Abstract
The lignin-degrading, biopulping white-rot fungus Physisporinus rivulosus secretes several laccases of distinct features such as thermostability, extremely low pH optima and thermal activation for oxidation of phenolic substrates. Here we describe the cloning, heterologous expression and structural and enzymatic characterisation of two previously undescribed P. rivulosus laccases. The laccase cDNAs were expressed in the methylotrophic yeast Pichia pastoris either with the native or with Saccharomyces cerevisiae α-factor signal peptide. The specific activity of rLac1 and rLac2 was 5 and 0.3 μkat/μg, respectively. However, mutation of the last amino acid in the rLac2 increased the specific laccase activity by over 50-fold. The recombinant rLac1 and rLac2 enzymes demonstrated low pH optima with both 2,6-dimethoxyphenol (2,6-DMP) and 2,2′-azino-bis(3-ethylbenzathiazoline-6-sulfonate). Both recombinant laccases showed moderate thermotolerance and thermal activation at +60 °C was detected with rLac1. By homology modelling, it was deduced that Lac1 and Lac2 enzymes demonstrate structural similarity with the Trametes versicolor and Trametes trogii laccase crystal structures. Comparison of the protein architecture at the reducing substrate-binding pocket near the T1-Cu site indicated the presence of five amino acid substitutions in the structural models of Lac1 and Lac2. These data add up to our previous reports on laccase production by P. rivulosus during biopulping and growth on Norway spruce. Heterologous expression of the novel Lac1 and Lac2 isoenzymes in P. pastoris enables the detailed study of their properties and the evaluation of their potential as oxidative biocatalysts for conversion of wood lignin, lignin-like compounds and soil-polluting xenobiotics.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Laccases (EC 1.10.3.2) are multicopper oxidases that catalyse the one-electron oxidation of a variety of phenolic compounds, aromatic amines and low-redox potential substrates with the concomitant reduction of O2 to H2O. Laccases are widely distributed in nature, occurring in fungi as well as in plants, insects, bacteria and archaea (Hoegger et al. 2006; Lundell et al. 2010; Giardina et al. 2010; Uthandi et al. 2010). Although fungal laccases have been repeatedly associated with the depolymerisation of lignin, their actual role in lignin and wood biodegradation is uncertain. When fungal laccases are combined with oxidation mediator substrate compounds, their enzymatic capacities are improved (Bourbonnais et al. 1997; Call and Mücke 1997; Koroleva et al. 2001; Martinez et al. 2004; Hatakka and Hammel 2011). Multiple laccase genes are recognized in the genomes of lignocellulose-decaying basidiomycetes as well as in mycorrhizal and plant-pathogenic fungi and in ascomycetous filamentous fungi (Kilaru et al. 2006; Courty et al. 2009; Martinez et al. 2008).
One of the most studied lignin-degrading white-rot fungi, Phanerochaete chrysosporium, was reported to be deficient in laccase-encoding genes (Martinez et al. 2004; Kersten and Cullen 2007), while one or several laccase isoenzymes are usually expressed by wood-decaying fungi (Hakala et al. 2005; Mäkelä et al. 2006; Saparrat et al. 2010). Due to the broad substrate specificity and oxidation capacity, there is potential to use laccases in industrial and environmental applications (Hildén et al. 2009; Tuomela and Hatakka 2011). For these applications, the research focus is on the molecular characterization of fungal laccases as well as on the improvement of the production levels by recombinant expression.
Physisporinus rivulosus (Berk. & Curt.) Ryvarden (synonym Ceriporiopsis rivulosa) is a selectively lignin-degrading, white-rot basidiomycete species, which is able to grow in a wide temperature range and efficiently decompose softwood lignin (Hakala et al. 2004). Pre-treatment of Norway spruce chips by cultivation of P. rivulosus strain T241i resulted in a 20 % decrease in the consumption of energy in mechanical pulping of wood (Hatakka et al. 2003). Under these biopulping conditions and with growth on spruce wood, P. rivulosus secreted, in addition to manganese peroxidases (MnPs) and oxalic acid, several laccases as multiple isoenzymes with low pI values (pI 3.5 to 4.8) (Hakala et al. 2005). Two of the isoenzymes demonstrated extremely low pH optima (below pH 3.5) for oxidation of phenolic substrate compounds with simultaneous thermostability and activation at elevated temperatures (Hildén et al. 2007). In defined liquid media, however, production of laccases by P. rivulosus is limited without lignocellulose, such as spruce wood sawdust as the sole carbon source (Hildén et al. 2007).
In the current study, we describe gene characteristics and heterologous expression in Pichia pastoris of two laccase-encoding genes (lac1 and lac2) of P. rivulosus T241i. The 3-D protein homology models of the predicted Lac1 and Lac2 enzymes illustrate a typical four copper laccase fold but indicate differences in protein topology at the T1-Cu and substrate-binding site (Bertrand et al. 2002; Piontek et al. 2002). The recombinant laccases demonstrate low pH optima (pH 3.5) and moderate thermotolerance for recombinant Lac1 with the laccase substrates 2,6-dimethoxyphenol (2,6-DMP) and ABTS (2,2′-azino-bis(3-ethylbenzathiazoline-6-sulfonate). Due to their unique biochemical properties, the recombinant P. rivulosus laccases may be utilized in biocatalytic applications, leading to conversion of lignin-like phenolic compounds, dyes or environmentally hazardous xenobiotics, under increased acidity or higher process temperature conditions. Via successful heterologous expression in P. pastoris, prominence for detailed molecular and functional characterisation of the low pH and thermotolerant laccases can be envisioned.
Materials and methods
Microbial strains
The white-rot fungus P. rivulosus (synonym C. rivulosa) strain T241i (FBCC949, DSM14618) is derived from a fruiting body found on a burned spruce wood log in a forest fire area in southern Finland (Hakala et al. 2004). The fungus is maintained on 2 % (wt/vol) malt extract agar (1.5 % wt/vol, Biokar, Beauvais, France) and deposited in the Fungal Biotechnology Culture Collection, University of Helsinki, Finland (fbcc@helsinki.fi). P. pastoris strain GS115 was purchased from Invitrogen (Gibco-BRL, Gaithersburg, USA).
Fungal culture conditions
The production of P. rivulosus laccase activity was followed in yeast extract–peptone (YEP) complex liquid medium and in wheat or rye bran-containing media. YEP medium contained 0.2 % (wt/vol) yeast extract (Oxoid Ltd, Basingstoke, UK), 0.5 % peptone (Oxoid Ltd) and 0.5 % (wt/vol) glucose (Fisher Scientific Ltd, Loughborough, UK) in distilled water. Bran media contained 2 % (wt/vol) wheat bran (Neal’s Yard, Burton upon Trent, UK) or 2 % (wt/vol) rye bran (Raisio, Finland) in distilled water. The pH was adjusted to 4.5 prior autoclaving in all media used. Three parallel flasks, each containing 100 ml of cultivation medium, were inoculated with three agar plugs (10 mm in diameter) covered with 14-day grown P. rivulosus mycelium on 2 % (wt/vol) malt extract agar plates. The cultures were incubated stationary at 28 °C for 21 days. Aliquots of 1 ml were taken twice a week for laccase activity measurement.
Assay of laccase activity and steady-state kinetic measurements
Laccase activity was determined spectrophotometrically at 420 and 476 nm by following the oxidation of ABTS (Sigma, St. Louis, USA) and 2,6-DMP (Aldrich, St. Louis, USA), respectively, at pH 3.0 in 50 mM sodium malonate buffer. A Shimadzu PharmaSpec UV-1700 spectrophotometer was used in all the measurements. Laccase activity is expressed as μkat l−1 (10-6 mol s−1 l−1) of the specific product formed upon oxidation of each substrate. Steady-state kinetic parameters (K m and k cat) were determined by using 2,6-DMP as reducing substrate. Triplicate reactions were initiated by the addition of 0.001–2.0 mM of 2,6-DMP. The K m and V max values were calculated based on Lineweaver–Burk plots and the k cat values were calculated as V max/rLac protein concentration. The protein concentrations were measured by BCA Protein Assay Reagent (Pierce, Thermo Fisher Scientific, Rockford, IL, USA) using bovine serum albumin as standard to determine specific laccase activities.
Isolation of nucleic acids and cDNA synthesis
Mycelia from the 7-, 14- and 21-day cultures were filtered through Miracloth (Calbiochem, Darmstadt, Germany), frozen to −80 °C and ground with a mortar and pestle in liquid N2. DNA was extracted by using a N-cetyl-N,N,N-trimethyl-ammonium bromide-based buffer and purified with chloroform (Hildén et al. 2005). Total RNA was extracted by Trizol (Invitrogen) following the manufacturer’s instructions. RNA samples were treated with RNase-free DNase (1 U/μg; Fermentas, Vilnius, Lithuania) at 37 °C for 90 min.
Prior to cDNA synthesis, the total RNA was used as a template for PCR to ensure the absence of genomic DNA in the RNA samples. Synthesis of cDNA from 1 μg of total RNA and rapid amplification of cDNA ends (RACE) and genome-walking PCR were performed as described previously (Hildén et al. 2005; Mäkelä et al. 2006).
Amplification and cloning of laccase cDNAs and genes
P. rivulosus DNA was used as a template for the amplification of a 230- and 231-bp fragment of lac1 and lac2 genes, respectively, with degenerate laccase primers (D’Souza et al. 1996). The 25-μl PCR reaction contained 0.5 μl DNA template, 0.3 mM dNTP mixture (Fermentas), 0.5 μM 5′ and 3′ primers, 1× Phusion HF buffer (Finnzymes, Espoo, Finland), 3 % DMSO and 0.6 U Phusion Hot Start DNA polymerase (Finnzymes). PCR was performed with initial denaturation at 98 °C for 30 s, then 40 cycles of denaturation at 98 °C for 10 s, annealing at 57 °C for 30 s, elongation at 72 °C for 30 s and final extension at 72 °C for 5 min.
A Smart Race cDNA Amplification kit (Clontech, Palo Alto, CA, USA) was used for amplification of the lac1 cDNA ends. The 5′ end of the transcript was amplified with the universal primer mix (UPM, Clontech) and a degenerate antisense laccase primer (Mäkelä et al. 2006) with cDNA extracted from P. rivulosus which was cultivated on wheat bran. In the nested PCR, an inner gene-specific antisense primer of lac1 (CAGAAGGTACCGGCTTGGT) was used with a nested universal primer (NUP, Clontech). The 3′ end of lac1 was amplified with a gene-specific sense primer (TTTGTGAACCAATGTCCGA) and UPM. In the second amplification round, nested gene-specific sense primer (CCCGAACTGGGTATTTTCGATCG) and NUP were used (Hildén et al. 2005). The full-length open reading frame (ORF) was amplified with the primer pair: sense CAAGCATGGCTCGCTTCC and antisense CACGTATCAGAAGGGTCAAGC.
The 5′ and 3′ ends of lac2 gene were amplified by the inverse PCR method (Halaouli et al. 2006). One microgram of DNA from P. rivulosus was digested with EcoRI or HindIII (New England Biolabs, Hitchin, UK). The digested DNA was circularised with 5 U of T4 ligase (New England Biolabs) and used as PCR template. The gene-specific primer pair (sense: GACTTCAGCGTCCCGGACCA, antisense: GACAAATGCGGGACCA) was used in the first round of PCR. In the second PCR round, the nested gene-specific primer pair (sense: ACACTCGCTGATATTGCGCACGA, antisense: CGATTGTCGTAGACTCGAGCA) was used. The full-length ORF was amplified with the primer pair: sense ATGCCTAACTTCCTGTCG and antisense TCACAGCTCATCGGAGGG. PCR reactions were conducted as described earlier.
The full-length ORF of laccase cDNA and gene fragments were cloned into the pJET1.2/blunt vector (Fermentas) and sequenced.
Protein characteristics and 3-D homology modelling
Protein sequence properties were predicted from the translated ORF sequences using Swissprot and EMBL web tools, the N-glycolysation software (http://www.cbs.dtu.dk/services/NetNGlyc/) and secretion and targeting signal peptide prediction (http://www.cbs.dtu.dk/services/SignalP and http://www.cbs.dtu.dk/services/TargetP/). Modelling of protein structures was performed with Modeller v9.4 software (Sali et al. 1995). Deduced crystal structure models of laccase from Trametes versicolor (PDB accession 1KYA and 1GYC) that showed the best homology to P. rivulosus Lac1 (70 % identity) were used as templates for homology modelling. Laccase 3-D structures from Trametes trogii (PDB accession 2HRH and 2HRG) were used as templates for P. rivulosus Lac2, showing 68 % identity. Verification of the structural models was performed with ProCheck v.3.4 (Laskowski et al. 1993) and ProSa2003 v.4.0 (Sippl 1993). The root mean square deviation between the main chain atoms of the modelled structures and templates was calculated to estimate the reliability of the 3-D models. Substrate-free models for Lac1 and Lac2 were overlaid onto the crystal structure of 1KYA containing one molecule of 2,5-xylidine, and 2HRG containing one molecule of p-methylbenzoate as a ligand in the reducing substrate-binding site, and using GRASP2 (Petrey and Honig 2003) and PyMol v.0.99 (DeLano 2002) software, which was also used for the calculation of molecular electrostatic potentials and visualization of the 3-D structural models.
Heterologous expression of P. rivulosus laccases
To construct plasmids for the heterologous expression in P. pastoris, the lac1 and lac2 encoding cDNAs were cloned into pPICZB together with their native N-terminal signal peptide sequences and into pPICZαA (Invitrogen) downstream of the α-mating factor signal peptide sequence from Saccharomyces cerevisiae. Primer pairs lac1 sense: GTGCTGGAATTCTCAAGCATGGCTCGC and lac1 antisense: TAACTAGGCGGCCGCCGACGTATCAG and lac2 sense: AACGAGGAATTCAATAATGCCTAACTTC and lac2 antisense: GAAAGCTGGCGGCCGCGCACAGCTCATC were used to clone cDNAs with the native signal sequence. Sense primers lac1: GGCGCAGAATTCGCCATCGGG and lac2: GCTGAAGCTGAATTCGCGATCGGCCCT with antisense primers as described above were used to clone cDNAs without the native signal sequence. An additional antisense primer (GAAAGCTGGCGGCCGCGCACACCTCATC) was designed to amplify a 3′-modified lac2 cDNA encoding the variant Lac2-L497V with mutated C-terminus in order to avoid possible endoplasmic reticulum (ER) retention. The resulting expression constructs were confirmed by sequencing. Expression of inserts in both vectors is controlled by the methanol-inducible AOX1 promoter. Both expression vectors contain C-terminal peptides containing the c-myc epitope and a polyhistidine (6× His) tag for detection and purification of recombinant fusion proteins. The plasmid constructs were linearized by SacI digestion and transformed into P. pastoris GS115-competent cells by electroporation. The transformants were selected on zeocin (100 μg/ml) containing YEP plates. Several zeocin-resistant clones were randomly picked and spotted onto buffered minimal medium (BMM, Invitrogen; pH 6.0) 2 % (wt/vol) agar plates containing 0.004 % histidine, 4 × 10−5 % biotin, 0.5 % methanol, 0.3 mM CuSO4 and 0.2 mM ABTS (Li et al. 2007). The plates were incubated at 28 °C for 4 days. Positive P. pastoris transformants were identified by the appearance of a green zone around laccase-expressing colonies.
According to the intensity of the green zone, ten of the positive transformant colonies were selected for laccase expression. The selected transformants were cultivated in liquid BMG medium (Invitrogen) containing 1 % glycerol at 28 °C in a rotary incubator until the OD600 was approximately 6–8. The cultures were centrifuged (1,200 g at 4 °C for 5 min) and washed with phosphate-buffered saline. The samples were re-centrifuged and the cell pellets were diluted with BMM medium supplemented with 0.3 mM CuSO4 and 0.8 % (wt/vol) alanine to an OD600 of 1 (Soden et al. 2002). Transformants were cultivated at 20 °C, with shaking at 200 rpm, and 0.5 % (vol/vol) methanol was added daily to maintain the induced production of the recombinant laccase. Cultivation time was 5–7 days and extracellular laccase activity was followed daily using the ABTS assay. The highest laccase activity-producing transformants of rLac1 and rLac2 were selected for the scale-up cultivations.
Purification of recombinant laccases
The culture fluids of P. pastoris were harvested on days 5–6 and ca. 2-l batches were concentrated using Spin-X UF tubes (Corning, Loughborough, UK) with 30-kDa cutoff membrane filter at 4 °C. The extracellular protein concentrates were stored at −20 °C. The concentrates were purified by Ni-affinity chromatography using the ÄKTA Explorer apparatus (GE Healthcare, Uppsala, Sweden). The concentrates were dialyzed against 20 mM phosphate buffer (pH 7) containing 500 mM NaCl and 20 mM imidazole (Sigma-Aldrich) with Vivaspin 2 centrifugal concentrators (10 kDa cutoff, Sartorius Stedim Biotech GmbH, Göttingen, Germany). Samples were fed into HisTrap™ FF Crude column (GE Healthcare) and eluted with 500 mM NaCl and 500 mM imidazole in phosphate buffer with stepwise gradient. The absorbance of the effluent was followed at 280, 405 and 610 nm.
Characterization of recombinant laccases
The purity of the enzymes was analysed by SDS-PAGE (10 % MiniProtean TGX gels; Bio-Rad, Hercules, USA). Ten micrograms of purified proteins was used for deglycosylation treatment by using protein deglycosylation mix (New England BioLabs). The proteins were visualized by PageBlue (Fermentas) gel staining reagent. Color Plus Protein Ladder (New England BioLabs) was used for the determination of the molecular mass.
The pH optima of the rLac1 and rLac2–L497V were estimated by measuring the oxidation of 2,6-DMP and ABTS as described earlier and the oxidation of syringaldazine (SGZ; 4-hydroxy-3,5-dimethoxybenzaldehyde; Apin Chemicals Ltd, Oxon, UK) at 525 nm. SGZ was dissolved in ethanol and other substrates in distilled water. The pH in the reactions was adjusted with sodium malonate buffer (50 mM) from pH 2.0 to 6.0 and with MES buffer (50 mM) from pH 6.0 to 7.5.
The thermotolerance of the recombinant laccases was determined over the range from 40 to 80 °C at the optimum pH by incubating the enzymes in 100-μl aliquots for 1 to 60 min. After incubation, the tubes were chilled on ice and the residual laccase activity was measured with 2,6-DMP as described earlier. Laccase activity at 25 °C was marked as 100 % for each substrate compound.
Nucleotide sequence accession numbers
The cDNA sequences are deposited at GenBank with accession numbers JQ027726 and JQ027727 for lac1 and lac2, respectively, and the internal transcribed spacer region with accession number JQ027728.
Results
Characterization of Lac1 and Lac2 polypeptides and respective genes
Two laccase-encoding genes and cDNAs of P. rivulosus were cloned and sequenced. As predicted from the full-length cDNA clones, lac1 and lac2 encode 519- and 518-aa-long polypeptides, respectively. The ORF of lac1 gene is interrupted by 11 introns of 51-93 nt in length, whereas the lac2 gene contains 12 introns of 50–68 nt in length. The exon–intron junctions of the laccase genes are shown in Fig. 1. The intron–exon organization of P. rivulosus lac1 and lac2 genes and the translated amino acid sequences of Lac1 and Lac2 showed the highest pairwise identity (86 and 91 % , respectively) to the Lcs-1 (gene lcs-1, AF053472) and the gene and protein model #137686 (http://genome.jgi-psf.org/Cersu1/Cersu1.home.html), respectively, from the taxonomically nearest-related polypore species Ceriporiopsis (Gelatoporia) subvermispora.
P. rivulosus laccases. Alignment of the predicted mature translated Lac1 and Lac2 amino acid sequences with their homologues from Cerioporiopsis subvermispora (Cs-lcs1 sequence accession AAC97074 and Cs-137686 protein model from the genome site at http://genome.jgi-psf.org/Cersu1/Cersu1.home.html, respectively). Boxed regions: L1–L4, laccase signature sequences containing the conserved copper atom ligand amino acid residues (Kumar et al. 2003); I–IV, variable loop sequence sites (Larrondo et al. 2003a, b). Exon–intron junctions in corresponding positions in the laccase genes are shaded and potential asparagines for N-glycosylation are underlined
The Lac1 and Lac2 protein sequences are preceded by 21-aa-long putative N-terminal leader peptides, and they show 74 % amino acid pairwise identity. The calculated pI values of predicted mature Lac1 and Lac2 are 4.5 and 4.1, respectively. The four conserved laccase copper-binding regions are conserved in both P. rivulosus laccases. Lac1 contains six potential N-glycosylation sites, while eight may be deduced in Lac2. Interestingly, the tripeptide (-DEL-) present in the C-terminal end of Lac2 may be an indication of a signal for protein retention in the ER.
Homology modelling of Lac1 and Lac2
The theoretical structural models for Lac1 and Lac2 were constructed by protein homology modelling. The 3-D models predict a typical protein fold for laccases and consist of three β-barrel cupredoxin-like domains, which form a globular structure (Fig. 2). Both enzymes contain a cluster of four copper atoms, which form the laccase catalytic site. In both laccases, the copper atom of the T1 site is coordinated by two histidines, cysteine and isoleucine (His396, His457, Cys452, Ile454 for Lac1 and His395, His456, Cys451, Ile453 for Lac2) and the three copper atoms of the T2/T3 site by eight histidines (His66, His68, His111, His113, His399, His401, His451, His453 for Lac1 and His64, His66, His109, His111, His398, His400, His450, His452 for Lac2). The T1 copper is linked with the T2/T3 copper cluster by the conserved His–Cys–His tripeptide (His451–Cys452–His453 for Lac1 and His450–Cys451–His452 for Lac2).
The modelled 3-D structures of Lac1 and Lac2 were superimposed near the vicinity of the T1 copper on the corresponding protein sites of the structurally characterized laccases from T. versicolor (PDB accession 1KYA) and T. trogii (PDB accession 2HRG) for determination of the amino acid residues playing a role in binding of the reducing organic substrates in these enzymes. The 3-D crystal structures 1KYA and 2HRG both contain a substrate-like ligand molecule (1KYA, 2,5-xylidine (2,5-dimethylaniline); 2HRG, p-methylbenzoate) (Fig. 3). At this substrate-binding site, the amino acid residues are in close proximity (maximum 5 Å distance) to the substrate molecules in 1KYA and 2HRG. The respective sites in the Lac1 and Lac2 models demonstrate four amino acid residues (Phe, His, Ile and Asp) that are highly conserved for all the four laccases.
Superimposition of active site amino acid residues of Lac1 (green) and Lac2 (red) onto the (a) 1KYA (light purple) and (b) 2HRG (light purple). The co-crystallized ligand (substrate analogues) 2,5-xylidine and p-methylbenzoate are illustrated (carbon skeleton in silver, nitrogen atoms in blue, oxygen atoms in red) in the figures
The reducing substrate-binding cavity is comprised of 17 and 18 amino acid residues in Lac1 and Lac2, respectively (Fig. 4). A comparison of the cavities showed that 12 amino acids were conserved, whereas the other five residues were substituted as follows: Pro209/Glu207, Asn266/Asp264, Asn267/Leu265, Leu340/Phe338 and Ile392/Val390 for Lac1/Lac2, respectively. Furthermore, an insertion of Gly392 was present in the substrate-binding site region of Lac2. The shape and volume of the substrate-binding cavity as well as the surface electrostatic charge vary between Lac1 and Lac2 (Fig. 4). The cavity of Lac2 is less bulky and more negatively charged than is observed in Lac1. In both laccases, the catalytically essential and conserved histidine (His457/His456 for Lac1/Lac2) and aspartic acid (Asp208/Asp206 for Lac1/Lac2) residues are located at the bottom of the cavity, with their side chain groups exposed to the surface of the enzymes.
Physico-chemical properties and kinetic parameters of the recombinant laccases
The Lac1 and Lac2 encoding genes were expressed in P. pastoris under the control of the tightly regulated AOX1 promoter, which was induced for expression by addition of methanol. Among the positive transformants, extracellular ABTS oxidation was observed with recombinant laccase constructs, harnessing either their native N-terminal signal peptides (rLac1/pPICZ and rLac2/pPICZ) or the α-factor signal peptide from S. cerevisiae (rLac1/pPICαZ and rLac2/pPICαZ). However, the latter constructs resulted in faster ABTS oxidation rates and demonstrated slightly higher oxidative activities within liquid cultures and were thereby selected for the scale-up studies (Fig. 5). For the rLac1/pPICZ, the specific laccase activity reached a maximum of 3.14 μkat/μg after 7 days of liquid expression cultures, whereas for rLac1/pPICZα the maximum of 4.9 μkat/μg was reached already after 2 days. For rLac2, both signal peptide variants, rLac2/pPICZ and rLac2/pPICZα, showed negligible laccase activity of 0.4 and 0.3 μkat/μg, respectively. In the liquid cultures, the transformant rLac2–L497V, which produced the one codon mutated variant, showed higher extracellular ABTS oxidation activities (16.5 μkat/μg) than either of the wild-type rLac2 variants and was thus selected for the enzyme characterisation studies (Fig. 5).
Time course of specific laccase activity in the extracellular medium of P. pastoris cultures producing recombinant laccases. Comparisons of rLac1 and rLac2 expression using vector with native (pPICZ) and α-factor secretion signal sequence (pPICZα). rLac1/pPICZ (open circle), rLac1/pPICZα (filled circle), rLac2/pPICZ (open square), rLac2/pPICZα (filled square) and rLac2–L497V /pPICZα (filled triangle). Activity was determined by ABTS oxidation. The mean values represent the specific laccase activities of three transformants of each type
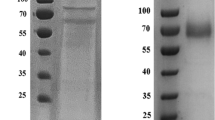
The molecular masses of the purified rLac1 and rLac2–L497V were 71 kDa, which was higher than the predicted masses of 54 and 53 kDa (Fig. 6). However, treatment of protein deglycosylation mix (PNGaseF, O-glycosidase, neuraminidase, β1-4 galactosidase, β-N-acetylglucosaminidase) showed that the molecular masses were not affected by deglycosylation.
SDS electrophoresis of purified laccases rLac1 and rLac2–L497V (a). Lane 1, rLac1; lane 2, protein standard; lane 3, rLac2–L497V; lane 4, P. pastoris culture extract. Deglycosylation of recombinant rLac1 and rLac2–L497V (b). Lane 1, rLac1; lane 2, rLac1 treated with deglycosylation mix; lane 3, protein mass marker; lane 4, rLac2–L497V; lane 5, rLac2–L497V treated with deglycosylation mix. The bands corresponding deglycosylation enzymes are marked. Approximately 10 μg of recombinant protein was applied to each lane
The rLac1 and rLac2–L497V recombinant laccases both showed acidic pH optima (around pH 3.5) for the oxidation of ABTS and 2,6-DMP (Fig. 7). With SGZ as reducing substrate, no oxidation activity was detected. The recombinant laccases were stable at temperatures below 50 °C (Fig. 8). The half-life of rLac1 for 2,6-DMP oxidation at pH 3.5 and at 60 °C was 60 min, whereas for rLac2–L497V a half-life of only 25 min at 60 °C was observed. Interestingly, rLac1 showed thermal activation since the 2,6-DMP oxidation rate after incubation for 10 min at 60 °C was increased up to 30 % of the original activity at room temperature prior to incubation. On the contrary, no such thermal activation was observed with rLac2–L497V enzyme.
Effect of pH on the activity of purified recombinant P. rivulosus laccases (rLac1 ABTS (filled square), rLac1 2,6-DMP (filled circle), rLac2 ABTS (open circle), rLac2 2,6-DMP (open square)). The measurements were carried out as duplicates and averages are plotted. The standard deviation did not exceed 1 %
Thermotolerance of the recombinant P. rivulosus rLac1 (a) and rLac2 (b) 40 °C (filled circle), 50 °C (filled diamond), 60 (filled square), 70 °C (filled triangle) and 80 °C (multiplication symbol) using 2,6-DMP as the substrate at pH 3.0. The measurements were carried out as duplicates and averages are plotted. The standard deviation did not exceed 1 %
The kinetic parameters (K m and k cat) were determined with 2,6-DMP as reducing substrate at pH 3.0. The V max of rLac1 and rLac2–L497V were 7 ± 0.01 and 57 ± 0.17 μkat/l, respectively. The K m values showed that rLac1 (39 ± 0.07 μM) has higher affinity towards 2,6-DMP than rLac2–L497V (206 ± 0.6 μM). The K cat values of rLac1 and rLac2–L497V were 5 ± 0.01/s and 34 ± 0.1/s, respectively. The catalytic efficiencies expressed as k cat/K m were 1.3 × 105 μM−1 s−1 for rLac1 and 1.6 × 105 μM−1 s−1 for rLac2-L497V.
Discussion
In this study, we cloned and heterologously expressed two laccases of the biopulping white-rot basidiomycete P. rivulosus. Biochemical characteristics showed that these novel laccases are acidic, thermotolerant and thermoactivated. The structural features were analysed by homology modelling. Based on our earlier work on cultivating P. rivulosus on spruce (Picea abies) wood chips, our hypothesis is that the acidic and thermotolerant laccases, together with the MnP isoenzymes (Hakala et al. 2005), provide P. rivulosus with an advantage in the decomposition of lignin (over cellulose) in coniferous wood.
In order to characterize the acidic laccases, the recombinant enzymes (rLac1 and rLac2–L497V) were expressed in the methylotrophic yeast P. pastoris, which has previously been applied as a host organism for a few basidiomycetous laccases, i.e. from T. versicolor, Rigidoporus microporus and Polyporus grammocephalus (Jönsson et al. 1997; Liu et al. 2003; Huang et al. 2011). In our study, increased specific activity levels of the recombinant P. rivulosus laccases were achieved with the S. cerevisiae α-factor signal sequence fusions when compared to the expression constructs harnessing the native laccase N-terminal signal peptides.
The enzymatic properties of the recombinant laccases demonstrated low pH optima (pH 3.5) for oxidation of ABTS and the phenolic compound 2,6-DMP. Previously characterized native laccases (Lac-3.5 and Lac-4.8) of P. rivulosus showed similar properties (Hildén et al. 2007). However, the correlation between recombinant and native ones (Lac-3.5 and Lac-4.8) has not been determined yet since P. rivulosus produces at least five different laccases (unpublished data). In contrast to the native P. rivulosus laccases, the recombinant enzymes were unable to oxidize the typical fungal laccase substrate SGZ. For the native P. rivulosus laccases, oxidation of SGZ occurred at a slow rate, with the highest activity peaking at pH 3.0 (Hildén et al. 2007). This is an unusually low pH for optimal activity of basidiomycetous laccases as the pH optima for oxidation of SGZ is commonly observed to be between pH 5 and 6 (Garzillo et al. 2001; Baldrian 2006) and implies significant differences at the organic substrate-binding cavity in the P. rivulosus laccases. It may be that the recombinant P. rivulosus laccases were even more restricted at the T1-Cu site.
Of the two acidic laccases of P. rivulosus, Lac1 demonstrates the highest protein sequence level identity with the only characterized laccase of C. subvermispora (Lcs-1; Karahanian et al. 1998; Larrondo et al. 2003a, b), while Lac2 shares the highest degree of sequence identity with one of the seven laccase protein and gene models (model #137686) recognized in the genome of C. subvermispora (http://genome.jgi-psf.org/Cersu1/Cersu1.home.html). The molecular mass of rLac1 corresponds to C. subvermispora laccase L1 of which N-terminus corresponds to the translated amino acid sequence of the lcs-1 gene (Fukushima and Kirk 1995; Karahanian et al. 1998). However, a deglycosylation assay did not alter the molecular mass of rLac1, indicating differences in glycosylation between rLac1 and L1. In Lac2 and gene model #137686, the C-terminus ends with SDEL, which we propose to potentially act as a remnant of the eukaryotic ER-retention signal (Pagny et al. 1999; Tonkin et al. 2006). This is supported by the fact that by changing the last amino acid of the C-terminal tripeptide (L497V), extracellular production of the recombinant rLac2 enzyme was facilitated by P. pastoris. In the phylogenetic tree of basidiomycetous laccases, the laccases of P. rivulosus and C. subvermispora cluster in the same branch (data not shown), which confirms the close phylogenetic relationship of the two species inferred from the nuclear and mitochondrial ribosomal RNA encoding gene and ITS sequences (Tomšovský et al. 2010; this work).
Analysis of the 3-D models for Lac1 and Lac2 showed their overall protein structures to be typical for fungal laccases consisting of three protein domains. As is conserved in the laccase architecture, the T1 copper site is in domain 3, and the T2/T3 trinuclear copper site is between domains 1 and 3 (Bertrand et al. 2002; Piontek et al. 2002). The reducing substrate-binding pocket is at the boundary between domains 2 and 3. The coordination sphere of the four copper ions is conserved in the fungal laccases and consists of ten histidine, one cysteine and one isoleucine or methionine residues (Giardina et al. 2010).
Superimposition of the substrate-binding pocket, occurring around the substrate ligands in the laccase 3-D structures of T. versicolor (Bertrand et al. 2002) and T. trogii (Matera et al. 2008), on the corresponding residues showed the presence of four conserved amino acids in both Lac1 and Lac2. Among them, the His residue (H457 in Lac1 and H456 in Lac2, respectively) coordinates the copper ion of the T1-Cu site and is exposed to the surface of the substrate-binding cavity. This histidine is assumed to play an important role in electron transfer from the reducing substrate molecule to the T1-Cu (Garzillo et al. 2001; Bertrand et al. 2002; Matera et al. 2008). The conserved acidic aspartate (Asp206 in T. versicolor LacIIIb) residue, which is normally deprotonated and thereby carrying a negative charge, is possibly involved in the binding and correct orientation of the substrate ligand molecule in the catalytic pocket (Garzillo et al. 2001; Bertrand et al. 2002).
Comparison of the substrate-binding pockets in the structural models of Lac1 and Lac2 indicated the presence of five amino acid substitutions. As a result of these substitutions, the size of the substrate-binding cavity in Lac2 is smaller compared with the Lac1 3-D protein model. Also, Lac2 contains the insertion of Gly392, which may affect the topology of the enzymatic cavity. The differences in the shape and volume of the substrate-binding site can be assumed to cause a distinction in the catalytic activity of the two P. rivulosus laccases. This is supported by the kinetic constant values of rLac1 and rLac2–S497V obtained with the phenolic substrate 2,6-DMP. Alteration of the oxidation ability with respect to bulky phenolic substrates was demonstrated for the structurally characterized laccase LacIIIb from T. versicolor using a series of site-directed mutations on the phenylalanines located in the enzymatic cavity, thus altering the shape of the substrate-binding pocket (Galli et al. 2011).
Considering the oxidation of the phenolic substrate 2,6-DMP, the rLac1 and rLac2–L497V showed acidic pH optima, which is in accordance with the properties of the native acidic laccases of P. rivulosus (Hildén et al. 2007). The recombinant laccases were stable at temperatures below 50 °C. The half-life of rLac1 at 60 °C was 60 min at pH 3.0 whereas rLac2–L497V showed a half-life of only 25 min at 60 °C. The native laccases of P. rivulosus tolerated higher temperatures than rLac1 and rLac2–L497V (Hildén et al. 2007).
Temperature activation for oxidation of phenolic substrates or ABTS has earlier been described only for a few laccases of fungal and bacterial origin (Hildén et al. 2009). Interestingly, rLac1 presented thermal activation, since the 2,6-DMP oxidation was increased up to 30 % (of the original activity) after 10 min incubation at 60 °C, which is in line with the thermal activation of the native Lac-4.8 of P. rivulosus at 50 °C (Hildén et al. 2007). In contrast, rLac2–L497V demonstrated no such thermal activation at any of the tested temperatures. These data imply that the rLac1 shares properties with the previously recognized Lac-4.8 isoenzyme. It may be proposed that the unusual thermal stability and activation for oxidation of the phenolic substrate (2,6-DMP) by rLac1 could be due to the more open architecture of the substrate-binding cavity in Lac1 than in Lac2, thus giving more flexibility for Lac1 for binding and oxidation of substrate molecules of variable sizes.
We present two novel recombinant laccases originating from the selective lignin-degrading and biopulping fungus P. rivulosus. By 3-D homology modelling, differences in the substrate-binding cavities near the T1-Cu site could be demonstrated in Lac1 and Lac2. The enzymatic properties described (acidity, thermotolerance and thermal activation) indicate that the novel recombinant Lac1 may be applied as a robust enzyme for oxidation and conversion of phenolic compounds under acidic conditions and at elevated temperatures. Heterologous expression of the novel Lac1 and Lac2 will facilitate the development of their use as oxidative biocatalysts for conversion of wood lignin, lignin-like compounds and soil-polluting xenobiotics.
References
Baldrian P (2006) Fungal laccases—occurence and properties. FEMS Microbiol Rev 30:215–242
Bertrand T, Jolivalt C, Briozzo P, Caminade E, Joly N, Madzak C, Mougin C (2002) Crystal structure of a four-copper laccase complexed with an arylamine: insights into substrate recognition and correlation with kinetics. Biochemistry 41:7325–7333
Bourbonnais R, Paice MG, Freiermuth B, Bodie E, Bornemann S (1997) Reactivities of various mediators and laccases with kraft pulp and lignin model compounds. Appl Environ Microbiol 63:4627–4632
Call HP, Mücke I (1997) History, overview and applications of mediated ligninolytic systems, especially laccase-mediator-systems (Lignozym®-process). J Biotechnol 53:163–202
Courty PE, Hoegger PJ, Kilaru S, Kohler A, Buée M, Garbaye J, Martin F, Kües U (2009) Phylogenetic analysis, genomic organization, and expression analysis of multi-copper oxidases in the ectomycorrhizal basidiomycete Laccaria bicolor. New Phytol 182:736–750
DeLano WL (2002) The PyMOL molecular graphics system. DeLano Scientific, San Carlos
D'Souza TM, Boominathan K, Reddy CA (1996) Isolation of laccase gene-specific sequences from white-rot and brown-rot fungi by PCR. Appl Environ Microbiol 62:3739–3744
Fukushima Y, Kirk TK (1995) Laccase component of the Ceriporiopsis subvermispora lignin-degrading system. Appl Environ Microbiol 61:872–876
Galli C, Gentili P, Jolivalt C, Madzak C, Vadala R (2011) How is the reactivity of laccase affected by single-point mutations? Engineering laccase for improved activity towards sterically demanding substrates. Appl Microbiol Biotechnol 91:123–131
Garzillo AM, Colao MC, Buonocore V, Oliva R, Falcigno L, Saviano M, Santoro AM, Zappala R, Bonomo RP, Bianco C, Giardina P, Palmieri G, Sannia G (2001) Structural and kinetic characterization of native laccases from Pleurotus ostreatus, Rigidoporus lignosus, and Trametes trogii. J Prot Chem 20:191–201
Giardina P, Faraco V, Pezzella C, Piscitelli A, Vanhulle S, Sannia G (2010) Laccases: a never-ending story. Cell Mol Life Sci 67:369–385
Hakala TK, Maijala P, Konn J, Hatakka A (2004) Evaluation of novel wood-rotting polypores and corticioid fungi for the decay and biopulping of Norway spruce (Picea abies) wood. Enzyme Microb Technol 34:255–263
Hakala TK, Lundell T, Galkin S, Maijala P, Kalkkinen N, Hatakka A (2005) Manganese peroxidases, laccases and oxalic acid from the selective white-rot fungus Physisporinus rivulosus grown on spruce wood chips. Enzyme Microb Technol 36:461–468
Halaouli S, Record E, Casalot L, Hamdi M, Sigoillot J-C, Asther M, Lomascolo A (2006) Cloning and characterization of a tyrosinase gene from the white-rot fungus Pycnoporus sanguineus, and overproduction of the recombinant protein in Aspergillus niger. Appl Genet Mol Biotechnol 70:580–589
Hatakka A, Hammel K (2011) Fungal biodegradation of lignocelluloses. In: Hofrichter M (ed) The Mycota: a comprehensive treatise on fungi as experimental systems for basic and applied research. X. Industrial applications, 2nd edn. Springer, Berlin, pp 319–340
Hatakka A, Maijala P, Hakala TK, Hauhio L, Ellmén J (2003) Novel white-rot fungus and use thereof in wood pretreatment. International patent application WO03/080812
Hildén SK, Martinez AT, Hatakka AI, Lundell TK (2005) Two novel manganese peroxidases from Phlebia radiata, a lignin-degrading fungus, are phylogenetically and structurally divergent. Fungal Genet Biol 42:403–419
Hildén K, Hakala TK, Maijala P, Lundell T, Hatakka A (2007) Novel thermotolerant laccases produced by the white-rot fungus Physisporinus rivulosus. Appl Microbiol Biotechnol 77:301–309
Hildén K, Hakala TK, Lundell T (2009) Thermotolerant and thermostable laccases. Biotech Lett 31:1117–1128
Hoegger PJ, Kilaru S, James TY, Thacker JR, Kües U (2006) Phylogenetic comparison and classification of laccase and related multicopper oxidase protein sequences. FEBS J 273:2308–2326
Huang SJ, Liu ZM, Huang XL, Guo LQ, Lin JF (2011) Molecular cloning and characterization of a novel laccase gene from a white-rot fungus Polyporus grammocephalus TR16 and expression in Pichia pastoris. Lett Appl Microbiol 52:290–297
Jönsson LJ, Saloheimo M, Penttilä M (1997) Laccase from the white-rot fungus Trametes versicolor: cDNA cloning of lcc1 and expression in Pichia pastoris. Curr Genet 32:425–430
Karahanian E, Corsini G, Lobos S, Vicuña R (1998) Structure and expression of a laccase gene from the ligninolytic basidiomycete Ceriporiopsis subvermispora. Biochim Biophys Acta 1443:65–74
Kersten P, Cullen D (2007) Extracellular oxidative systems of the lignin-degrading basidiomycete Phanerochaete chrysosporium. Fungal Genet Biol 44:77–87
Kilaru S, Hoegger PJ, Kües U (2006) The laccase multi-gene family in Coprinopsis cinerea has seventeen different members that divide into two distinct subfamilies. Curr Genet 50:45–60
Koroleva OV, Stepanova EV, Binukov VI, Timofeev VP, Pfeil W (2001) Temperature-induced changes in copper centers and protein conformation of two fungal laccases from Coriolus hirsutus and Coriolus zonatus. Biochim Biophys Acta 1547:397–407
Kumar SVS, Prashant SP, Durani S, Wangikar PP (2003) Combined sequence and structure analysis of the fungal laccase family. Biotechnol Bioeng 83:386–394
Larrondo LF, Avila M, Salas L, Cullen D, Vicuña R (2003a) Heterologous expression of laccase cDNA from Ceriporiopsis subvermispora yields copper-activated apoprotein and complex isoform patterns. Microbiology 149:1177–1182
Larrondo LF, Salas L, Melo F, Vicuña R, Cullen D (2003b) A novel extracellular multicopper oxidase from Phanerochaete chrysosporium with ferroxidase activity. Appl Environ Microbiol 69:6257–6263
Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK—a program to check the stereochemical quality of protein structures. J Appl Cryst 26:283–291
Li JF, Hong YZ, Xiao YZ, Xu YH, Fang W (2007) High production of laccase B from Trametes sp. in Pichia pastoris. World J Microbiol Biotechnol 23:741–745
Liu W, Chao Y, Liu S, Bao H, Qian S (2003) Molecular cloning and characterization of a laccase gene from the basidiomycete Fome lignosus and expression in Pichia pastoris. Appl Microb Biotechnol 63:174–181
Lundell TK, Mäkelä MR, Hildén K (2010) Lignin-modifying enzymes in filamentous basidiomycetes—ecological, functional and phylogenetic review. J Basic Microbiol 50:5–20
Mäkelä MR, Hildén K, Hakala TK, Hatakka A, Lundell T (2006) Expression and molecular properties of a new laccase of the white rot fungus Phlebia radiata grown on wood. Curr Genet 50:323–333
Martinez D, Larrondo LF, Putnam N, Sollewijn Gelpke MD, Huang K, Chapman J, Helfenbein KG, Ramaya P, Detter JC, Larimer F, Coutinho PM, Henrissat B, Berka R, Cullen D, Rokhsar D (2004) Genome sequence of the lignocellulose degrading fungus Phanerochaete chrysosporium strain RP78. Nat Biotechnol 22:695–700
Martinez D, Berka RM, Henrissat B, Saloheimo M, Arvas M, Baker SE, Chapman J, Chertkov O, Coutinho PM, Cullen D, Danchin EGJ, Grigoriev IV, Harris P, Jackson M, Kubicek CP, Han CS, Ho I, Larrondo LF, Lopez de Leon A, Magnuson JK, Merino S, Misra M, Nelson B, Putnam N, Robbertse B, Salamov AA, Schmoll M, Terry A, Thayer N, Westerholm-Parviainen A, Schoch CL, Yao J, Barabote R, Nelson MA, Detter C, Bruce D, Kuske CR, Xie G, Richardson P, Rokhsar D, Lucas SM, Rubin ER, Dunn-Coleman N, Ward M, Brettin TS (2008) Genome sequencing and analysis of the biomass-degrading fungus Trichoderma reesei (syn. Hypocrea jecorina). Nat Biotechnol 26:553–560
Matera I, Gullotto A, Tilli S, Ferraroni M, Scozzafava A, Briganti F (2008) Crystal structure of the blue multicopper oxidase from the white-rot fungus Trametes trogii complexed with p-toluate. Inorg Chim Acta 361:4129–4137
Pagny S, Lerouge P, Faye L, Gomord V (1999) Signals and mechanisms for protein retention in the endoplasmic reticulum. J Exp Bot 50:157–164
Petrey D, Honig B (2003) GRASP2: visualization, surface properties, and electrostatics of macromolecular structures and sequences. Methods Enzymol 374:492–509
Piontek K, Antorini M, Choinowski T (2002) Crystal structure of a laccase from the fungus Trametes versicolor at 1.90-Å resolution containing a full complement of coppers. J Biol Chem 277:37663–37669
Sali A, Potterton L, Yuan F, van Vlijmen H, Karplus M (1995) Evaluation of comparative protein modelling by MODELLER. Proteins 23:318–326
Saparrat M, Balatti PA, Martínez MJ, Jurado M (2010) Differential regulation of laccase gene expression in Coriolopsis rigida LPSC No. 232. Fungal Biol 114:11–12
Sippl MJ (1993) Recognition of errors in three-dimensional structures of proteins. Proteins 17:355–362
Soden DM, O’Callaghan J, Dobson ADW (2002) Molecular cloning of a laccase isozyme gene from Pleurotus sajor-caju and expression in the heterologous Pichia pastoris host. Microbiology 148:4003–4014
Tomšovský M, Menkis A, Vasaitis R (2010) Phylogenetic relationships in European Ceriporiopsis species inferred from nuclear and mitochondrial ribosomal DNA sequences. Fungal Biol 114:350–358
Tonkin CJ, Struck NS, Mullin KA, Stimmler LM, McFadden GI (2006) Evidence for Golgi-independent transport from the early secretory pathway to the plastid in malaria parasites. Mol Microbiol 61:614–630
Tuomela M, Hatakka A (2011) Oxidative fungal enzymes for bioremediation. In: Moo-Young M, Agathos S (eds) Comprehensive biotechnology, vol 6, Environmental biotechnology and safety. Elsevier, London, pp 183–196
Uthandi S, Saad B, Humbard MA, Maupin-Furlow JA (2010) LccA, an archaeal laccase secreted as a highly stable glycoprotein into the extracellular medium by Haloferax volcanii. Appl Environ Microbiol 76:733–743
Acknowledgements
This study was supported by grants #129869 (RFBR-Finland 08-04-91763 AF_a), #138331 and #1133022 from the Academy of Finland and by the EC Marie Curie actions PIEF-GA-2009-236714 and PERG08-GA-2010-276794 and Finnish Cultural Foundation for K.H. Dr. Graham Whyteside is acknowledged for help in Pichia pastoris expression work.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Hildén, K., Mäkelä, M.R., Lundell, T. et al. Heterologous expression and structural characterization of two low pH laccases from a biopulping white-rot fungus Physisporinus rivulosus . Appl Microbiol Biotechnol 97, 1589–1599 (2013). https://doi.org/10.1007/s00253-012-4011-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-012-4011-6