Abstract
We demonstrate glutamate production from β-glucan using endoglucanase (EG)-expressing Corynebacterium glutamicum. The signal sequence torA derived from Escherichia coli K12, which belongs to the Tat pathway, was suitable for secreting EG of Clostridium thermocellum using C. glutamicum as a host. Using the torA signal sequence, endoglucanase from Clostridium cellulovorans 743B was successfully expressed, and the secreted EG produced 123 mg of reducing sugar from 5 g of β-glucan at 30 °C for 72 h, which is the optimal condition for C. glutamicum growth. Subsequently, glutamate fermentation from β-glucan was carried out with the addition of Aspergillus aculeatus β-glucosidase produced by recombinant Aspergillus oryzae. Using EG-secreting C. glutamicum, 178 mg/l of glutamate was produced from 15 g of β-glucan. This is the first report of glutamate fermentation from β-glucan using endoglucanase-secreting C. glutamicum.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Corynebacterium glutamicum is a non-pathogenic, non-sporulating, non-motile gram-positive soil bacterium belonging to the order Actinomycetales, which includes Corynebacteria, Nocardia, Rhodococci, and other related microorganisms (George 2001). C. glutamicum is an important industrial microorganism for its high production of amino acids such as glutamate and lysine (Hermann 2003; Leuchtenberger et al. 2005; Tateno et al. 2007). Amino acids are widely used in medicine, in fodder, and as supplements; they are also necessary as building blocks of other chemicals (Bourke and Kohn 2003; Fukuoka et al. 2004; US Department of Energy 2004). Engineered C. glutamicum is also superior for producing various kinds of organic compounds, such as ethanol as a bio-fuel (Sakai et al. 2007), cadaverine as a component of bio-based nylon (Mimitsuka et al. 2007; Tateno et al. 2009), and some organic acids under oxygen deprivation conditions (Inui et al. 2004; Okino et al. 2008).
Cellulose is a promising renewable carbon source that is composed of glucose units linked together by β-1,4-glycosidic linkages, as it is the most abundant biomass in the world. Since cellulose is unavailable for amino acid production by C. glutamicum, a saccharification process is needed prior to fermentation. Enzymatic degradation of crystalline cellulose requires several types of cellulolytic enzymes. Endoglucanase (EG) randomly digests the amorphous region of the cellulose chain. Exocellobiohydrolase (CBH) acts on the resulting reducing and non-reducing ends of crystalline cellulose. By the synergistic action of both enzymes, cellulose is efficiently degraded to soluble cellooligosaccharides. Finally, β-glucosidase degrades these oligosaccharides to glucose. Since the production cost of these cellulolytic enzymes is very high, a strategy that can reduce the amount of enzyme used needs to be developed.
Expression of heterologous cellulase genes in useful microorganisms is a promising way to achieve cost-effective production processes. For example, direct ethanol fermentation from amorphous cellulose by using a recombinant yeast strain that co-expresses three types of cellulolytic enzymes has been demonstrated (Fujita et al. 2002; 2004). Lactic acid production using EG-expressing lactic acid bacteria has also been reported recently (Okano et al. 2010). Although C. glutamicum is superior for producing amino acids from glucose, the production of amino acids from cellulosic material using engineered C. glutamicum has not yet been attempted.
In this study, we demonstrated glutamate production from barley β-glucan using an EG-secreting strain of C. glutamicum (ATCC 13032). An engineered C. glutamicum having a suitable secretion signal sequence and high endoglucanase activity was constructed. Using the endoglucanase-secreting strain, we demonstrated glutamate production from β-glucan in the presence of an added β-glucosidase solution that was produced by recombinant Aspergillus oryzae.
Materials and methods
Bacterial strains and media
The bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli was grown in LB medium (10 g/l tryptone, 5 g/l yeast extract, and 5 g/l sodium chloride) containing 50 mg/l kanamycin at 37 °C. C. glutamicum ATCC 13032 and the recombinant strains were grown in BY medium (10 g/l peptone, 10 g/l meat extract, 5 g/l yeast extract, and 5 g/l sodium chloride) containing 25 mg/l kanamycin at 30 °C (Katsumata et al. 1984). MM-β-glucan medium, which contained 10 g/l (NH4)2SO4, 3 g/l K2HPO4, 1 g/l KH2PO4, 2 g/l urea, 0.3 g/l MgSO4·7H2O, 0.01 g/l FeSO4·7H2O, 0.01 g/l MnSO4·5H2O, 1 mg/l thiamine, 0.03 mg/l biotin, 5 g/l Tween-40, 15 g/l β-d-glucan, and 0.1% (v/v) of BGL solution (final enzyme activity was 0.30 U/ml), was utilized in glutamate fermentation.
Construction of plasmids
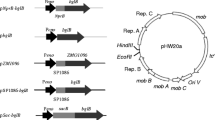
All genetic manipulations were done using Escherichia coli SCS110, and all polymerase chain reactions (PCRs) were conducted using KOD-plus DNA polymerase (Toyobo, Osaka, Japan). The oligonucleotide primers used for the construction of plasmids are listed in Table 2. The C. glutamicum–E. coli shuttle vectors with the cspB promoter, pCC and pCCS, were constructed in our previous study (Tateno et al. 2007).
To find the appropriate signal sequence for EG expression using C. glutamicum as a host, the following plasmids were constructed. The signal sequences of cmt2 and rpf2 were amplified by PCR from C. glutamicum ATCC 14020 using the primer pairs ss-cmt2_F and ss-cmt2_R, and ss-rpf2_F and ss-rpf2_R, respectively. The signal sequences of hyaA and torA were amplified by PCR from E. coli K12 using the primer pairs ss-hyaA_F and ss-hyaA_R, and ss-torA_F and ss-torA_R, respectively. These amplified fragments were digested with BamHI and EcoRV, and digested fragments were introduced into the same sites of pCC. The resultant plasmids were named as pCCC (pCC containing cmt2 signal sequence), pCCR (pCC containing rpf2 signal sequence), pCCH (pCC containing hyaA signal sequence), and pCCT (pCC containing torA signal sequence). We also prepared modified cspB signal sequences (Peyret et al. 1993). The cspB signal sequence containing several amino acids of N-terminal mature CspB was amplified by PCR using the following primer pairs: ss-cspB_F and ss-cspBN10_R, or ss-cspB_F and ss-cspBN20_R, respectively. The amplified fragments were digested with BamHI and EcoRV and inserted into the same sites of pCC. The resultant plasmids were named as pCCS-N10 (pCC containing cspB signal sequence and 10 amino acids from cspB N terminus) and pCCS-N20 (pCC containing cspB signal sequence and 20 amino acids from cspB N terminus).
An EG gene, celA from Clostridium thermocellum, was amplified by PCR from genomic DNA of C. thermocellum ATCC 27405 using primer pairs Cth-celA_F and Cth-celA_R. The NCBI accession is YP_001036436.1, and Gene ID is 4810536. The amplified fragment was digested with SacI and XhoI and introduced into pCCS, pCCC, pCCR, pCCH, pCCT, pCCS-N10, and pCCS-N20. The resultant plasmids were named pCCS-Cth-celA, pCCC-Cth-celA, pCCR-Cth-celA, pCCH-Cth-celA, pCCT-Cth-celA, pCCS-N10-Cth-celA, and pCCS-N20-Cth-celA, respectively.
Other putative EG genes were amplified by PCR from various kinds of genomic DNAs. We chose five kinds of putative EGs: Clocel3242 from Clostridium cellulovorans 743B (NCBI accession YP_003844692.1 and Gene ID 9610136), Clocel2821 from C. cellulovorans 743B (NCBI accession YP_003844281.1 and Gene ID 9609710), Sde3237 from Saccharophagus degradans 2–40 (NCBI accession YP_528706.1 and Gene ID 3965710), Tfu2712 from Thermobifida fusca YX (NCBI accession YP_290768.1 and Gene ID 3580691), and ZMO1086 from Zymomonas mobilis ZM4 (NCBI accession YP_162821.1 and Gene ID 3188168). A FLAG-tag sequence was introduced at the C terminus of each EG gene and their original signal sequences were removed. The amplified Clocel3242 and Clocel 2821 fragments were digested with SacI and XhoI. The amplified Sde3237 and ZMO1086 fragments were digested with NheI and XhoI. The amplified Tfu2712 fragments were digested with NcoI and NheI. Each digested fragment was inserted into the same respective sites of pCCT. The resultant plasmids were named as pCCT-Clocel3242, pCCT-Clocel2821, pCCT-Sde3237, pCCT-ZMO1086, and pCCT-Tfu2712, respectively. All PCR-amplified DNA fragments were confirmed by DNA sequencing analysis using ABI PRISM 3100 Genetic Analyzer (Applied Biosystems).
The 12 plasmids described above were introduced into C. glutamicum ATCC 13032. The transformation was conducted by electroporation with a 2.5-kV, 200-Ω, 25-μF electric pulse in a 0.2-cm cuvette using a Gene Pulser (Bio-Rad, Richmond, CA, USA). The resultant C. glutamicum strains were named as C. glutamicum-pCCS-Cth-celA, C. glutamicum-pCCC-Cth-celA, C. glutamicum-pCCR-Cth-celA, C. glutamicum-pCCH-Cth-celA, C. glutamicum-pCCT-Cth-celA, C. glutamicum-pCCS-N10-Cth-celA and C. glutamicum-pCCS-N20-Cth-celA, C. glutamicum-pCCT-Clocel3242, C. glutamicum-pCCT-Clocel2841, C. glutamicum-pCCT-Sde3237, C. glutamicum-pCCT-ZMO1086, and C. glutamicum-pCCT-Tfu2712, respectively.
Endoglucanase assay
First, we tested endoglucanase activity using E. coli SCS110 as a host. All plasmids were introduced into E. coli SCS110 and EG activity was evaluated by a halo assay using carboxy methyl cellulose (CMC) plates (20). LB agar plates containing 0.75% CMC (Nacalai Tesque, Kyoto, Japan) (w/v), 0.03% trypan blue (Nacalai Tesque), and 50 mg/l kanamycin (LB-CMC plate) were used. E. coli-harboring plasmids were incubated on LB-CMC plates at 37 °C for 24 h and their EG activities were detected by halo formation. We also tested EG activity using C. glutamicum ATCC 13032 as a host. The transformants of C. glutamicum were incubated on BY plates containing 0.75% CMC, 0.03% trypan blue, and 25 g/l kanamycin (BY-CMC plate) at 30 °C for 48 h.
The activity of secreted EG was evaluated using the amount of total reducing sugars derived from barley β-d-glucan (Sigma, St. Louis, MO, USA). All transformants were precultivated in 5 ml of BY medium containing 25 mg/l kanamycin in a test tube. C. glutamicum-harboring pCC was used as a negative control. After precultivation at 30 °C for 24 h, all culture medium was inoculated into 20 ml of BY medium containing 25 mg/l kanamycin in a 200-ml flask. After cultivation at 30 °C for 24 h, the supernatant was collected by centrifugation at 12,000 rpm for 5 min at 4 °C. Then 1.8 ml of supernatant was mixed with 2 ml of 1% (w/v) β-d-glucan and 0.2 ml of 1 M citrate buffer (pH 6.0). In the case of C. glutamicum-pCCS-Cth-celA, C. glutamicum-pCCC-Cth-celA, C. glutamicum-pCCR-Cth-celA, C. glutamicum-pCCH-Cth-celA, C. glutamicum-pCCT-Cth-celA, C. glutamicum-pCCS-N10-Cth-celA, and C. glutamicum-pCCS-N20-Cth-celA, the mixtures were incubated for 48 h at 60 °C, which was the optimal temperature for CelA, and others were incubated for 72 h at 30 °C. After incubation, the amount of reducing sugar was determined by the Somogyi–Nelson method (Somogyi 1952).
Glutamate production from glucan
Glutamate fermentation from barley β-glucan was performed as follows.
C. glutamicum-pCCT-Clocel3242 was precultivated in 5 ml of BY medium containing 25 μg/ml of kanamycin at 30 °C for 20 h. The preculture (0.2 ml) was transferred to 20 ml of BY medium containing kanamycin. After 24 h cultivation, 2 ml of the culture was inoculated into 20 ml of MM-β-glucan medium. The β-glucosidase solution was prepared using recombinant strains of A. oryzae (Adachi et al. 2008). To assist the β-glucan degradation, 300 μl of preculture supernatant was added. The fermentation was carried out at 30 °C, and OD600 and the glutamate concentration of culture supernatants were monitored. Glutamate concentration was measured by the amino acid analyzer system Prominence (Shimadzu) using a Shim-pack Amino-Li column (0.5 μm, 100 mm × 6.0 mm I.D.)
Results
Signal peptide optimization for endoglucanase expression using C. glutamicum
Several secretion signal sequences were selected as candidates for EG expression using C. glutamicum as a host. One set of signal sequences, cspB, cmt2, and rpf2, belong to the Sec pathway, while the other sequences, hyaA and torA, belong to the Tat pathway. We also prepared slightly modified cspB signal sequences that included 10 or 20 amino acids derived from N-terminal mature cspB. These signal sequences were fused to the N terminus of EG from C. thermocellum (Cth-celA) and expressed using C. glutamicum.
The transformants of C. glutamicum strains, C. glutamicum-pCCS-Cth-celA, C. glutamicum-pCCC-Cth-celA, C. glutamicum-pCCR-Cth-celA, C. glutamicum-pCCH-Cth-celA, C. glutamicum-pCCT-Cth-celA, C. glutamicum-pCCS-N10-Cth-celA, and C. glutamicum-pCCS-N20-Cth-celA, were cultivated in BY medium. The culture supernatant was incubated with β-glucan as a substrate at 60°C and the EG activity was evaluated by the amount of produced reducing sugar. After a 72-h reaction, the torA signal sequence fused to EG showed the highest endoglucanase activity and produced 890 mg/l of reducing sugar from β-glucan (Table 3). Modified cspB sequences fused to EG produced about half the amount of reducing sugar compared to that of torA. Other signal sequences fused to EG showed no activity in the culture supernatant. Therefore, the torA signal sequence was used in following experiments.
Screening for EG having high activity under C. glutamicum growing conditions
At first we simply checked the EG activities using E. coli as a host. Halo assays on LB plates containing 0.75% CMC were used to detect EG activity. Five kinds of EGs showed clear halos (data not shown). These EG genes are as follows: from C. cellulovorans (Clocel3242, Clocel2841), C. thermocellum (Cth-celA), S. degradans (Sde3237), T. fusca (Tfu2712), and Z. mobilis (ZmO1086). Plasmids carrying EG genes were introduced into C. glutamicum ATCC 13032.
The EG activities of EG-expressing C. glutamicum were similarly evaluated by Halo assays described above, and all six recombinants of C. glutamicum showed EG activity halos on CMC plates (data not shown). Then the enzyme activity of the culture supernatant was evaluated by the amount of produced reducing sugar using β-glucan as a substrate at 30 °C, which is the optimal temperature for C. glutamicum growth. As shown in Fig. 1, Clocel3242 (EG from C. cellulovorans) showed the highest EG activity, and 123 mg/l of reducing sugar was released from 5 g/l of β-glucan at 30 °C after 72 h. The activities of CelA and Sde3237 were about half of the activity of Closel3242. ZMO1086, Clocel2842, and Tfu2712 produced only a small amount of the reducing sugar.
The amount of reducing sugar after incubation with β-glucan and C. glutamicum strains harboring endoglucanase expression vectors. Symbols show C. glutamicum strains harboring pCC (triangles), pCCT-Cth-celA (open circles), pCCT-Clocel3242 (closed circles), pCCT-ZMO1086 (squares), and pCCT-Sde3237 (diamonds). The culture supernatant was mixed with 0.5% of β-glucan, and the amount of reducing sugar was measured after 48 h incubation at 30 °C. Data points represent the mean and standard error of three independent experiments
Glutamate fermentation from β-glucan
Encouraged by these findings, we proceeded to carry out glutamate fermentation from barley β-glucan using EG Clocel3242-secreting C. glutamicum. To assist the degradation of β-glucan, a β-glucosidase solution produced by recombinant A. oryzae that expresses A. aculeatus BGL1 was added. C. glutamicum-harboring pCC was used as a control. As shown in Fig. 2a, following the addition of the β-glucosidase solution, the OD600 of EG Clocel3242-secreting C. glutamicum was increased after fermentation, while that of pCC-harboring C. glutamicum did not increase at all. The amount of produced glutamate is shown in Fig. 2b. After 72 h fermentation, EG Clocel3242-secreting C. glutamicum successfully produced 178 mg/l of glutamate from 15 g/l of β-glucan, which was 5-fold higher than that of pCC-harboring C. glutamicum. In the absence of BGL solution, no glutamate was produced from glucan using Clocel3242-secreting C. glutamicum (Fig. 2b). This result demonstrated successful production of glutamate from β-glucan using EG-secreting C. glutamicum supplemented with a β-glucosidase solution, and the data support synergism between EG and BGL1 activities.
Fermentation from β-glucan using C. glutamicum harboring pCCT-Clocel3242 (circles) or pCC (triangles) in the presence (closed symbols) or in the absence (open symbols) of β-glucosidase solution produced by recombinant A. oryzae. a OD600. b Glutamate concentration. Data points represent the mean and standard error of three independent experiments
Discussion
The goal of this study was to achieve direct amino acid production from cellulosic materials, the most abundant biomass in the world, using C. glutamicum as a host. In order to simplify the manufacturing process and to reduce the production cost of amino acids from cellulose, simultaneous saccharification and fermentation of cellulose has been attempted. Although a cellulase-expressing Corynebacterium was reported previously (Paradis et al. 1987; Adham et al. 2001), there has been no report demonstrating direct glutamate production from cellulosic materials using endoglucanase-expressing C. glutamicum.
We chose endoglucanase from C. thermocellum (Cth-celA) to elucidate a suitable signal sequence for secretion using C. glutamicum as a host. Heterologous expression of EG (Cth-celA) using lactic acid bacteria as a host has been previously reported (Okano et al. 2010). However, in the case of C. glutamicum, EG activity was not detected in the supernatant when the cspB signal sequence, which is suitable for amylase secretion, was used to express EG Cth-celA. Western blotting analysis using FLAG-tag showed that Cth-celA was located in the intracellular, not extracellular, fraction (data not shown). This suggested a failure of secretion, and hence we focused on optimization of the secretion signal sequence.
Most bacteria possess two types of protein export pathways, the secretory (Sec) pathway and the twin-arginine translocation (Tat) pathway (Settles and Martienssen. 1998; Berks et al. 2000). C. glutamicum possesses both the Sec and Tat pathways (Kikuchi et al. 2009). Heterologous protein expression has been achieved using C. glutamicum through both pathways (Date et al. 2003; Kikuchi et al. 2006, 2009). Here, we selected Sec pathway-dependent signal sequences (cspB, cmt2, and rpf2) and Tat pathway-dependent signal sequences (hyaA and torA). We also employed two types of modified signal sequence of cspB. Although EG activity was not detected when cmt2 or rpf2 signal sequence was used, the use of modified cspB signal sequences (CspB-N10 and CspB-N20) improved the secreted EG activity (Table 3). Alternatively, when using the torA signal sequence, EG activity significantly improved, resulting in about 4-fold higher reducing sugar production compared to that using modified cspB signals. These results showed that the torA signal sequence, which is included in the Tat pathway, is suitable for expression of secreted EG using C. glutamicum as a host. In our previous report, the cspB signal sequence allowed a high level of expression of secreted α-amylase (Tateno et al. 2007), which may indicate that the appropriate signal sequence depends on the target protein.
To achieve more efficient degradation of cellulose, we evaluated several kinds of EG activity and glutamate production at 30 °C (Fig. 1), which is an appropriate condition for C. glutamicum growth. EG Clocel3242, an endoglucanase derived from C. cellulovorans, showed the highest activity at 30 °C, and three times the amount of reducing sugar was produced compared to that of Cth-celA.
In an effort to test further applications of EG-secreting C. glutamicum, we performed glutamate fermentation with barley β-glucan (Fig. 2). Glutamate fermentation was successfully achieved only when both EG-secreting C. glutamicum and a β-glucosidase solution were used, and 178 mg/l of glutamate was produced under growth conditions for C. glutamicum. Although the optimal temperature of EG activity is higher than the optimal temperature for C. glutamicum growth (around 30 °C), our result showed efficient β-glucan degradation and glutamate fermentation under suitable conditions for C. glutamicum. This successful result also can be explained by the synergistic action of both endoglucanase and BGL1 produced by recombinant A. oryzae. Although the yield of glutamate from 15g/l of β-glucan (178 mg/l) using Clocel3242-secreting Corynebacterium was not enough compared to that from glucose (Yao et al. 2009), increasing both cellulase activities (EG and BGL) leads to a more efficient degradation of β-glucan and improved glutamate yield.
In conclusion, we have achieved for the first time glutamate fermentation from cellulosic materials using EG-secreting C. glutamicum. As a next step, it is necessary to express several kinds of BGL using C. glutamicum as a host. Further studies are needed to co-express BGL and EG in C. glutamicum for efficient glutamate fermentation from cellulose.
References
Adachi T, Ito J, Kawata K, Kaya M, Ishida H, Sahara H, Hata Y, Ogino C, Fukuda H, Kondo A (2008) Construction of an Aspergillus oryzae cell-surface display system using a putative GPI-anchored protein. Appl Microbiol Biotechnol 81:711–719
Adham SA, Honrubia P, Díaz M, Fernández-Abalos JM, Santamaría RI, Gil JA (2001) Expression of the genes coding for the xylanase Xys1 and the cellulase Cel1 from the straw-decomposing Streptomyces halstedii JM8 cloned into the amino-acid producer Brevibacterium lactofermentum ATCC13869. Arch Microbiol 177:91–97
Berks BC, Sargent F, Palmer T (2000) The Tat protein export pathway. Mol Microbiol 35:260–274
Bourke SL, Kohn J (2003) Polymers derived from the amino acid L-tyrosine: polycarbonates, polyarylates copolymers with poly(ethylene glycol). Adv Drug Deliv Rev 25:447–466
Date M, Yokoyama K, Umezawa Y, Matsui H, Kikuchi Y (2003) Production of native-type Streptoverticillium mobaraense transglutaminase in Corynebacterium glutamicum. Appl Environ Microbiol 69:3011–3014
Fujita Y, Takahashi S, Ueda M, Tanaka A, Okada H, Morikawa Y, Kawaguchi T, Arai M, Fukuda H, Kondo A (2002) Direct and efficient production of ethanol from cellulosic material with a yeast strain displaying cellulolytic enzymes. Appl Environ Microbiol 68:5136–5141
Fujita Y, Ito J, Ueda M, Fukuda H, Kondo A (2004) Synergistic saccharification, and direct fermentation to ethanol, of amorphous cellulose by use of an engineered yeast strain codisplaying three types of cellulolytic enzyme. Appl Environ Microbiol 70:1207–1212
Fukuoka T, Uyama H, Kobayashi S (2004) Polymerization of polyfunctional macromolecules: synthesis of a new class of high molecular weight poly(amino acid)s by oxidative coupling of phenol-containing precursor polymers. Biomacromolecules 5:977–983
George MG (2001) Bergey’s manual of systematic bacteriology, 2nd edn. Springer, New York
Hermann T (2003) Industrial production of amino acids by coryneform bacteria. J Biotechnol 104:155–172
Inui M, Murakami S, Okino S, Kawaguchi H, Vertès AA, Yukawa H (2004) Metabolic analysis of Corynebacterium glutamicum during lactate and succinate productions under oxygen deprivation conditions. J Mol Microbiol Biotechnol 7:182–196
Katsumata R, Ozaki A, Oka T, Furuya A (1984) Protoplast transformation of glutamate-producing bacteria with plasmid DNA. J Bacteriol 159:306–311
Kikuchi Y, Date M, Itaya H, Matsui K, Wu LF (2006) Functional analysis of the twin-arginine translocation pathway in Corynebacterium glutamicum ATCC 13869. Appl Environ Microbiol 72:7183–7192
Kikuchi Y, Itaya H, Date M, Matsui K, Wu LF (2009) TatABC overexpression improves Corynebacterium glutamicum Tat-dependent protein secretion. Appl Environ Microbiol 75:603–607
Leuchtenberger W, Huthmacher K, Drauz K (2005) Biotechnological production of amino acids and derivatives: current status and prospects. Appl Microbiol Biotechnol 69:1–8
Mimitsuka T, Sawai H, Hatsu M, Yamada K (2007) Metabolic engineering of Corynebacterium glutamicum for cadaverine fermentation. Biosci Biotechnol Biochem 71:2130–2135
Okano K, Zhang Q, Yoshida S, Tanaka T, Ogino C, Fukuda H, Kondo A (2010) D-lactic acid production from cellooligosaccharides and β-glucan using genetically modified L-lactate dehydrogenase gene-deficient and endoglucanase-secreting Lactobacillus plantarum. Appl Microbiol Biotechnol 85:643–650
Okino S, Noburyu R, Suda M, Jojima T, Inui M, Yukawa H (2008) An efficient succinic acid production process in a metabolically engineered Corynebacterium glutamicum strain. Appl Microbiol Biotechnol 81:459–464
Paradis FW, Warren RA, Kilburn DG, Miller RC (1987) The expression of Cellulomonas fimi cellulase genes in Brevibacterium lactofermentum. Gene 61:199–206
Peyret JL, Bayan N, Joliff G, Gulik-Krzywicki T, Mathieu L, Schechter E, Leblon G (1993) Characterization of the cspB gene encoding PS2, an ordered surface-layer protein in Corynebacterium glutamicum. Mol Microbiol 9:97–109
Sakai S, Tsuchida Y, Nakamoto H, Okino S, Ichihashi O, Kawaguchi H, Watanabe T, Inui M, Yukawa H (2007) Effect of lignocellulose-derived inhibitors on growth of and ethanol production by growth-arrested Corynebacterium glutamicum R. Appl Environ Microbiol 73:2349–2353
Settles AM, Martienssen R (1998) Old and new pathways of protein export in chloroplasts and bacteria. Trends Cell Biol 8:494–501
Somogyi M (1952) Notes on sugar determination. J Biol Chem 195:19–23
Tateno T, Fukuda H, Kondo A (2007) Direct production of L-lysine from raw corn starch by Corynebacterium glutamicum secreting Streptococcus bovis alpha-amylase using cspB promoter and signal sequence. Appl Microbiol Biotechnol 77:533–541
Tateno T, Okada Y, Tsuchidate T, Tanaka T, Fukuda H, Kondo A (2009) Direct production of cadaverine from soluble starch using Corynebacterium glutamicum coexpressing α-amylase and lysine decarboxylase. Appl Microbiol Biotechnol 82:115–121
US Department of Energy (2004) Top value added chemicals from biomass, volume I—results of screening for potential candidates from sugars and synthesis gas. T. Werpy and G. Petersen, the Pacific Northwest National Laboratory (PNNL)
Yao W, Deng X, Zhong H, Liu M, Zheng P, Sun Z, Zhang Y (2009) Double deletion of dtsR1 and pyc induce efficient L-glutamate overproduction in Corynebacterium glutamicum. J Ind Microbiol Biotechnol 36:911–921
Acknowledgments
This work was partially supported by a Special Coordination Funds for Promoting Science and Technology, Creation of Innovation Centers for Advanced Interdisciplinary Research Areas (Innovative Bioproduction Kobe) from the Ministry of Education, Culture, Sports, Science and Technology of Japan. This work was also partially supported by a Grant-in-Aid for JSPS Fellows (20859 to T. Tateno).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Tsuchidate, T., Tateno, T., Okai, N. et al. Glutamate production from β-glucan using endoglucanase-secreting Corynebacterium glutamicum . Appl Microbiol Biotechnol 90, 895–901 (2011). https://doi.org/10.1007/s00253-011-3116-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-011-3116-7