Abstract
The oleaginous yeast Rhodosporidium toruloides, which belongs to the Pucciniomycotina subphylum in the Basidiomycota, has attracted strong interest in the biofuel community recently due to its ability to accumulate more than 60% of dry biomass as lipid under high-density fermentation. A 3,543-nucleotide (nt) DNA fragment of the glyceraldehyde-3-phosphate dehydrogenase gene (GPD1) was isolated from R. toruloides ATCC 10657 and characterized in details. The 1,038-nt mRNA derived from seven exons encodes an open reading frame (ORF) of 345 amino acids that shows high identity (80%) to the Ustilago maydis homolog. Notably, the ORF is composed of codons strongly biased towards cytosine at the Wobble position. GPD1 is transcriptionally regulated by temperature shock, osmotic stress, and carbon source. Nested deletion analysis of the GPD1 promoter by GFP reporter assay revealed that two regions, −975 to −1,270 and −1,270 to −1,429, upstream from the translational start site of GPD1 were important for responses to various stress stimuli. Interestingly, a 176-bp short fragment maintained 42.2% promoter activity of the 795-bp version in U. maydis whereas it was reduced to 17.4% in R. toruloides. The GPD1 promoter drove strong expression of a codon-optimized enhanced green fluorescent protein gene (RtGFP) and a codon-optimized hygromycin phosphotransferase gene (hpt-3), which was critical for Agrobacterium tumefaciens-mediated transformation in R. toruloides.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rhodosporidium toruloides is an oleaginous yeast belonging to the Pucciniomycotina subphylum in the Basidiomycota (Sampaio et al. 2003). Strikingly, it can be cultured to extremely high cell density (>100 g/l dry cell mass) and accumulate more than 60% biomass as triglycerides, making it a good host for the production of oil for biodiesel and many other applications from biomass (Liu et al. 2009; Turcotte and Kosaric 1988; Zhao et al. 2010). To date, it is hardly possible to genetically modify the lipid biosynthesis pathway in R. toruloides. The only transformation method reported is the PEG-mediated transformation of protoplast (Tully and Gilbert 1985). Unfortunately, this method is severely limited by the low efficiency, instable chromosomal integration, and auxotrophic selection.
Strong promoters are critical for establishing efficient gene expression and transformation systems. One of the most frequently used promoters derives from the glyceraldehyde-3-phosphate dehydrogenase (GPD, EC 1.2.1.12) encoding gene, the expression of which is often regarded as strong and constitutive (Tristan et al. 2011). The mRNA transcripts of GPD can reach 2–5% of the total poly(A) + RNA pool in Saccharomyces cerevisiae (Holland and Holland 1978) and account for 5% of the soluble intracellular proteins in Aspergillus nidulans (Redkar et al. 1998). Consequently, GPD promoters have been isolated and exploited for gene expression and transformation in numerous fungal species (Halaouli et al. 2006; Ji et al. 2010; Kilaru et al. 2006; Nitta et al. 2004; Vassileva et al. 2001; Wolff and Arnau 2002).
We report here the isolation and characterization of a GPD1 homolog in R. toruloides, and the comparison of its regulatory activities in both R. toruloides and Ustilago maydis, a well-studied basidiomycetous fungus (reviewed in Brefort et al. 2009). We report, for the first time, the successful transformation of R. toruloides by Agrobacterium tumefaciens-mediated transformation (ATMT) by dominant selection.
Materials and methods
Strains, media, and culture conditions
R. toruloides strain ATCC 10657, Rhodosporidium glutinis ATCC 90781, and Rhodotorula glutinis ATCC 204091 were obtained from ATCC (USA). Rhodotorula graminis strain WP1 and Sporobolomyces roseus FGSC 10293 (IAM13481) were obtained from Fungal Genetics Stock Center (University of Missouri, USA). U. maydis strain L8 is as described previously (Ji et al. 2010). R. toruloides and U. maydis strains were grown at 28 °C in YPD broth (1% yeast extract, 2% peptone, 2% glucose, w/v) or on potato-dextrose agar (PDA). A. tumefaciens strain AGL1 (Lazo et al. 1991) was grown at 28 °C in either liquid or solid 2YT medium (1.6% tryptone, 1% yeast extract, 0.5% NaCl). Escherichia coli XL1-Blue was cultured in Luria–Bertani (LB) broth or on LB agar and used for routine DNA manipulations.
Binary plasmids
Oligonucleotides used are listed in Table 1. All restriction and modification enzymes were from New England Biolabs (NEB, MA, USA). Plasmid pEX2 is a pEX1 derivative (Liu et al. 2011) with multiple cloning site added at the T-DNA left border (LB) (Supplementary Material Fig. S1a). Plasmid pMF2-3c (Brachmann et al. 2004) is a generous gift from Prof. Dr. Michael Feldbrügge (Institute for Microbiology, Heinrich-Heine-University Düsseldorf, Düsseldorf, Germany) (plasmid map and sequence can be found at http://www-public.rz.uni-duesseldorf.de/~mikrobio/plasmids/plasmid_pmf23c.htm). pEX2 was digested with PstI (blunt-ended) and SpeI to remove the hygromycin-resistant gene-cauliflower mosaic virus 35S gene terminator cassette (hpt::T 35S ), and ligated with a NcoI (blunt-ended)–SpeI digested fragment of plasmid pMF2-3c containing the carboxin resistance gene cassette to create plasmid pEC2 (Supplementary Material Fig. S1b). pEC3 (Supplementary Material Fig. S1c) was made by self-ligation of NcoI–PmeI double-digested and blunt-ended pEC2. pEC3GPD-EGFP (Supplementary Material Fig. S1d) was generated by ligation of SacI–PacI double-digested and blunt-ended pEC3 with the EGFP expression cassette (Pgpd::EGFP::Tnos) that was amplified using pEX1GPD–EGFP as the template (Liu et al. 2011) and Pgap-Sf and Tnos-Pmr as primers, in which Pgpd is the 595-bp gpd1 promoter from U. maydis (Smith and Leong 1990), EGFP is the enhanced green fluorescent protein encoding gene, and Tnos is the nopaline synthase gene terminator of A. tumefaciens.
For nested deletion analysis of the GPD1 promoter, the 1,429-bp upstream sequence of R. toruloides GPD1 (RtGPD1) was amplified from genomic DNA using oligos Rt011S and Rt012N (Table 1) and PfuUltra DNA polymerase (Stratagene, CA, USA). The PCR product was digested with various restriction enzymes at the 5′ end (SpeI, HindIII, MspA1I, EarI, MseI, BstNI, SalI, and PstI, respectively) (Fig. 4a). After being blunt-ended with T4 DNA polymerase at the 5′ ends followed by NcoI digestion, the DNA fragments were inserted into pEC3GPD-EGFP or pRH203 (Supplementary Material Fig. S1e) at the SpeI and NcoI sites to create pEC3Pxxx-EGFP and pRH203Pxxx-RtGFP, respectively (Fig. 4a, Supplementary Material Fig. S1f and S1g). xxx refers to the length of the RtGPD1 promoters and pRH203 contains both the hygromycin resistance gene expression cassette (Pgpd::hpt-3::Tnos) and the RtGFP gene expression cassette (Pgpd::RtGFP::T 35S ), in which hpt-3 and RtGFP were codon-optimized based on the preferred codon usage table for R. toruloides as listed in the Codon Usage Database at the Kazusa DNA Research Institute (http://www.kazusa.or.jp/codon/).
Transformation vector series pEX2Pxxx-HPT (Supplementary Material Fig. S1h) was created by fusing three fragments, the 8.7-kb PmeI–SmaI fragment of pEX2, the 795 EarI (blunt-ended)–NcoI or 176-bp PstI (blunt-ended)–NcoI fragment of RtGPD1 promoter, and the SphI (blunt-ended)–BspHI fragment containing the hpt::T 35S cassette obtained by PCR using pEX2 as the template and HygU-BspHI and 35STer as primers (Table 1). pEC3Pxxx-HPT3 series (Supplementary Material Fig. S1i) was created by replacing the EGFP fragment in pEC3Pxxx-EGFP with the hpt-3.
Extraction of genomic DNA and total RNA
Extraction of genomic DNA and total RNA from R. toruloides was adapted from the method for U. maydis (Ji et al. 2010) with some modifications. For DNA extraction, cell culture (2 ml) at exponential stage was collected, washed with 1 M sorbitol, and suspended in 0.5 ml of SCS buffer (1 M sorbitol, 20 mM sodium citrate, pH 5.8). After adding 100 μg glass beads (1 mm in diameter; Sigma-Aldrich, MO, USA), vigorous vortexing was applied to the solution for 10 min. Total DNA was precipitated by ethanol after extraction with phenol/chloroform (1:1, v/v, pH 8.0). For RNA extraction, cells were collected and suspended in 350 μl AE buffer (50 mM sodium acetate, 10 mM EDTA, pH 5.3) followed with 35 μl SDS (10%, w/v) and 425 μl AE buffer-saturated phenol (pH 5.3). Cells were ruptured by vigorous vortexing of glass beads at high temperature (75 °C for 15 min) and freeze–thawing (−20 °C for at least 1 h). Total RNA was purified by phenol–chloroform extraction (1:1 v/v, pH 4.8) and precipitation with ethanol. The concentrations of DNA or RNA samples were determined with NanoDrop® ND-1000 Spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA) and the integrity of the extracted nucleic acids were checked by agarose gel electrophoresis.
Degenerate PCR
Partial cDNA of RtGPD1 was amplified using degenerate primers Rtgpdf and Rtgpdr (Table 1) that were designed according to the conserved motifs among the known GPD proteins (Fig. 1). PCR was carried out using Easy-A High-Fidelity PCR Cloning Enzyme (Stratagene) in a PTC-200TM Programmable Thermal Controller (Bio-Rad Laboratories, CA, USA). The PCR product was cloned into pGEM-T Easy (Promega, WI, USA) followed by sequencing using oligos M13FP and M13RP (Table 1).
Sequence alignment of glyceraldehyde-3-phosphate dehydrogenases. The sequences and origins can be found in the following protein identifiers in GenBank: A. niger (XP001397496), A. thaliana (NP187062), E. coli (NP753744), H. sapiens (NP002037), U. maydis (XP758638), and R. toruloides ATCC 10657 (JN208861). Substrate binding sites are boxed and marked with S. Residues essential for NAD binding sites (aspartic acid) are indicated by triangle, and catalytic reaction (cysteine and histidine) by vertical arrows. Putative phosphorylation sites are boxed and marked with P. The horizontal arrows indicate the highly conserved motifs used for the design of degenerate primers
Inverse PCR
The genomic DNA sequences flanking the degenerate PCR fragment were identified by inverse PCR (Ochman et al. 1988). Genomic DNA was individually digested with BamHI, EcoRI, or PstI before self-ligation. Nested PCR was performed using oligos Rtgpd-IP2f/Rtgpd-IP1r (Table 1) for the first round PCR and Rtgpd-IP2f/Rtgpd-IP2r (Table 1) for the second round. The amplified DNA was cloned into pGEM-T Easy and sequenced. To obtain sufficient length of upstream sequence information of RtGPD1, a second inverse PCR was performed using the PstI-digested genomic DNA and oligos Rt008 and Rt009 (Table 1).
Rapid amplification of cDNA ends (RACE)
The 5′ and 3′ end of GPD1 cDNA sequences were obtained by 5′ RACE and 3′ RACE using BD SMARTerTM RACE cDNA Amplification Kit (Clontech, CA, USA) according to the manufacturer’s instruction. Oligo Rt007 and Rtgpd-IP1r (Table 1) was used as the specific primer for 5′ RACE and 3′ RACE, respectively. The full-length cDNA was cloned by reverse transcription PCR using DNase I-treated total RNA as the template and Rt013 and Rt005 as primers (Table 1).
Southern and northern blot analysis
For Southern blot analysis, genomic DNA (10 μg) was digested with BamHI or EcoRI and separated by electrophoresis in 0.8% agarose gel. For northern blot analysis, RNA samples (10 μg) were separated in a 1.2% formaldehyde agarose gel. Southern and northern hybridizations were performed using DIG High Prime DNA Labeling and Detection Starter Kit (Roche Diagnostics, IN, USA) (Liu et al. 2011), and the probe used was RtGPD1 fragment amplified using oligos Rt001 and Rt006 (Table 1). For temperature shock, R. toruloides was cultured in YPD broth at 28 °C for 24 h before being transferred to high temperature (37 °C) or low temperature (4 °C). Other stress treatments included a 6-h treatment in YPDK broth (YPD broth supplemented with 1 M KCl) and YPG broth (glucose in YPD replaced by the same concentration of glycerol).
Analysis of promoter activity
The constructs pEC3Pxxx-EGFP were linearized with SspI and integrated as a single copy at the ip locus (encoding the iron-sulfur protein subunit of succinate dehydrogenase) of U. maydis via PEG-mediated protoplast transformation (Loubradou et al. 2001). Constructs pRH203Pxxx-RtGFP were introduced to R. toruloides ATCC 10657 via ATMT.
Green fluorescence intensity was determined as described previously (Liu et al. 2011) with some modifications. Strains transformed with Pxxx::EGFP or Pxxx::RtGFP were cultured to mid-exponential phase, diluted to 0.2 OD600 in YPD medium, and cultured at 30 °C for 30 min. A fraction of the cultures (100 μl) was loaded to a well in 96-well flat-bottom transparent plates (Nunc, Roskilde, Denmark) and FluoroNunc plates (Thermo Fisher Scientific, Langenselbold, Germany) to determine the optical density and green fluorescence intensity, respectively. The data was acquired using the Infinite M200 plate reader (Tecan, Salzburg, Austria). Cell optical density was read at 600 nm while GFP fluorescence was measured with excitation and emission wavelength at 488 nm and 508 nm, respectively. All experiments were performed in statistical triplicates.
Transformation and identification of transformants
PEG-mediated protoplast transformation and A. tumefaciens-mediated transformation of U. maydis were performed as described previously (Ji et al. 2010; Loubradou et al. 2001). ATMT of R. toruloides was adapted from the method for U. maydis (Ji et al. 2010). Briefly, A. tumefaciens AGL1 strain harboring a binary plasmid was cultured in liquid minimal medium (Hooykaas et al. 1979) supplemented with rifampicin (50 μg/ml) and spectinomycin (50 μg/ml) until saturation. The culture was diluted to approximately 0.1 OD600 in Induction Medium supplemented with 100 μM of acetosyringone (Cangelosi et al. 1991) and cultured for approximately 6 h. The induced cell culture (100 μl) was mixed with a R. toruloides culture (100 μl) and spread on a Hybond N+ membrane (GE Healthcare, Uppsala, Sweden). After co-cultured on an Induction Medium plate with acetosyringone (100 μM) at 24 °C for 2 days, the membrane was transferred to a YPD agar plate supplemented with cefotaxime (300 μg/ml) and hygromycin B (150 μg/ml), and incubated at 28 °C for 3–5 days.
To identify true transformants, fungal colony PCR was performed. Briefly, hygromycin B-resistant colonies were inoculated in YPD broth and cultured for several hours. Colony PCR was performed in a PTC-200TM Programmable Thermal Controller (Bio-Rad Laboratories, Hercules, CA, USA) for 35 cycles using 1 μl of the cultures as the templates and HptRU/HptRSL2 (Table 1) as primers. Strains exhibiting the 581-bp fragment were identified as true transformants.
Microscopy
Colonies were observed using a Nikon SMZ 800 fluorescence microscope equipped with Plan Apo WD70 objective (Nikon, Tokyo, Japan) and a GFP-L filter (GFP Band pass, Ex 480/40 DM 505 BA 510). Images were acquired with a Nikon DS-5M camera.
Gene nomenclature and GenBank accession numbers
As recommended (Ianiri et al. 2011), gene and protein nomenclatures for the Rhodosporidium genus adopt the system used in S. cerevisiae and S. roseus. All GPD1 sequences have been submitted to GenBank under the following accession numbers: JN208861 for R. toruloides ATCC 10657, JQ806385 for R. glutinis ATCC 204091, JQ806386 for R. graminis WP1, and JQ806384 for S. roseus. GenBank accession number for the codon-optimized sequence of hpt-3 and RtGFP is JQ806387 and JQ806388, respectively.
Results
Isolation and characterization of GPD-encoding gene in R. toruloides
GPD sequences from divergent species, Aspergillus niger, Arabidopsis thaliana, E. coli, Homo sapiens, and U. maydis, were aligned and two highly conserved motifs (NGFGRIGR and TGAAKAVG) were identified (Fig. 1). Degenerate primers Rtgpdf and Rtgpdr (Table 1) were designed to amplify the coding sequence between the two conserved motifs. An approximate 0.6-kb fragment was amplified by RT–PCR using total RNA of R. toruloides as the template (Supplementary Material Fig. S2a). Sequencing of the cloned fragment revealed a 633-bp sequence that could potentially encode a polypeptide with high homology to GPDs (Fig. 1). PCR using the same primers also produced a 1,225-bp fragment when genomic DNA was used as the template (Supplementary Material Fig. S2a).
To obtain the full-length GPD1 gene sequence, oligo pairs Rtgpd-IP2f/Rtgpd-IP1r and Rtgpd-IP2f/Rtgpd-IP2r (Table 1) were designed for inverse PCR according to the partial genomic DNA sequence. A 2.1-, 2.8-, and 0.6-kb PCR product was amplified from the genomic DNA template digested with BamHI, PstI, and EcoRI, respectively (Supplementary Material Fig. S2b). Sequence assembly yielded a 2,278-bp continuous DNA sequence. To extend the sequence at the 5′ end, a second inverse PCR was performed using the PstI-digested DNA as the template and oligos Rt008 and Rt009 (Table 1) as primers, which resulted in a 1.2-kb PCR product (Supplementary Material Fig. S2b). Assembly of all sequences yielded a 3,543-bp DNA fragment that could potentially encode a full-length GPD homolog (Supplementary Material Fig. S3a).
The 5′ RACE and 3′ RACE yielded a cDNA fragment of approximately 0.9 and 0.7 kb, respectively (Supplementary Material Fig. S2c). Using oligo pair Rt013 and Rt005 (Table 1), the full-length cDNA of RtGPD1 was successfully amplified by RT–PCR (Supplementary Material Fig. S2d). The 1,166-nt full-length mRNA contains a 1,038 nt ORF, 49 nt 5′ UTR (untranslated region), and 94 nt 3′ UTR. The ORF is very GC rich with a GC content of 61.2%. Comparison between the cDNA and genomic sequences revealed seven exons separated by six introns (Supplementary Material Fig. S3a). All splicing junctions and putative branching point sequences are highly similar to other fungi (Supplementary Material Fig. S3b). Similar to other species in the Pucciniomycotina, the introns of RtGPD1 are distributed mostly in the 5′ regions (Fig. 2b and Supplementary Material Fig. S3a).
Phylogenetic relationship and intron distributions in basidiomycetous GPDs. a Phylogenetic analysis of basidiomycetous GPD1 proteins. A neighbor-joining tree derived from MEGA5 analysis (Tamura et al. 2011) of GPDs in 16 species is shown. The bootstrap values are shown above the branches. GenBank accession numbers and annotations are as follows: Sporisorium reilianum (CBQ71234); U. maydis (CAA30726); Malassezia globosa (XP001731693); P. graminis (PGTG04956.4 from Puccinia Group Database, http://www.broadinstitute.org/annotation/genome/puccinia_group/MultiHome.html); P. triticina (PTTG08295.1 from Puccinia Group Database); Rhodotorula glutinis (JQ806385); R. toruloides (AEK26271); R. graminis (JQ806386); S. roseus (JQ806384); Microbotryum violaceum (MVLG_06238, partial sequence obtained from M. violaceum genome database, http://www.broadinstitute.org/annotation/genome/Microbotryum_violaceum/MultiHome.html); Cryptococcus curvatus (AAD25080); Xanthophyliomyces dendrorhous (CAA69652); Armillaria tabescens (CAD29456); Pycnoporus coccineus (BAD69793); Phanerochaete chrysosporium (AAA33732); Agaricus bisporus II (AAA32634). b Illustration of intron positions in Pucciniomycotina GPD1 genes. Intron positions are marked according to their positions in the multiple sequence alignment of GPD1 proteins and the total numbers of introns are marked on the right. R. toruloides, R. glutinis, and R. graminis represents R. toruloides strain ATCC 10657, R. glutinis strain ATCC 90781, and R. graminis strain WP1, respectively
The predicted ORF of RtGPD1 encodes a protein of 345 amino acids (aa), exhibiting high sequence homology to other known GPDs, especially those of Basidiomycetes. The highest identity was found with the U. maydis homolog (80%, GenBank accession number XP758638). The highly conserved substrate binding site (ASCTTNCL), amino acid residues that are critical for catalytic reaction (C156 and H183), and NAD binding (D39 and N320) (Harris and Waters 1976) could be found in the product of RtGPD1. The protein could be potentially phosphorylated at positions 107–111 (TTKEK), 188–190 (TQK), 196–199 (SAKD), 245–247 (SVV), 250–255 (TVRLEK), 297–300 (SIFD), 314–317 (SWYD), and 326–334 (RRVVDLLVF) (Fig. 1 and Supplementary Material Fig. S3b). Analysis of the unrooted phylogenetic tree based on the GPD amino acid sequences of 16 species in the Basidiomycetes revealed that the RtGPD1 is clearly clustered within the Pucciniomycotina and distinct from the Agaricomycotina and Ustilaginomycotina clades (Fig. 2a).
Only 38 out of 61 possible codons were found in RtGPD1 (Table 2), and the codons show a clear and strong preference to cytosine at the Wobble position with the single exception of serine, which predominantly adopts the UCG codon (15/346) over UCC codon (5/346). Similar to many other fungi, the UAG codon was used as the translation termination codon in RtGPD1. Sequence analysis of the 3′ region of RtGPD1 promoter revealed one putative CAAT box (CCAAT) at −461 from the putative translational start codon. However, no TATA box could be found (Supplementary Material Fig. S3b). A pyrimidine-rich region (ct box) (−74 to −50 bp) was identified. Other conserved motifs identified in the Ascomycota counterparts such as gpd, pgk, qut, and qa boxes (Punt et al. 1990) could not be found in RtGPD1. RtGPD1 contains no canonical polyadenylation signal (AATAAA) in the 3′ UTR. However, a short region with TG repeats could be found 22 nt upstream of the polyadenylation site, similar to what was reported in the rabbit β-globin gene (Levitt et al. 1989) (Supplementary Material Fig. S3b).
Southern blot analysis revealed a single band in either BamHI or EcoRI digestion, indicating that a single GPD1 gene is present in the R. toruloides genome (Fig. 3a). Based on the sequence of RtGPD1, the homologous DNA sequence was isolated by PCR from R. glutinis ATCC 90781. Both sequences and splicing patterns were highly conserved, with only three nucleotide substitutions (313 G > A, 457 T > C, and 649 T > C) in the coding region (−49 to +1,719), all being silent mutations located at the Wobble positions of the respective codons (E28, P40, and I64) (Supplementary Material Fig. 3b).
Copy number of GPD1 and gene expression in R. toruloides. a Southern blot analysis of RtGPD1. Genomic DNA (10 μg) of R. toruloides were digested with BamHI (marked as B) or EcoRI (E) and probed with a digoxigenin-labeled GPD1 DNA fragment. b Northern blot analysis of GPD1 expression in R. toruloides. Total RNA samples (10 μg) of R. toruloides from various treatments were blotted to membrane and probed with a digoxigenin-labeled GPD1 DNA fragment. The blotted Hybond N+ membrane was stained with methylene blue to monitor the loading and blotting the rRNAs
Transcription of GPD1 mRNA in R. toruloides
Conditions that affect the transcription of GPD1 mRNA in R. toruloides were investigated by northern blot analysis. The transcripts were detected at high levels under all conditions tested. Although comparable mRNA levels were detected in cells cultured at 28 °C and 37 °C in YPD medium, RtGPD1 transcription was significantly enhanced by osmotic stress and repressed by cold treatment (4 °C) or when glycerol was used as the sole carbon source (Fig. 3b).
Analysis of RtGPD1 promoter by GFP reporter assay
In the absence of a reliable transformation method for R. toruloides, we made use of U. maydis for the analysis of the isolated RtGPD1 promoter in consideration of the close phylogenetic relationship between the two species. RtGPD1 promoter and various nested deletions were fused with EGFP in a transformation vector series (Supplementary Material Fig. S1e). To avoid positional effects of the inserted genes, all reporter constructs were integrated at the ip locus of U. maydis (Loubradou et al. 2001) as single copies by homologous recombination, which was confirmed by Southern blot analysis (data not shown). Whereas no fluorescence could be observed in transformants of the promoter-less construct, transformants of all other constructs displayed strong green fluorescence (Supplementary Material Fig. S4). Quantification of the cell fluorescence revealed that the 795-bp fragment was among the strongest promoters, reaching a level similar to the 595 bp U. maydis endogenous gpd1 (Umgpd1) promoter. Deletions from −1,429 to −612 showed only marginal decrease in promoter activity (Fig. 4b). A small 176-bp fragment functioned as the minimal promoter as it displayed approximately 40% promoter activity of the full-length version. The region between −612 and −441 appeared to contain a second strong enhancer of transcription as the GFP intensity was more than doubled when the region was included.
Analysis of GPD1 promoters in U. maydis (vector series pEC3Pxxx-EGFP) and R. toruloides (vector series pRH203Pxxx-RtGFP). a Schematic illustration of EGFP reporter constructs. The length of the promoters refers to the number of nucleotides from the translational start codon. The restriction enzyme sites and putative enhancer elements are shown. P—PstI; H—HindIII; M1—MspA1I; B—BstNI; E—EarI; M2—MseI; S—SalI; Um—gpd1 promoter of U. maydis (595 bp); P550 and P174—predicted promoter enhancer elements; CAAT box—potential binding site for the RNA transcription factor; ct box—pyrimidine-rich region. b Activities of truncated RtGPD1 promoters in U. maydis. c Activities of truncated RtGPD1 promoters in R. toruloides. The highest normalized green fluorescence intensity is set at 100%. Results are derived from triplicates
The GFP reporter assay was repeated after an efficient transformation method became available for R. toruloides (see the section below). As the above reporter cassettes showed no GFP fluorescence when transformed to R. toruloides by ATMT, a new EGFP gene variant (RtGFP) that follows the codon preference of R. toruloides was synthesized, fused to the promoter series described above, and transformed to R. toruloides by ATMT. The 795-bp fragment was the strongest promoter and all truncated RtGPD1 promoters tested showed significantly higher activity than the heterologous 595-bp Umgpd1 promoter, including the 176-bp version that exhibited 2.4-fold higher promoter activity than the Umgpd1 promoter (Fig. 4c and Supplementary Material Fig. S4).
Relative activities of truncated promoters under stress conditions
Quantification of GFP fluorescence of the above reporter strains revealed that the 975-bp version RtGPD1 promoter was the most sensitive to stress in R. toruloides followed by the 795-bp and 612-bp versions (Fig. 5a, b). The full-length and the short 176-bp versions were the most resistant to stress. In other words, the region between −975 and −1,429 was important for suppression of gene expression under osmotic pressure, heat shock, and cold shock. Surprisingly, the patterns of promoter activity showed no similarity between the two hosts (Fig. 5a, b). In U. maydis, the region from −975 and −1,429 was responsible for down-regulation by stress stimuli whereas this was mainly attributed to the region between −975 and −1,429 in R. toruloides.
Effects of various stress stimuli on RtGFP expression under control of RtGPD1 promoter fragments. a In U. maydis with vector series pEC3Pxxx-EGFP. All reporter constructs were integrated as single copy at the ip locus in U. maydis via PEG-mediated protoplast transformation. b In R. toruloides with vector series pRH203Pxxx-RtGFP. The transformants were obtained by ATMT. To minimize positional effect of reporter gene expression, at least 500 transformants on the membrane were collected to create a mixed population of transformants, which were cultured and assayed for the strength of GFP fluorescence. The name of the promoter refers to the nucleotide position from the translational start codon. The highest normalized green fluorescence intensity is set at 100% and results are average of triplicates. Osmotic: 1 M KCl; carbon: carbon source shift from glucose to glycerol; cold: 4 °C; heat: 37 °C
Application of RtGPD1 promoter for genetic transformation
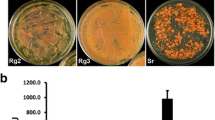
To confirm that RtGPD1 promoter is able to drive the expression of selection markers to enable dominant selection for transformants, hygromycin resistance gene hpt was fused with the 795-bp and 176-bp RtGPD1 promoters. When transformed to U. maydis via ATMT, the 795-bp version of RtGPD1 promoter showed comparable transformation efficiency (∼1,000 transformants per 106 U. maydis receptor cells) to the native Umgpd1 promoter whereas the 176-bp version promoter yielded about 20% as many transformants as the 795-bp version (Supplementary Material Fig. S5). However, no transformants could be obtained in R. toruloides in repeated trials although false positives were unusually high as confirmed by colony PCR and Southern blot analysis (data not shown). After realizing the strong codon bias in RtGPD1 and a number of other genes listed in the GenBank, a codon-optimized hygromycin resistance gene (hpt-3) based on the codon usage bias of R. toruloides was synthesized and fused with the 1,429-bp, 795-bp, and 176-bp RtGPD1 promoters as well as the 595-bp Umgpd1 promoter. As expected, transformants were successfully obtained using the new T-DNA constructs (Fig. 6). The 1,429-bp version showed the highest transformation efficiency with ∼1,000 transformants per plate, while the 795-bp and 179-bp versions yielded ∼400 and ∼200 transformants per plate, respectively (Fig. 6). Southern blot analysis confirmed the T-DNAs were integrated into the genome (Supplementary Fig. S6).
ATMT of R. toruloides using truncated GPD1 promoters from R. toruloides (vector series pEC3Pxxx-HPT3). The codon-optimized hygromycin resistance gene (hpt-3) is driven by R. toruloides GPD1 promoters of various lengths (labeled as 1429, 795, and 176, respectively), and the 595-bp U. maydis gpd1 promoter (Um-595)
Discussion
Similar to Agaricomycotina and Ustilaginomycotina (Harmsen et al. 1992; Kilaru and Kües 2005), Pucciniomycotina GPDs are highly variable in the numbers and positions of their introns, ranging from 6 to 11 (Fig. 2b). The different Pucciniomycotina GPD introns are between 68 and 110 nt long (average values calculated from introns at conserved positions; Table 3), which is similar to the other basidiomycetous counterparts (49–110 nt on average) excluding that of U. maydis (Supplementary Materials Table S1). In Pucciniomycotina, GPD introns of Rhodosporidium and Rhodotorula genera are larger (103–110 nt on average) than those of Puccinia and Sporobolomyces genera (68–76 nt on average; Table 3). The intron positions at the 5′ part of GPDs are more conserved among the six Pucciniomycotina species examined (Fig. 2b). Two intron positions (the first and third) are conserved among the four species in the Microbotryomycetes class (R. toruloides, R. glutinis, R. graminis, and S. roseus). The third, fourth, and fifth introns are shared by all species tested except S. roseus and R. graminis, which has lost the fourth and fifth intron found in R. toruloides, respectively (Fig. 2b). Meanwhile, the GPD genes of the Pucciniomycotina class (Puccinia triticina and Puccinia graminis) lack the first intron and their ORFs do not have the codons for the first 6 aa that are present in Microbotryomycetes. Altogether, the intron data suggest a common evolutionary origin between the GPD genes in Microbotryomycetes and Pucciniomycetes.
The first intron adjacent to the start codon is essential for gene expression in Agaricomycotina GPDs (Burns et al. 2005; Ma et al. 2001). However, this was not the case in Umgpd1 (Ji et al. 2010) nor in RtGPD1 (Figs. 4, 5, and Supplementary Material Fig. S4). It is surprising that a 176-bp fragment immediately upstream of the translational start codon retained 42% promoter activity in U. maydis and 17.4% in R. toruloides compared with the 795-bp fragment (Fig. 4b, c), and remained responsive to various stress stimuli (Fig. 5a, b). Similar results have been reported in a few other species, e.g., Schizophyllum commune (130 bp) (Schuren and Wessels 1994), rat (154 bp) (Tsuchiya et al. 2004), and A. thaliana (176 bp) (Chan et al. 2001). The short promoter is more convenient to use although the strength is considerably compromised. The region between −612 and −441 of RtGPD1 appeared to contain a second strong enhancer of transcription as the GFP intensity was more than doubled. This is consistent with the prediction of a CAAT box at −461 and a potential enhancer element (P550) located from −550 to −500 (Fig. 4a).
The draft genome sequence of Rhodotorula graminis WP1 (http://genome.jgi-psf.org/Rhoba1_1/Rhoba1_1.home.html) and genome scaffolds of R. glutinis ATCC 204091 (GenBank accession no. GL989638.1) have been released recently. A comparison of the promoter sequences revealed that R. graminis WP1 homolog shares only 52.2% identity with the RtGPD1, which is comparable to the 48.1% identity between promoters of Umgpd1 and RtGPD1, whereas the promoter sequences are identical between R. glutinis ATCC 204091 and R. toruloides ATCC 10657. Interestingly, a 1.0-kb promoter of R. graminis GPD1 could also drive strong expression of RtGFP in R. toruloides (data not shown).
Transformation of Pucciniomycotina has been very difficult. Only auxotrophic selection has been succeeded, e.g., in P. triticina (Webb et al. 2006), Melampsora lini (Lawrence et al. 2010), and S. roseus (Ianiri et al. 2011). Here, the combination of strong promoters and the codon-optimized antibiotic resistance gene (hpt-3) has led to a breakthrough in the use of dominant selection in Pucciniomycotina as exemplified in R. toruloides. The vector has been successfully used for ATMT of S. roseus (data not shown). It is possible that the promoters, selection markers, and transformation method reported herein will have much broader applications in the biotechnological applications in other species in Pucciniomycotina.
References
Brachmann A, König J, Julius C, Feldbrügge M (2004) A reverse genetic approach for generating gene replacement mutants in Ustilago maydis. Mol Genet Genomics 272:216–226
Brefort T, Doehlemann G, Mendoza-Mendoza A, Reissmann S, Djamei A, Kahmann R (2009) Ustilago maydis as a pathogen. Annu Rev Phytopathol 47:423–445
Burns C, Gregory KE, Kirby M, Cheung MK, Riquelme M, Elliott TJ, Challen MP, Bailey A, Foster GD (2005) Efficient GFP expression in the mushrooms Agaricus bisporus and Coprinus cinereus requires introns. Fungal Genet Biol 42:191–199
Cangelosi GA, Best EA, Martinetti G, Nester EW (1991) Genetic analysis of Agrobacterium. Methods Enzymol 204:384–397
Chan CS, Guo L, Shih MC (2001) Promoter analysis of the nuclear gene encoding the chloroplast glyceraldehyde-3-phosphate dehydrogenase B subunit of Arabidopsis thaliana. Plant Mol Biol 46:131–141
Halaouli S, Record E, Casalot L, Hamdi M, Sigoillot JC, Asther M, Lomascolo A (2006) Cloning and characterization of a tyrosinase gene from the white-rot fungus Pycnoporus sanguineus, and overproduction of the recombinant protein in Aspergillus niger. Appl Microbiol Biotechnol 70:580–589
Harmsen MC, Schuren FH, Moukha SM, van Zuilen CM, Punt PJ, Wessels JG (1992) Sequence analysis of the glyceraldehyde-3-phosphate dehydrogenase genes from the basidiomycetes Schizophyllum commune, Phanerochaete chrysosporium and Agaricus bisporus. Curr Genet 22:447–454
Harris JI, Waters M (1976) Glyceraldehyde 3 phosphate dehydrogenase. In: Boyer PD (ed) The enzymes. Academic Press, New York, pp 1–49
Holland MJ, Holland JP (1978) Isolation and identification of yeast messenger ribonucleic acids coding for enolase, glyceraldehyde-3-phosphate dehydrogenase, and phosphoglycerate kinase. Biochemistry 17:4900–4907
Hooykaas PJJ, Roobol C, Schilperoort RA (1979) Regulation of the transfer of Ti-plasmids of Agrobacterium tumefaciens. J Gen Microbiol 110:99–109
Ianiri G, Wright SA, Castoria R, Idnurm A (2011) Development of resources for the analysis of gene function in Pucciniomycotina red yeasts. Fungal Genet Biol 48:685–695
Ji L, Jiang ZD, Liu Y, Koh CM, Zhang LH (2010) A simplified and efficient method for transformation and gene tagging of Ustilago maydis using frozen cells. Fungal Genet Biol 47:279–287
Kilaru S, Kües U (2005) Comparison of gpd genes and their protein products in basidiomycetes. Fungal Genet Newsl 52:18–23
Kilaru S, Hoegger P, Majcherczyk A, Burns C, Shishido K, Bailey A, Foster G, Kües U (2006) Expression of laccase gene lcc1 in Coprinopsis cinerea under control of various basidiomycetous promoters. Appl Microbiol Biotechnol 71:200–210
Lawrence GJ, Dodds PN, Ellis JG (2010) Transformation of the flax rust fungus, Melampsora lini: selection via silencing of an avirulence gene. Plant J 61:364–369
Lazo GR, Stein PA, Ludwig RA (1991) A DNA transformation-competent Arabidopsis genomic library in Agrobacterium. Nat Biotechnol 9:963–967
Levitt N, Briggs D, Gil A, Proudfoot NJ (1989) Definition of an efficient synthetic poly(A) site. Genes Dev 3:1019–1025
Liu H, Zhao X, Wang F, Li Y, Jiang X, Ye M, Zhao ZK, Zou H (2009) Comparative proteomic analysis of Rhodosporidium toruloides during lipid accumulation. Yeast 26:553–566
Liu Y, Koh CM, Sun L, Ji L (2011) Tartronate semialdehyde reductase defines a novel rate-limiting step in assimilation and bioconversion of glycerol in Ustilago maydis. PLoS One 6:e16438
Loubradou G, Brachmann A, Feldbrügge M, Kahmann R (2001) A homologue of the transcriptional repressor Ssn6p antagonizes cAMP signalling in Ustilago maydis. Mol Microbiol 40:719–730
Ma B, Mayfield MB, Gold MH (2001) The green fluorescent protein gene functions as a reporter of gene expression in Phanerochaete chrysosporium. Appl Environ Microbiol 67:948–955
Nitta Y, Miyazaki Y, Nakamura M, Iimura Y, Shishido K, Kajita S, Morohoshi N (2004) Molecular cloning of the promoter region of the glyceraldehyde-3-phosphate dehydrogenase gene that contributes to the construction of a new transformation system in Coriolus versicolor. Mycoscience 45:131–136
Ochman H, Gerber AS, Hartl DL (1988) Genetic applications of an inverse polymerase chain reaction. Genetics 120:621–623
Punt PJ, Dingemanse MA, Kuyvenhoven A, Soede RD, Pouwels PH, van den Hondel CA (1990) Functional elements in the promoter region of the Aspergillus nidulans gpdA gene encoding glyceraldehyde-3-phosphate dehydrogenase. Gene 93:101–109
Redkar RJ, Herzog RW, Singh NK (1998) Transcriptional activation of the Aspergillus nidulans gpdA promoter by osmotic signals. Appl Environ Microbiol 64:2229–2231
Sampaio JP, Gadanho M, Bauer R, Weiß M (2003) Taxonomic studies in the Microbotryomycetidae: Leucosporidium golubevii sp. nov., Leucosporidiella gen. nov. and the new orders Leucosporidiales and Sporidiobolales. Mycol Prog 2:53–68
Schuren FH, Wessels JG (1994) Highly-efficient transformation of the homobasidiomycete Schizophyllum commune to phleomycin resistance. Curr Genet 26:179–183
Smith TL, Leong SA (1990) Isolation and characterization of a Ustilago maydis glyceraldehyde-3-phosphate dehydrogenase-encoding gene. Gene 93:111–117
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Tristan C, Shahani N, Sedlak TW, Sawa A (2011) The diverse functions of GAPDH: views from different subcellular compartments. Cell Signal 23:317–323
Tsuchiya K, Tajima H, Yamada M, Takahashi H, Kuwae T, Sunaga K, Katsube N, Ishitani R (2004) Disclosure of a pro-apoptotic glyceraldehyde-3-phosphate dehydrogenase promoter: anti-dementia drugs depress its activation in apoptosis. Life Sci 74:3245–3258
Tully M, Gilbert HJ (1985) Transformation of Rhodosporidium toruloides. Gene 36:235–240
Turcotte G, Kosaric N (1988) Biosynthesis of lipids by Rhodosporidium toruloides ATCC 10788. J Biotechnol 8:221–237
Vassileva A, Chugh DA, Swaminathan S, Khanna N (2001) Expression of hepatitis B surface antigen in the methylotrophic yeast Pichia pastoris using the GAP promoter. J Biotechnol 88:21–35
Webb CA, Szabo LJ, Bakkeren G, Garry C, Staples RC, Eversmeyer M, Fellers JP (2006) Transient expression and insertional mutagenesis of Puccinia triticina using biolistics. Funct Integr Genomics 6:250–260
Wolff AM, Arnau J (2002) Cloning of glyceraldehyde-3-phosphate dehydrogenase-encoding genes in Mucor circinelloides (syn. racemosus) and use of the gpd1 promoter for recombinant protein production. Fungal Genet Biol 35:21–29
Zhao X, Wu S, Hu C, Wang Q, Hua Y, Zhao ZK (2010) Lipid production from Jerusalem artichoke by Rhodosporidium toruloides Y4. J Ind Microbiol Biotechnol 37:581–585
Acknowledgments
This work was financially supported by the Singapore Economic Development Board and Temasek Trust. We thank especially Prof. Dr. Ursula Kües, Molecular Wood Biotechnology and Technical Mycology, Büsgen-Institute, Georg-August-University Göttingen, Göttingen, Germany, for many helpful suggestions for this manuscript, and Prof. Dr. Michael Feldbrügge, Institute for Microbiology, Heinrich-Heine-University Düsseldorf, Düsseldorf, Germany, for the gift of plasmid pMF2-3c.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOCX 6.41 mb)
Rights and permissions
About this article
Cite this article
Liu, Y., Koh, C.M.J., Sun, L. et al. Characterization of glyceraldehyde-3-phosphate dehydrogenase gene RtGPD1 and development of genetic transformation method by dominant selection in oleaginous yeast Rhodosporidium toruloides . Appl Microbiol Biotechnol 97, 719–729 (2013). https://doi.org/10.1007/s00253-012-4223-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-012-4223-9