic acid in E. coli by establishing a dual biosynthesis pathway that used both coumaric acid and l-DOPA as intermediates (Lin and Yan 2012). Choi et al. biosynthesized caffeic acid in an effort to make phenylpropanoid products in E. coli (Choi et al. 2011). In addition, Streptomyces fradiae has also been utilized as a heterologous host for caffeic acid biosynthesis (Berner et al. 2006). However, the reported caffeic acid titers in these studies remain low, as the highest titer achieved so far was 50 mg/L in E. coli (Lin and Yan 2012). In the presented paper, two parallel biosynthesis routes leading from tyrosine to caffeic acid were reconstituted in an E. coli tyrosine over-producer, and the effects of different production media and biosynthetic gene copy number were investigated to improve the production. These different engineering approaches resulted in the highest reported titers of caffeic acid from glucose and the first report of caffeic acid production from xylose.
Materials and methods
Caffeic acid tolerance assay
In order to study the toxicity of caffeic acid on E. coli, a previously constructed E. coli strain rpoA14(DE3) was grown in LB medium supplemented with 0, 10, 100, and 1,000 mg/L caffeic acid standard. Twenty microliters of overnight LB culture of rpoA14(DE3) was inoculated into 1 mL of such prepared medium. After 24 h of growth at 250 rpm at 37 °C, optical density (OD) at 600 nm was measured for individual cultures.
Bacterial cultivation conditions
All strains used in this study were cultivated at 250 rpm at 37 °C. For caffeic acid production in test tube, 40 μL of overnight LB culture was inoculated into 2 mL MOPS, synthetic or rich medium and grown for 3 days. To avoid undesired formation of caffeic acid-derived o-quinone and poly-aromatics induced by light, the test tubes were wrapped with aluminum foil. The ingredient of the media used in this work is as follows: 1 L MOPS medium contained 1× MOPS minimal medium salt (Teknova), 2.84 g Na2HPO4, and 5 g glucose or xylose; 1 L synthetic medium contained 2 g NH4Cl, 5 g (NH4)2SO4, 2.993 g KH2PO4, 7.315 g K2HPO4, 8.372 g MOPS, 0.5 g NaCl, 2.4 g MgSO4, and 5 g glucose or xylose; and 1 L rich medium contained 10 g tryptone, 5 g yeast extract, 10 g NaCl, and 5 g glucose or xylose. Trace elements containing 0.4 mg/L Na2EDTA, 0.03 mg/L H3BO3, 1 mg/L thiamine, 0.94 mg/L ZnCl2, 0.5 mg/L CoCl2, 0.38 mg/L CuCl2, 1.6 mg/L MnCl2, 3.77 mg/L CaCl2, and 3.6 mg/L FeCl2 were also supplemented to the synthetic and rich media. For cell cultivation, 0.1 mM IPTG and appropriate antibiotics were added at inoculation. The working concentrations of antibiotics were: 100 mg/L ampicillin, 50 mg/L kanamycin, 50 mg/L streptomycin, and 34 mg/L chloramphenicol.
Construction of strains and plasmids
Strain rpoA14(DE3) (E. coli K12 ΔpheA ΔtyrR lacZ::PLtetO-1-tyrAfbraroGfbr tyrR::PLtetO-1-tyrAfbraroGfbr hisH(L82R) carrying pHACM-rpoA14) used in this study was previously constructed for tyrosine overproduction (Santos et al. 2012). Plasmid pTrc-RgTALsyn, pTrc-RgTALsyn–Pc4CLsyn and pCDF-trc-RgTALsyn-Pc4CLsyn were constructed in our previous study (Santos et al. 2011). Plasmid pTrc-RgTALsyn (pCA1) contained a codon-optimized (for E. coli) Rhodotorula glutinis tyrosine ammonia lyase (RgTALsyn; Electronic supplementary material (ESM) 1). Plasmid pTrc-RgTALsyn-Pc4CLsyn (pCA2) contained RgTALsyn and a codon-optimized Petroselinum crispus 4-coumarate:CoA ligase (Pc4CLsyn; ESM 2) pCA1 and pCA2 were both derived from plasmid pTrcHis2B (Invitrogen) with a low plasmid copy number (15–25 copies per cell). pCDF-trc-RgTALsyn (pCA3) and pCDF-trc-RgTALsyn-Pc4CLsyn (pCA4) were derived from pCDFDuet-1 (Novagen) and contained a relatively higher copy number than pCA1 and pCA2 (20–40 copies per cell). pCA3 was constructed by removal of Pc4CLsyn gene from pCA4 using restriction enzyme SalI digestion followed by a self-ligation. All four plasmids contain a trc promoter for gene expression. The sam5 gene encoding a 4-coumarate 3-hydroxylase (Coum3H) from Saccharothrix espanaensis (Genbank accession number, DQ357071.1) was codon-optimized and synthesized by Genewiz (South Plainfield, NJ, USA; ESM 3). The Coum3H gene flanked by an NdeI and XhoI restriction sites was then subcloned into plasmid pRSFDuet-1 by NdeI and XhoI digestion followed by ligation of appropriate fragments. The resulting plasmid was named pCA5. The constructed plasmids were transformed into E. coli rpoA14(DE3) with different combinations to generate strains RPC1, RPC2, RPC3, and RPC4. The description of strains and plasmids used in this study is summarized in Table 1. All enzymes used for cloning were purchased from New England Biolabs.
Analytical methods
For caffeic acid and coumaric acid quantification, 0.75 mL culture samples were extracted with ethyl acetate, air dried, and resuspended in 0.15 mL methanol containing 20 mg/L protocatechuic acid internal standard. Extract samples were then analyzed by an Applied Biosystems API2000 LC/MS/MS system equipped with a Waters symmetry 5 μm C8 column. Caffeic acid and coumaric acid were eluted using a linear gradient of 95 % acetonitrile and 5 % water (0 min) to 35 % acetonitrile and 65 % water (10 min) at a flow rate of 0.3 mL/min. For the quantification of l-tyrosine, cell-free culture supernatants were filtered through 0.2 μm pore-size polytetrafluoroethylene membrane syringe filters (VWR International) and 100 μL such-treated culture supernatant was mixed with 10 μL 1 g/L dopamine internal standard. Prepared samples were then analyzed by an Applied Biosystems API2000 LC/MS/MS system equipped with a Waters Spherisorb 5 μm C18 column. Tyrosine was eluted using an isocratic elution with 20 mM ammonium acetate (pH adjusted to 4 by formic acid) at a flow rate of 0.3 mL/min.
Bioreactor production
Twenty milliliters overnight LB culture of E. coli rpoA14(DE3) harboring pCA1 and pCA2 (RPC1) was inoculated into a 2–L BioFlo110 modular fermentor system (New Brunswick Scientific) containing 1 L synthetic medium supplemented with appropriate antibiotics and 5 g/L glucose. The whole bioreactor vessel was wrapped with aluminum foil to avoid exposure of culture to light. Culture conditions were maintained as follows: air flow rate, 1 L/min; pH value, 7.6; temperature, 37 °C; and agitation, 350 rpm. Experimental samples were withdrawn every 24 h for OD600 measurements and product quantification.
Results
Caffeic acid tolerance of E. coli
In order to study the effect of caffeic acid on E. coli growth, strain rpoA14(DE3) was grown in LB medium supplemented with caffeic acid at final concentrations of 0, 10, 100, and 1,000 mg/L. After growth at 37 °C for 24 h, the cultures’ cell densities (OD600) were measured for comparison. As shown in Fig. 2, cell growth was slightly inhibited as caffeic acid concentration increased, though the presence of 1,000 mg/L caffeic acid only caused a 20 % final cell density drop, compared to the control culture without caffeic acid. The result of this assay shows that E. coli can tolerate high titer caffeic acid and thus has potential as a heterologous host for caffeic acid overproduction.
Caffeic acid production on glucose
In order to make caffeic acid in E. coli, a previously constructed tyrosine overproducer rpoA14(DE3) was transformed with plasmids pCA1 and pCA5, resulting in E. coli RPC1 carrying TAL and Coum3H genes (Fig. 1). Glucose was fed to the RPC1 culture at a final concentration of 5 g/L. After a 3-day cultivation at 37 °C, the engineered E. coli strain produced the expected caffeic acid product, as confirmed by LC/MS/MS analysis (ESM 4, Fig. S1). Therefore, a caffeic acid biosynthesis pathway was successfully reconstituted in E. coli, which was able to utilize inexpensive sugar substrate glucose to make high-value product caffeic acid.
We next set out to improve the caffeic acid production by a combination of different engineering approaches. First, a potential alternative route for making caffeic acid through coumaroyl-CoA and caffeoyl-CoA was introduced into E. coli. Specifically, a 4-coumarate:CoA ligase (4CL) was used to convert p-coumaric acid to coumaroyl-CoA, followed by Coum3H-mediated hydroxylation to make caffeoyl-CoA (Kneusel et al. 1989). De-esterification of caffeoyl-CoA to yield caffeic acid used E. coli’s native promiscuous thioesterase activity (Fig. 1).
Second, two sets of plasmids with relatively low and high copy numbers were constructed to vary TAL and 4CL gene doses for caffeic acid production. The transformation of these plasmids into the tyrosine overproducer rpoA14(DE3) yielded four E. coli strains, as listed in Table 1. Lastly, the constructed strains were grown in three different media, including a MOPS medium, a synthetic medium, and a rich medium, to further improve caffeic acid production on glucose.
Figure 3 shows caffeic acid, coumaric acid, and tyrosine production by different E. coli strains cultivated in the three media. As shown in Fig. 3a, caffeic acid was produced in E. coli RPC1, RPC3, and RPC4, but not in RPC2. The failure of caffeic acid biosynthesis in RPC2 was later found out by a qPCR analysis to be due to the lack of TAL and 4CL gene transcription (ESM 5 Fig. S2(a) and (b)). Similar phenomenon was observed when the same plasmid was used in our previous study, although the mechanism behind this is still unclear (Santos et al. 2011). Comparison of RPC3 and RPC4 clearly shows that addition of coumaroyl-CoA/caffeoyl-CoA biosynthesis route did not help improve the caffeic acid production on glucose. Instead, it introduced extra metabolic burden on the E. coli host and resulted in even lower production. Similarly, RPC1 produced more caffeic acid than RPC3 in all three media, suggesting that increasing copy number of TAL gene decreased the caffeic acid production due to the unnecessary metabolic burden. Overall, caffeic acid production in the rich medium was higher than in the other two media, which could be associated with higher cell density in rich medium as shown in Fig. 3d. In addition, glucose substrate was depleted in rich medium, but there was a big portion of un-used glucose in all E. coli strains grown in both the MOPS and synthetic media (data not shown). The results here indicate that engineered E. coli is better able to grow and thus make more caffeic acid on glucose in the rich medium.
Figure 3b shows that large amounts of coumaric acid was accumulated in the MOPS and synthetic media, which clearly suggests that, in these media, the production is limited by coumaric acid to caffeic acid conversion catalyzed by Coum3H. In contrast, there was significantly less coumaric acid accumulation in the rich medium, indicating that coumaric acid to caffeic acid conversion is higher in the rich medium. Notably, it was also found that a big portion of produced coumaric acid was released to the culture broth, in addition to the accumulation inside the cells. Among the four constructed strains, RPC1 produced the most coumaric acid in both the MOPS and synthetic media.
Tyrosine titers are shown in Fig. 3c. It should be noted that the rich medium contains yeast extract and thus contains around 220 mg/L tyrosine. RPC2 was found to produce a large amount of tyrosine, but it could not further use it to make coumaric acid. In the MOPS and synthetic media, RPC3 and RPC4 produced similarly low levels of tyrosine. In comparison, RPC1 made more tyrosine than RPC3 and RPC4, which translates into high caffeic acid production.
The highest caffeic acid titer of 88 mg/L was achieved in strain RPC1 grown in the rich medium. The highest specific titer, defined as caffeic acid titer divided by cell density, reached 38 mg/L/OD in RPC1 grown in the synthetic medium. These results imply that engineered E. coli is more efficient for making caffeic acid in the synthetic medium than in the rich medium. In addition, RPC1 was the best producer among all four strains grown on glucose.
Caffeic acid production on xylose
Xylose is an important component of renewable biomass feedstocks. Its utilization for producing industrially valuable chemicals is of current research interest. As E. coli natively is able to consume xylose, we studied whether xylose can be used by our engineered E. coli strains to make caffeic acid. Similar to production on glucose, strains containing different gene copy numbers were grown in three media supplemented with xylose to optimize caffeic acid production.
Figure 4a shows that the rich medium produced high concentrations of caffeic acid in all four strains except RPC2. Notably, RPC3 produced a slightly higher level of caffeic acid than RPC1, whereas RPC1 produce a higher level of caffeic acid than RPC3 when grown on glucose. It is therefore suggested that the effect of increasing TAL gene copy number is sugar dependent. However, mRNA quantification analysis showed that the use of different sugars did not have a significant impact on TAL gene transcription (ESM 5, Fig. S2(a)). Therefore, the improvement of caffeic acid production on xylose in RPC3 did not directly result from upregulation of TAL gene transcription. In the case of synthetic medium, RPC1 produced the most caffeic acid, whereas in the MOPS medium RPC1 and RPC4 produced similarly level of caffeic acid. Generally, caffeic acid production on xylose was lower than on glucose.
As shown in Fig. 4b, engineered E. coli strains, except RPC2, accumulated high titers of intermediate coumaric acid in all three media. However, coumaric acid accumulated in these media was not well converted to caffeic acid, since intermediate coumaric acid concentrations were much higher than final product caffeic acid. Figure 4c further shows that RPC1 made much more tyrosine in the synthetic medium than in the MOPS medium. Similar results were observed for RPC3 and RPC4. RPC2 was able to produce high level of tyrosine but not coumaric acid and caffeic acid. Similar to growth on glucose, cell growth on xylose in the rich medium was much better than in the other two media, as shown in Fig. 4d. The highest caffeic acid titer of 70 mg/L and the highest specific titer of 25 mg/L/OD were both achieved in strain RPC3 grown in the rich medium. Overall, the caffeic acid production on glucose was better than on xylose because glucose is a better carbon source for E. coli metabolism.
Bioreactor production
After establishing caffeic acid production in test tubes, we next evaluated production in a 2-L bioreactor. Glucose was chosen as the substrate as it produced a higher level of caffeic acid than xylose. E. coli RPC1 was the best producer on glucose, and thus it was used in this bioreactor production study. The synthetic medium was chosen as the preferred production medium (a) because it contains only inexpensive components and thus is economically attractive, and (b) because the highest specific production of 38 mg/L/OD was achieved in the synthetic medium. Although the cell density was low in test tubes, using bioreactor technique to increase cell density was expected to further boost the caffeic acid titer.
As shown in Fig. 5, production of caffeic acid was detected at day 1 and peaked at day 4. A gradual decrease in caffeic aid titer was observed after day 4, which should be associated with the formation of undesired o-quinone and its polyaromatic derivatives. The highest OD value of the bioreactor culture was 1.7, higher than the test tube culture (data not shown). The peak caffeic acid titer was 106 mg/L, in comparison with 51 mg/L achieved in a test tube in the same medium. The highest specific production of 75 mg/L/OD at day 4 was also 97 % higher than the result of the shake flask experiment. The production profiles of coumaric acid and tyrosine followed the same trend with that of caffeic acid. Notably, the cell accumulated high levels of these two compounds during the fermentation. In particular, coumaric acid titer was one order of magnitude higher than caffeic acid, highlighting the need to increase conversion of this intermediate to the final product.
Discussion
High level caffeic acid biosynthesis is an attractive research subject due to the valuable pharmacological properties of this aromatic compound. In the present study, E. coli was selected as a surrogate biosynthesis host due to its established advantages for heterologous natural product production (Zhang et al. 2008). A caffeic acid biosynthesis pathway was reconstituted in engineered E. coli and successfully converted sugar-derived tyrosine to caffeic acid by using a tyrosine ammonia lyase (TAL) and a 4-coumarate 3-hydroxylase (Coum3H). However, introduction of the alternative biosynthesis route through coumaroyl-CoA and caffeoyl-CoA did not increase production. It was therefore suggested that only the biosynthesis route involving TAL and Coum3H was active for caffeic acid production in the constructed E. coli. The failure of the alternative route could be due to an inactive or low activity of an E. coli thioesterase against caffeoyl-CoA. To further investigate the utility of this biosynthesis route, improving E. coli native thioestarase activity or introducing a heterologous caffeoyl-CoA thioestarase activity should be conducted.
Varying copy numbers of TAL and 4CL genes resulted in altered caffeic acid production on both glucose and xylose. Low copies of TAL gene was found to be sufficient for high titer production, whereas introduction of additional 4CL gene or increasing TAL gene copy number decreased the production in most cases, implying that a balanced biosynthetic gene copy number is crucial for final product biosynthesis. Strain RPC2 containing low copies of TAL and 4CL genes did not produce caffeic acid due to the lack of the corresponding gene transcription. A similar finding was reported in our previous study (Santos et al. 2011), yet the mechanism of this effect is not clear. Notably, although large amounts of tyrosine and coumaric acid were accumulated in all engineered strains, caffeic acid production remained relatively low. These results therefore suggest that Coum3H activity needs to be enhanced in order to improve caffeic acid biosynthesis from coumaric acid. Since the copy number of CoumH gene used in this study was high (~100 copies per cell), fine-tuning Coum3H gene copy number or utilizing Coum3H from other organisms with higher 4-coumarate 3-hydroxylase activity should be pursued to increase the coumaric acid utilization in future studies.
The rich medium was found to make the highest titer of caffeic acid because its rich nutrient content helped improve cell growth. However, since the carbon source in the rich medium was mainly consumed for the cell growth, instead of caffeic acid production, specific production level in the rich medium was lower than the synthetic medium which contained only defined salts and glucose. Among the three media, the MOPS medium showed the lowest caffeic acid production. This can be explained by the fact that it was not supplemented with trace elements and thus lacked needed metal cofactors for heterologous enzyme activity. For example, 4-coumarate 3-hydroxylase has been reported to require copper to carry out the enzymatic reaction (Nambudiri and Bhat 1972), but the MOPS medium did not contain sufficient copper to support the enzyme activity.
Caffeic acid production was further improved by cultivating RPC1 cell in a bioreactor containing the synthetic medium and glucose. Better pH control and oxygen supply were considered to help the cell growth and finally improve the caffeic acid titer to 106 mg/L, which is higher than any previous studies. However, undesired formation of o-quinone and its derivative led to a decrease in caffeic acid titer after a 4-day fermentation. For future studies, oxidation of caffeic acid and formation of polyaromatics need to be reduced in order to prevent the loss of caffeic acid product. In addition, metabolic engineering of the above identified pathway steps could further improve the caffeic acid heterologous production in E. coli.
References
Berner M, Krug D, Bihlmaier C, Vente A, Müller R, Bechthold A (2006) Genes and enzymes involved in caffeic acid biosynthesis in the actinomycete Saccharothrix espanaensis. J Bacteriol 188(7):2666–2673
Celik S, Erdogan S, Tuzcu M (2009) Caffeic acid phenethyl ester (CAPE) exhibits significant potential as an antidiabetic and liver-protective agent in streptozotocin-induced diabetic rats. Pharmacol Res 60(4):270–276
Cheng JT, Liu IM, Tzeng TF, Chen WC, Hayakawa S, Yamamoto T (2003) Release of beta-endorphin by caffeic acid to lower plasma glucose in streptozotocin-induced diabetic rats. Horm Metab Res 35(4):251–258
Choi O, Wu CZ, Kang SY, Ahn JS, Uhm TB, Hong YS (2011) Biosynthesis of plant-specific phenylpropanoids by construction of an artificial biosynthetic pathway in Escherichia coli. J Ind Microbiol Biotechnol 38(10):1657–1665
Chuu CP, Lin HP, Ciaccio MF, Kokontis JM, Hause RJ Jr, Hiipakka RA, Liao S, Jones RB (2012) Caffeic acid phenethyl ester suppresses the proliferation of human prostate cancer cells through inhibition of p70S6K and Akt signaling networks. Cancer Prev Res (Phila) 5(5):788–797
Gosset G (2009) Production of aromatic compounds in bacteria. Curr Opin Biotechnol 20(6):651–658
Gross GG, Zenk MH (1974) Isolation and properties of hydroxycinnamate:CoA ligase from lignifying tissue of Forsythia. Eur J Biochem 42(2):453–459
Grunberger D, Banerjee R, Eisinger K, Oltz EM, Efros L, Caldwell M, Estevez V, Nakanishi K (1988) Preferential cytotoxicity on tumor cells by caffeic acid phenethyl ester isolated from propolis. Experientia 44(3):230–232
Hudson EA, Dinh PA, Kokubun T, Simmonds MS, Gescher A (2000) Characterization of potentially chemopreventive phenols in extracts of brown rice that inhibit the growth of human breast and colon cancer cells. Cancer Epidemiol Biomark Prev 9(11):1163–1170
Ikeda K, Tsujimoto K, Uozaki M, Nishide M, Suzuki Y, Koyama AH, Yamasaki H (2011) Inhibition of multiplication of herpes simplex virus by caffeic acid. Int J Mol Med 28(4):595–598
Juminaga D, Baidoo EE, Redding-Johanson AM, Batth TS, Burd H, Mukhopadhyay A, Petzold CJ, Keasling JD (2012) Modular engineering of l-tyrosine production in Escherichia coli. Appl Environ Microbiol 78(1):89–98
Jung UJ, Lee MK, Park YB, Jeon SM, Choi MS (2006) Antihyperglycemic and antioxidant properties of caffeic acid in db/db mice. J Pharmacol Exp Ther 318(2):476–483
Kneusel RE, Matern U, Nicolay K (1989) Formation of trans-caffeoyl-CoA from trans-4-coumaroyl-CoA by Zn2+-dependent enzymes in cultured plant cells and its activation by an elicitor-induced pH shift. Arch Biochem Biophys 269(2):455–462
Leonardis AD, Macciola V, Domenico ND (2005) A first pilot study to produce a food antioxidant from sunflower seed shells (Helianthus annuus). Eur J Lipid Sci Technol 107(4):220–227
Lin Y, Yan Y (2012) Biosynthesis of caffeic acid in Escherichia coli using its endogenous hydroxylase complex. Microb Cell Fact 11:42
Lutke-Eversloh T, Stephanopoulos G (2008) Combinatorial pathway analysis for improved l-tyrosine production in Escherichia coli: identification of enzymatic bottlenecks by systematic gene overexpression. Metab Eng 10(2):69–77
Maurya DK, Devasagayam TP (2010) Antioxidant and prooxidant nature of hydroxycinnamic acid derivatives ferulic and caffeic acids. Food Chem Toxicol 48(12):3369–3373
McKenna R, Nielsen DR (2011) Styrene biosynthesis from glucose by engineered E. coli. Metab Eng 13(5):544–554
Mori H, Iwahashi H (2009) Antioxidant activity of caffeic acid through a novel mechanism under UVA irradiation. J Clin Biochem Nutr 45:49–55
Morton LW, Croft KD, Puddey IB, Byrne L (2000) Phenolic acids protect low density lipoproteins from peroxynitrite-mediated modification in vitro. Redox Rep 5(2–3):124–125
Nakagawa A, Minami H, Kim JS, Koyanagi T, Katayama T, Sato F, Kumagai H (2011) A bacterial platform for fermentative production of plant alkaloids. Nat Commun 2:326
Nambudiri AMD, Bhat JV (1972) Conversion of p-coumarate into caffeate by Streptomyces nigrifaciens. Purification and properties of the hydroxylating enzyme. Biochem J 130(2):425–433
Natarajan K, Singh S, Burke TR Jr, Grunberger D, Aggarwal BB (1996) Caffeic acid phenethyl ester is a potent and specific inhibitor of activation of nuclear transcription factor NF-KB. Proc Natl Acad Sci USA 93(17):9090–9095
Qi WW, Vannelli T, Breinig S, Ben-Bassat A, Gatenby AA, Haynie SL, Sariaslani FS (2007) Functional expression of prokaryotic and eukaryotic genes in Escherichia coli for conversion of glucose to p-hydroxystyrene. Metab Eng 9(3):268–276
Rajendra-Prasad N, Karthikeyan A, Karthikeyan S, Reddy BV (2011) Inhibitory effect of caffeic acid on cancer cell proliferation by oxidative mechanism in human HT-1080 fibrosarcoma cell line. Mol Cell Biochem 349(1–2):11–19
Ramirez-AhumadaMdel C, Timmermann BN, Gang DR (2006) Biosynthesis of curcuminoids and gingerols in turmeric (Curcuma longa) and ginger (Zingiber officinale): identification of curcuminoid synthase and hydroxycinnamoyl-CoA thioesterases. Phytochemistry 67(18):2017–2029
Rosler J, Krekel F, Amrhein N, Schmid J (1997) Maize phenylalanine ammonia-lyase has tyrosine ammonia-lyase activity. Plant Physiol 113(1):175–179
Santos CN, Stephanopoulos G (2008) Melanin-based high-throughput screen for l-tyrosine production in Escherichia coli. Appl Environ Microbiol 74(4):1190–1197
Santos CN, Koffas M, Stephanopoulos G (2011) Optimization of a heterologous pathway for the production of flavonoids from glucose. Metab Eng 13(4):392–400
Santos CN, Xiao W, Stephanopoulos G (2012) Rational, combinatorial, and genomic approaches for engineering l-tyrosine production in Escherichia coli. Proc Natl Acad Sci USA 109(34):13538–13543
Satoh Y, Tajima K, Munekata M, Keasling JD, Lee TS (2012) Engineering of a tyrosol-producing pathway, utilizing simple sugar and the central metabolic tyrosine, in Escherichia coli. J Agric Food Chem 60(4):979–984
Song F, Zhuang Z, Finci L, Dunaway-Mariano D, Kniewel R, Buglino JA, Solorzano V, Wu J, Lima CD (2006) Structure, function and mechanism of the phenylacetate pathway hotdog-fold thioesterase PAAI. J Biol Chem 281(16):11028–11038
Sud'ina GF, Mirzoeva OK, Pushkareva MA, Korshunova GA, Sumbatyan NV, Varfolomeev SD (1993) Caffeic acid phenethyl ester as a lipoxygenase inhibitor with antioxidant properties. FEBS Lett 329(1–2):21–24
Takeda H, Tsuji M, Inazu M, Egashira T, Matsumiya T (2002) Rosmarinic acid and caffeic acid produce antidepressive-like effect in the forced swimming test in mice. Eur J Pharmacol 449(3):261–267
Verhoef S, Ruijssenaars HJ, de Bont JAM, Wery J (2007) Bioproduction of p-hydroxybenzoate from renewable feedstock by solvent-tolerant Pseudomonas putida S12. J Biotechnol 132:49–56
Wang J, Lu D, Zhao H, Ling X, Jiang B, Ouyang P (2009) Application of response surface methodology optimization for the production of caffeic acid from tobacco waste. Afr J Biotechnol 8(8):1416–1424
Ye JC, Hsiao MW, Hsieh CH, Wu WC, Hung YC, Chang WC (2010) Analysis of caffeic acid extraction from Ocimum gratissimum Linn. by high performance liquid chromatography and its effects on a cervical cancer cell line. Taiwan J Obstet Gynecol 49(3):266–271
Zhang H, Wang Y, Pfeifer BA (2008) Bacterial hosts for natural product production. Mol Pharm 5(2):212–225
Zhuang Z, Song F, Zhao H, Li L, Cao J, Eisenstein E, Herzberg O, Dunaway-Mariano D (2008) Divergence of function in the hot dog fold enzyme superfamily: the bacterial thioesterase YciA. Biochemistry 47(9):2789–2796
Acknowledgments
This study was supported by ARPA-E (DE-AR0000059) provided by US Department of Energy and the Singapore MIT Alliance.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
DOCX 116 kb
Rights and permissions
About this article
Cite this article
Zhang, H., Stephanopoulos, G. Engineering E. coli for caffeic acid biosynthesis from renewable sugars. Appl Microbiol Biotechnol 97, 3333–3341 (2013). https://doi.org/10.1007/s00253-012-4544-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-012-4544-8