Abstract
The yeast Yarrowia lipolytica is one of the most intensively studied “non-conventional” yeast species. Its ability to secrete various organic acids, like pyruvic (PA), citric, isocitric, and alpha-ketoglutaric (KGA) acid, in large amounts is of interest for biotechnological applications. We have studied the effect of the alpha-ketoglutarate dehydrogenase (KGDH) complex on the production process of KGA. Being well studied in Saccharomyces cerevisiae this enzyme complex consists of three subunits: alpha-ketoglutarate dehydrogenase, dihydrolipoyl transsuccinylase, and lipoamide dehydrogenase. Here we report the effect of overexpression of these subunits encoding genes and resulting increase of specific KGDH activity on organic acid production under several conditions of growth limitation and an excess of carbon source in Y. lipolytica. The constructed strain containing multiple copies of all three KGDH genes showed a reduced production of KGA and an elevated production of PA under conditions of KGA production. However, an increased activity of the KGDH complex had no influence on organic acid production under citric acid production conditions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Due to its ability to utilize a wide spectrum of different substrates, like hydrophobic substrates, ethanol, glycerol and glucose, and its capability to secrete high amounts of metabolites and proteins, the ascomycetous yeast Yarrowia lipolytica is an important microorganism with relevance for biotechnological applications. Protein expression, dimorphism, genetics, and physiology as well as hydrophobic substrate utilization and biotechnological applications were well reviewed (Barth and Gaillardin 1996, 1997; Barth et al. 2003; Fickers et al. 2005; Finogenova et al. 2005; Bankar et al. 2009). The yeast Y. lipolytica is unique in its ability to secrete high amounts of a broad range of organic acids (up to 250 g L−1) including intermediates of the tricarboxylic acid cycle (TCA), citric (CA), isocitric (ICA), and alpha-ketoglutaric acid (KGA), as well as pyruvic acid (PA). The production of these acids depends on the availability of the carbon source (e.g. plant oils, fats, glycerol, ethanol, molasses and starch hydrolysates, alkanes, and glucose) and nutrition factors, like N-source, thiamine, or the mineral salt components P, S or Mg. Under excess of carbon source nitrogen exhaustion triggers secretion of CA and ICA, while limitation by thiamine at low pH values results in the secretion of mainly alpha-ketoglutarate and pyruvate (Stottmeister et al. 1982; Barth and Gaillardin 1996, 1997; Aurich et al. 2003; Mauersberger et al. 2003; Finogenova et al. 2005; Kamzolova et al. 2008). Previous studies revealed that changes in enzyme activities of TCA enzymes can influence the amounts and ratios of produced organic acids in Y. lipolytica. Particularly with regard to the production of CA and ICA, the involvement of different TCA and glyoxylate cycle enzymes in the product formation was shown for selected mutants (Akiyama et al. 1973; Stottmeister et al. 1982; Ermakova et al. 1986; Finogenova et al. 1986, 1991, 2002, 2005; Il'chenko et al. 2002; Kamzolova et al. 2008). In previous studies, we have demonstrated that a high-level expression of aconitase (ACO) and isocitrate lyase (ICL) caused by amplification of the encoding genes and vice versa a loss of the ICL changes the CA/ICA ratio on different carbon sources (Förster et al. 2007a, b; Holz et al. 2009). Therefore we investigated the influence of further TCA and glyoxylate cycle enzymes with regard to the production of organic acids by Y. lipolytica. Here we present the results of our study on the effects of changes in activity of alpha-ketoglutarate dehydrogenase (KGDH) which catalyzes the oxidative decarboxylation of KGA to succinyl-coenzyme A and carbon dioxide. In Saccharomyces cerevisiae, KGDH is a mitochondrial macromolecular multienzyme complex consisting of three components: alpha-ketoglutarate dehydrogenase (Kgd1p), dihydrolipoyl transsuccinylase (Kgd2p), and lipoamide dehydrogenase (Lpd1p). Each of these components is present in multiple copies in this macromolecular complex (Ross et al. 1988; Repetto and Tzagoloff 1989, 1990, 1991; Huh et al. 2003). Whereas Kgd1p and Kgd2p are exclusive components of KGDH, Lpd1p forms also an essential component of the glycine decarboxylase and pyruvate dehydrogenase complexes (Pronk et al. 1996). Repetto and Tzagoloff (1991) examined the assembly of the KGDH complex and revealed that a regulated and balanced production of Kgd1p and Kgd2p is relevant for attaining the correct subunit stoichiometry during complex assembly.
In this study, we wanted to reveal how overexpression of all subunits together influences enzyme activity of the KGDH complex in Y. lipolytica. To understand whether the alpha-ketoglutarate dehydrogenase activity plays a role in organic acid production in Y. lipolytica we studied enzyme activity of KGDH and organic acid production in wild-type and transformant strains harboring multiple copies of all three genes.
Methods and materials
Strains, media and growth conditions
The Y. lipolytica strains H222 (MATA, wild type; Barth and Gaillardin 1996), the recipient strain H222-S4 (MATA ura3-302::pXPR2-ScSUC2; Mauersberger et al. 2001), the strain E150 (MATB his-1 leu2-270 ura3-302 xpr2-322; Barth and Gaillardin 1996), and the newly constructed KGD1, KGD2, LPD1 multicopy transformant H222-MH1 (MATA ura3-302::ScSUC2 ura3d4 KGD1mc, KGD2mc, LPD1mc, mc for multicopy) as well as for cloning the E. coli strain DH5αc were used in this study. For cultivation 500-ml shaking flasks were used. The strains were grown at 28 °C and 220 rpm in 20 to 30 ml complete medium YPD (Barth and Gaillardin 1996) or in 100 to 200-ml minimal medium Mg (Mauersberger et al. 2003), MpC [medium for citric acid production] (Mauersberger et al. 2003) or MpA [medium for alpha-ketoglutaric acid production] (Weissbrodt et al. 1989) containing (a) mineral salts: 5 g L−1 NH4Cl, 2 g L−1 KH2PO4, 1 g L−1 MgSO4 × 7 H2O, 10 mg L−1 FeSO4 × 7 H2O, 44 mg L−1 ZnSO4 × 7 H2O, 60 mg L−1 CaCl2; (b) trace elements: 0.04 mg L−1 CuSO4 × 5 H2O, 0.4 mg L−1 MnSO4 × 4 H2O, 0.2 mg L−1 FeCl3 × 6 H2O, 0.2 mg L−1 Na2MoO4 × 2 H2O, 0.4 mg L−1 ZnSO4 × 7 H2O, 0.1 mg L−1 KI and 0.5 mg L−1 H3BO3; (c) vitamin: 3.5 μg L−1 thiamine hydrochloride (0.05 mg L−1 in preculture) added in 0.5 μg L−1 aliquot in intervals of 24 h. The pH of the final medium M was 3.5–4.0. The concentration of the carbon sources was 1–2% (in Mg) and 10% (in Mp). During cultivation, the pH was adjusted using 10 N NaOH.
For cultivation in a bioreactor (1.4 L Multifors, Infors HT, Bottmingen Switzerland) the strains were grown at 30 °C in 600-ml production medium (Weissbrodt et al. 1989, modified) containing: (a) mineral salts: 6,17 g L−1 (NH4)2SO4, 2 g L−1 KH2PO4, 1 g L−1 MgSO4 × 7 H2O, 10 mg L−1 FeSO4 × 7 H2O, 44 mg L−1 ZnSO4 × 7 H2O, 60 mg L−1 CaCl2; (b) trace elements as described above; (c) vitamin: 0.020 mg L−1 thiamine hydrochloride. After a growth phase the production phase was started by changing pH from pH 5.0 to 3.8 and oxygen saturation rate from 50% to 10%. During cultivation the pH was regulated with 32% NH4OH. The concentration of glycerol was 12% at the beginning of fermentation.
Vector construction
Based on the multicopy integrative plasmid p64PT (Gerber 1999) the vectors p64KGD1, p64KGD2, and p64LPD1 were constructed for multiple integration of all three genes of alpha-ketoglutaric acid dehydrogenase (KGD1, KGD2, LPD1) in the yeast genome. Therefore p64PT was treated with ApaI and religated after elimination of the 2-kb ApaI-fragment containing the major part of pICL1D. The new vector, called p64T, served as basic vector for vector construction. DNA from Y. lipolytica strain E150 was used as template for PCR amplification (CombiZyme DNA Polymerase, Invitek GmbH, Berlin). A 5.77-kb fragment of the KGD1 gene with promoter and terminator regions was amplified by PCR with the primer pair KGD1for (ATATAAGATCTGCATGTACGTGTCATCTGTCC, creating the new underlined BglII restriction site on one end of the fragment) and KGD1rev (ACCACTCTACCTGCACAACC, located directly downstream of the BglII site in the terminator). The primer pair KGD2for (ATATAAGATCTATCGCTCCTGGTACTGTACG, creating the new underlined BglII restriction site on one end of the fragment) and KGD2rev (ATATAAGATCTATGTCCGACTACCACGATGC, creating the new underlined BglII restriction site on the other end of the fragment) were used for amplification of a 3.16-kb fragment of the KGD2 gene with promoter and terminator regions. A 3.78-kb fragment of the LPD1 gene with promoter and terminator regions was amplified with the primer pair LPD1for (ATATACATATGCTAACTCGGTCTCCTGGTTC, creating the new underlined NdeI restriction site on one end of the fragment) and LPD1rev (ATATAACGCGTGCTTAACACGCAGAACGTCG, creating the new underlined MluI restriction site on the other end of the fragment). The BglII-treated KGD1 and KGD2 fragments were each ligated with the purified vector fragment obtained from p64T after BglII digestion. The NdeI- and MluI-treated LPD1 fragment was ligated with the purified vector fragment obtained from p64T after NdeI and MluI digestion. For each vector plasmids without any amino acid exchange in the respective ORF were selected. These multicopy vectors p64KGD1, p64KGD2, and p64LPD1 contained the promoter truncated, defective ura3d4 allele as multicopy selection marker, a rDNA sequence as integration target sequence and the complete expression cassette for either KGD1, KGD2, or LPD1, as previously described for the respective ICL1 multicopy vectors (Juretzek et al. 2001; Förster et al. 2007a, b).
Transformation
Integrative transformation of the Y. lipolytica strain H222-S4 was performed by the lithium acetate method (Barth and Gaillardin 1996). The plasmids p64KGD1, p64KDG2, and p64LPD1 were digested by SacII before transformation.
Chromosomal DNA preparation and southern blotting
Chromosomal DNA was prepared according to Hoffmann and Winston (1987) and digested with XbaI. For Southern blotting, 800-ng digested and separated DNA samples were analyzed after vaccum blotting to Hybond-N membranes using the Gene ImagesTM CDP-StarTM Detection Module and HyperfilmTM ECLTM (GE Healthcare Life Sciences, Germany). The KGD1 DNA probe containing the Van91I fragment of the KGD1 gene (1.7 kb) was prepared from plasmid p64KGD1, KGD2 DNA probe from p64KGD2 (2.5-kb KGD2-containing BglII/XbaI fragment), and LPD1 DNA probe from p64LPD1 (3.5-kb LPD1-containing MluI/XbaI fragment). All DNA probes were labeled with flourescein using the Gene ImagesTM Random-Prime Labelling Module (GE Healthcare Life Sciences, Germany).
Enzyme assay
Activity of alpha-ketoglutaric acid dehydrogenase was determined in cell-free supernatant fractions by a spectrophotometric assay according to Brown and Perham (1976). Cell-free supernatants were prepared as described in Holz et al. (2009), using 0.5 M potassium phosphate buffer, pH 7.4, as cell disruption buffer. Protein content was determined by the method of Bradford (1976), using bovine serum albumin as standard.
Analytical determination of organic acids
The determination of organic acid content in cell-free samples of the fermentation broth was carried out using the DX320 ion chromatography system (column IonPac AS19, Dionex, Sunnyvale, CA, USA). The ion separation was carried out under following conditions: sample injection volume, 10 μl; eluent flow rate, 0.3 ml min−1; KOH eluent gradients, 5-mM hold for 25 min, 5–38 mM in 9 min, 38-mM hold for 2 min, 38–5 mM in 6 min. The organic acids were quantified using calibration curves with the software Chromeleon 6.8 (Dionex, Sunnyvale).
Determination of glycerol was performed using a commercial enzymatic test kit (No. 10148270035, R-Biopharm, Germany).
Results
Bioinformatical analysis of alpha-ketoglutarate dehydrogenase genes in Y. lipolytica
In S. cerevisiae, as well as in Escherichia coli, the alpha-ketoglutarate dehydrogenase complex consists of three subunits (Repetto and Tzagoloff 1989). Alpha-ketoglutarate dehydrogenase subunit is encoded by the gene KGD1 (YIL125w) and dihydrolipoyl transsuccinylase by KGD2 (YDR148c; Repetto and Tzagoloff 1989, 1990, 1991; yeastgenome.org). Lpd1p, which is also part of pyruvate dehydrogenase complex and glycine decarboxylase in S. cerevisiae, is encoded by the gene LPD1 (YFL018c; Pronk et al. 1996; yeastgenome.org). These three subunits are characterized as mitochondrial-located proteins (Huh et al. 2003; Reinders et al. 2007).
In Y. lipolytica, three genes with highly similar sequences to KGD1, KGD2 and LPD1 were found after analysis of Y. lipolytica genome database (Consortium Génolevures: http://cbi.labri.fr/Genolevures/; Casaregola et al. 2000; Dujon et al. 2004). The intron-containing gene YALI0E33517g encodes a putative Kgd1p gene product of 1,004 amino acids, exhibiting 67% homology to ScKgd1. A putative Kgd2p gene product of 447 amino acids is encoded by the gene YALI0E16929g, which shows 60% homology to ScKgd2. Furthermore, there is the intron-containing gene YALI0D20768g encoding a putative gene product of 499 amino acids and exhibiting a 63% homology to ScLpd1. According to the specific analysis program MitoProt (Claros and Vincens 1996), all deduced proteins contain N-terminal mitochondrial-targeting signal sequences of 29 (Kgd1p), 61 (Kgd2p), or 32 amino acids (Lpd1p). These Y. lipolytica genes were chosen for the overexpression of alpha-ketoglutarate dehydrogenase in Y. lipolytica and referred to as KGD1, KGD2 and LPD1 hereafter.
Overexpression of alpha-ketoglutarate dehydrogenase
To examine whether an increase of KGDH expression level influences the enzyme activity and the production of organic acids in Y. lipolytica, a recombinant Y. lipolytica strain was constructed containing multiple copies of KGD1, KGD2 and LPD1. As described in Material and methods, the integrative multicopy vectors p64KGD1, p64KGD2, and p64LPD1 were constructed by integration of the PCR-amplified ORF of the appropriate gene as well as 5′ upstream and 3′ downstream areas of approximately 1 kb each in the recipient vector p64T. For transformation in the recipient Y. lipolytica strain H222-S4 (ura3-302), these vectors were linearized by SacII. Afterwards, the prototrophic transformant H222-MH1 was obtained. Southern blot analysis evidenced the multiple integration of each vector, additionally to the single genomic copies of KGD1, KGD2 and LPD1 which are present in all strains (Fig. 1a, b). Due to similar intensities of detected vector-related signals we assume that an equal copy number of each plasmid was integrated in the genome of recipient strain H222-S4.
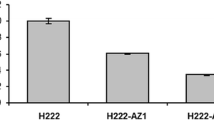
Southern blot analysis (A and B) and KGDH activity (C) of Y. lipolytica wild-type strain H222 and the KGD1, KGD2, LPD1 multicopy integrative transformant H222-MH1 evidenced high-copy integration of the KGD1, KGD2, and LPD1 containing vector in the multicopy transformant. Genomic DNA was digested with XbaI and probed with specific fragments of KGD1 (A), KGD2 and LPD1 (B). 1 Molecular weight standard, λ-DNA EcoRI/HindIII digested, 2 H222, 3 H222-MH1. 16.2-kb fragment of single copy genomic KGD1 (a); 8.5-kb KGD1 containing fragment of the integrated vector p64KGD1 (b); 5.1-kb fragment of single copy genomic KGD2 (d); 2.7-kb KGD2 containing fragment of the integrated vector p64KGD2 (f); 8.0-kb fragment of single copy genomic LPD1 (c); 4.1-kb LPD1 containing fragment of the integrated vector p64LPD1 (e). C KGDH activity during cultivation in minimal medium Mg with 2% glucose in wild-type strain H222 and KGD1, KGD2, LPD1 multicopy transformant H222-MH1 after 8 h of cultivation in shaking flasks
Furthermore, alpha-ketoglutarate dehydrogenase activity was determined in the transformant H222-MH1 and wild-type strain H222 to prove whether the multiple integration of KGD1, KGD2 and LPD1 have an influence on KGDH expression level. The strains were cultivated on 2% glucose or glycerol as carbon sources in minimal medium Mg under conditions for normal growth (no thiamine-exhaustion). Measured KGDH activities after 8 h of cultivation are shown in Fig. 1c. During growth in glucose 1.8- to 2.1-fold higher KGDH activities in transformant H222-MH1 were determined in comparison to the wild-type strain (Fig. 1c). The highest KGDH activities were measured in the early growth phase between 8 and 14 h for H222-MH1 and the wild-type strain (0.2 U mg−1 in H222, 0.4 U mg−1 in H222-MH1). However, the maximal KGDH activities of glycerol-grown cells were comparable to the activities in glucose-cells of H222-MH1 and H222, respectively.
Organic acid production
To examine whether the elevated KGDH expression level also influences the production of organic acids in Y. lipolytica, the KGD1, KGD2, LPD1 multicopy transformant H222-MH1, and the wild-type strain H222 were cultivated in shaking flasks and in bioreactors under the conditions for CA or KGA production. The production of KGA was initiated by thiamine-exhaustion (starting concentration 20 μg L−1 in bioreactors) under excess of the carbon source glycerol (Weissbrodt et al. 1989). In Fig. 2, a normal course of KGA production during cultivation in bioreactors is shown. KGA and PA production started when cells passed from logarithmic to stationary growth phase after shift of pH to 3.8 and pO2 to 10%. The production of PA rose faster than KGA production at the beginning of organic acid production, but after 46 h of cultivation PA amounts declined until the end of fermentation. The most striking results of these KGA production experiments were the reduction of produced KGA and the increase of produced PA amounts for multicopy transformant H222-MH1 in contrast to the wild-type strain H222. KGA amounts up to 97 g L−1 could be determined in the cultivation broth of H222 and up to 72 g L−1 for the transformant H222-MH1 after 114 h of fermentation (Fig. 2). The volumetric rate of product formation (QKGA) was reduced to 76% (0.9 g L−1 h−1 for H222 and 0.7 g L−1 h−1 for H222-MH1) and the specific rate of product formation (qKGA) for KGA to 74% in the transformant (0.035 g g−1 h−1) compared to the wild-type strain (0.047 g g−1 h−1). Furthermore, PA production was increased up to 128% in H222-MH1 compared to H222. We determined PA amounts of 66 g L−1 and 3.95 g g−1 for H222-MH1 and 52 g L−1 and 3.06 g g−1 for H222. The PA amounts detected in the cultivation broth of H222-MH1 rose faster at the beginning of fermentation and declined slower until the end than these of H222. At the end of fermentation, the detected PA in the cultivation broth of H222-MH1 was still 1.7 times higher than PA amounts of H222. We also determined KGDH activity during cultivation in bioreactor. In Fig. 3, the specific KGDH activity of H222 and H222-MH1 after 12 (growth phase) and 61 h (production phase) of fermentation are shown. Comparable to the detected KGDH activities under normal growth conditions (medium Mg) H222-MH1 developed 1.7 to 1.9 times higher activities than H222. After 61 h of cultivation, specific KGDH activities in both strains were approximately 5 times higher than KGDH activities after 12 h.
Kinetics of organic acid production (KGA and PA) for the wild-type strain H222 and the KGD1, KGD2, LPD1 multicopy transformant H222-MH1 with glycerol as substrate. Cultivation in 1.4-L bioreactors with 600 ml production medium for AKG production. Arrow indicates shift of pH and pO2 values, carried out after 12 h cultivation associated with start of KGA and PA accumulation
Apart from the influence on KGA production, we also examined whether there is any influence on CA and ICA production by the elevated KGDH expression level. Wild-type strain H222 and transformant H222-MH1 were cultivated in shaking flasks under the condition of CA and ICA production (medium MpC) from excess of the carbon source (10%) initiated by N-exhaustion (Mauersberger et al. 2003; Förster et al. 2007a, b; Holz et al. 2009). Comparable to our previous studies a CA/ICA ratio of 89–90% CA and 10–11% ICA on glucose as carbon source was detected for the wild-type strain H222. Maximal CA amounts of 42 to 45 g L−1 and 5.6 to 6.1 g L−1 ICA were produced by H222 after 216 to 240 h of cultivation. Transformant H222-MH1 showed comparable amounts of CA and ICA (42 to 44 g L−1 CA and 4.6 to 5.3 g L−1 ICA) and CA/ICA ratio (89–90%) after 216 to 264 h of cultivation.
Discussion
Overexpression of alpha-ketoglutarate dehydrogenase
In this study, we have demonstrated that the gene products of KGD1 (YALI0E33517g), KGD2 (YALI0E16929g), and LPD1 (YALI0D20768g) showing the highest homology to the S. cerevisiae alpha-ketoglutarate dehydrogenase genes ScKGD1 (YIL125w), ScKGD2 (YDR148c), and ScLPD1 (YFL018c) are obviously encoding a KGDH complex functioning in the mitochondrial TCA cycle in Y. lipolytica. The increased copy numbers of these three genes in the multicopy transformant H222-MH1 obtained in this study caused a 1.7- to 2.1-fold increase in the total KGDH activity due to a gene–dose effect, thus leading to the conclusion that these genes actually encode the KGDH complex. In previous studies, the overexpression of Y. lipolytica genes encoding aconitase (ACO1) and isocitrate lyase (ICL1) through integrative transformation of the gene-containing multicopy plasmid resulted in 8 to 15 times higher enzyme activities (Förster et al. 2007b; Holz et al. 2009). As described in Juretzek et al. (2001), at least 10–12 copies of the defective ura3d4 marker gene, which is part of the integrative plasmid, are necessary for full complementation of the ura3 gene disruption in the recipient strain. In contrast to the overexpression of ACO1 and ICL1, KGDH overexpression requires a simultaneous overexpression of the genes KGD1, KGD2 and LPD1 being located on separate plasmids. For complementation of the uracil auxotrophy a lower copy number of each multicopy plasmid-containing ura3d4 is sufficient which leads to a lower increase in enzyme activity in comparison to previous overexpression experiments. Furthermore, the complete size of KGD1, KGD2 and LPD1 promoter areas in Y. lipolytica is not known until now. In this study, a 5′ upstream region harboring the putative promoters with a length of approximately 1 kb was chosen for overexpression of KGDH genes. In previous studies it was shown that 5′ upstream regions of approximately 1 kb can be sufficient for gene expression in Y. lipolytica, e. g., for ACO1 (Holz et al. 2009) or XPR2 (Nicaud et al. 1989), but it was also demonstrated for ICL1 that larger upstream regions are necessary for full expression (Juretzek et al. 2001). To achieve a possible improvement of expression of KGDH genes, an intensive study of their promoter elements would be necessary. Another possible explanation for the only slight increase of KGDH activity would be an unbalanced rate of produced KGDH subunits. As described in Repetto and Tzagoloff (1990, 1991), the assembly of the KGDH complex seems to be dependent on a regulated and balanced production of Kgd1p and Kgd2p and the correct subunit stoichiometry during complex assembly. They revealed that overproduction of Kgd2p relative to Kgd1p resulted in a predominance of incompletely assembled complexes leading to a decrease of KGDH activity whereas an excess of Kgd1p leads to increased enzyme activity. It is not clear if this assembly mechanism in Y. lipolytica is similar to that in S. cerevisiae, but it should still be considered that an overexpression of KGD1 and KGD2 by integration of separate plasmids in the genome of Y. lipolytica may cause an imbalance in Kgd1p and Kgd2p molecule amounts and thus might also lead to a disturbed complex assembly and effects on enzyme activity.
Organic acid production
Furthermore, we demonstrated in this study that the selective increase in KGDH activity in KGD1, KGD2, LPD1 multicopy transformant Y. lipolytica H222-MH1 significantly changes the produced amounts of KGA and PA. In comparison to wild-type strain Y. lipolytica H222, the KGA production was decreased by 25.8%, but PA production was increased 1.3-fold. The changes in organic acid production observed in this study are possibly related to an influence of the thiamine availability on account of the overproduction of thiamine-dependent enzyme complex KGDH in H222-MH1. As described in several publications dealing with KGA production in Y. lipolytica, this yeast is unable to synthesize thiamine and therefore requires the addition of the vitamin to the medium. Under conditions of thiamine deficit, Y. lipolytica produces KGA and PA. This effect is traced to the thiamine-dependence of the PA- and KGA-metabolizing enzymes pyruvate dehydrogenase (PDH) and KGDH. A deficit of thiamine presumably leads to a reduction of enzyme activity and thus to an overflow of PA and KGA (Stottmeister et al. 1982; Barth and Gaillardin 1996; Kim 1999; Chernyavskaya et al. 2000; Mauersberger et al. 2003; Finogenova et al. 2005). In this study, the overexpression of KGDH genes KGD1, KGD2 and LPD1 along with an enhanced amount of functional KGDH molecules possibly caused a consumption of thiamine in favor of the KGDH complex molecules and to the disadvantage of the PDH complex molecules. Consequently, the amount of produced PA increased due to its decreased metabolization by PDH and the amount of produced KGA decreased possibly due to the minimized carbon flux from PA to KGA and due to the increased specific activity of KGDH complex. In the yeast Torulopsis glabrata, it was already shown that large amounts of PA can be accumulated, when the PA-converting enzymes PDH, pyruvate decarboxylase (PDC), and pyruvate carboxylase (PC) are limited due to the multi-vitamin auxotrophy of T. glabrata (Hua et al. 1999; Miyata and Yonehara 1999; Li et al. 2001; Huang et al. 2006; Zhang et al. 2009). However, carbon flux in T. glabrata can be redistributed from PA to KGA through manipulating the specific activity of related enzymes by changing the contents of metabolic co-factors thiamine and biotin or by overexpression of the PDC1 gene (Huang et al. 2006; Liu et al. 2007; Zhang et al. 2009). In Y. lipolytica, there was hitherto a lot of emphasis on improving KGA production. Several groups were able to increase KGA production by isolation of suitable mutants and by varying cultivation conditions (thiamine content, carbon source, pH, pO2, nitrogen concentration) (Chernyavskaya et al. 2000; Il'chenko et al. 2002; Finogenova et al. 2005). But depending on the carbon sources used in cultivation-varying amounts of PA were detected. A maximal PA production of 50–61 g L−1 and a proportion of produced organic acid of up to 90% PA was described for Y. lipolytica strains cultivated with glucose or glycerol as carbon sources and whereas cultivation on n-alkane- and ethanol-containing media results in a prevailing production of KGA. (Stottmeister et al. 1982; Stottmeister and Hoppe 1991; Kim 1999; Chernyavskaya et al. 2000; Il'chenko et al. 2002; Finogenova et al. 2005). The Y. lipolytica strain H222-MH1 obtained in this study is due to the reduced KGA production obviously not suitable for an improvement of KGA production, but for a further focus on PA production with Y. lipolytica, this strain could be interesting because of the increased PA amounts. However, the main problems concerning PA production in Y. lipolytica are the re-uptake of produced PA during cultivation and the efficient manipulation of PA-converting enzymes. As described in this study, the overproduction of KGDH complex resulted in an increase of produced PA amounts, but KGDH overproduction might be improved by optimization of KGD1 and KGD2 copy numbers, the ratio of Kgd1p and Kgd2p molecules and variation of promoter lengths and thus may result in an enhanced shift of organic acid production from KGA to PA. On the other hand, research should be focussed on the enzymes PDH and PC which convert PA to acetyl coenzyme A and oxaloacetate. In T. glabrata, PDH and PC, but also PDC which seems not to be present in Y. lipolytica (no analogous gene was found in Y. lipolytica genome database), play an important role for PA production and can be influenced by variation of thiamine (PDH) and biotin (PC) contents (Hua et al. 1999; Miyata and Yonehara 1999; Li et al. 2001; Huang et al. 2006; Zhang et al. 2009). It must be regarded that Y. lipolytica shows thiamine auxotrophy, but in contrast to T. glabrata it is prototroph for biotin (Barth and Gaillardin 1997; Chernyavskaya et al. 2000; Il'chenko et al. 2002; Finogenova et al. 2005). Whereas PDH activity could be affected by optimization of thiamine contents, a change of PC activity might be only possible by manipulation of the PC-encoding gene PYC1 (YALI0C24101g). Flores and Gancedo (2005) already reported the cloning and characterization of the PC-encoding gene PYC1 in Y. lipolytica and the effects of PYC1 deletion on growth. It was shown that the absence of PC activity does not inhibit growth in glucose-ammonium medium, but organic acid production was not examined until now.
We also demonstrated in this study that the selective increase in KGDH activity in KGD1, KGD2, LPD1 multicopy transformant Y. lipolytica H222-MH1 does not change the produced amounts of CA, ICA and CA/ICA ratios with glucose or glycerol as carbon sources in comparison to the wild-type strain H222. In the study of Il'chenko et al. (2002), central metabolism enzymes of TCA and glyoxylate cycles under conditions of KGA and CA production were determined. Only low activities of KGDH were described on ethanol, glucose, and n-alkanes as carbon sources under nitrogen deficiency. A doubling of KGDH activity under these conditions might result in a still very low activity and therefore does not affect CA and ICA production significantly. For a better understanding of the processes in cells of Y. lipolytica H222-MH1 under N-limitation conditions, enzyme activities of further TCA and glyoxylate enzymes should be measured in further studies.
References
Akiyama S, Suzuki T, Sumino Y, Nakao Y, Fukuda H (1973) Induction and citric acid productivity of fluoroacetate-sensitive mutant strains of Candida lipolytica. Agr Biol Chem 37(4):879–884, 885–888
Aurich A, Förster A, Mauersberger S, Barth G, Stottmeister U (2003) Citric acid production from renewable resources by Yarrowia lipolytica. Biotechnol Adv 21:454–455
Bankar AV, Kumar AR, Zinjarde SS (2009) Environmental and industrial applications of Yarrowia lipolytica. Appl Microbiol Biotechnol 84:847–865
Barth G, Gaillardin C (1996) Yarrowia lipolytica. In: Ke W (ed) Nonconventional yeasts in biotechnology. Springer-Verlag, Berlin, pp 313–388
Barth G, Gaillardin C (1997) Physiology and genetics of the dimorphic fungus Yarrowia lipolytica. FEMS Microbiol Rev 19(4):219–237
Barth G, Beckerich JM, Dominguez A, Kerscher S, Ogrydziak D, Titorenko V, Gaillardin C (2003) Functional genetics of Yarrowia lipolytica. In: de Winde JH, Hohmann S (eds) Functional genetics of industrial yeasts. Springer-Verlag, Berlin, pp 236–239
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Brown JP, Perham RN (1976) Selective inactivation of the transacylase components of the 2-oxo acid dehydrogenase multienzyme complexes of Escherichia coli. Biochem J 155(2):419–427
Casaregola S, Neuveglise C, Lepingle A, Bon E, Feynerol C, Artiguenave F, Wincker P, Gaillardin C (2000) Genomic exploration of the hemiascomycetous yeasts: 17. Yarrowia lipolytica. FEBS Lett 487(1):95–100
Chernyavskaya OG, Shishkanova NV, Il'chenko AP, Finogenova TV (2000) Synthesis of alpha-ketoglutaric acid by Yarrowia lipolytica yeast grown on ethanol. Appl Microbiol Biotechnol 53(2):152–158
Claros MG, Vincens P (1996) Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem 241(3):779–786
Dujon B, Sherman D, Fischer G, Durrens P, Casaregola S, Lafontaine I, De Montigny J, Marck C, Neuveglise C, Talla E et al (2004) Genome evolution in yeasts. Nature 430(6995):35–44
Ermakova IT, Shishkanova NV, Melnikova OF, Finogenova TV (1986) Properties of Candida lipolytica mutants with the modified glyoxylate cycle and their ability to produce citric and isocitric acid. I. Physiological, biochemical and cytological characteristics of mutants grown on glucose or hexadecane. Appl Microbiol Biotechnol 23:372–377
Fickers P, Benetti PH, Wache Y, Marty A, Mauersberger S, Smit MS, Nicaud JM (2005) Hydrophobic substrate utilisation by the yeast Yarrowia lipolytica, and its potential applications. FEMS Yeast Res 5(6–7):527–543
Finogenova TV, Shishkanova NV, Ermakova IT, Kataeva IA (1986) Properties of Candida lipolytica mutants with the modified glyoxylate cycle and their ability to produce citric and isocitric acid. II. Synthesis of citric and isocitric acid by Candida lipolytica mutants and peculiarities of their enzyme systems. Appl Microbiol Biotechnol 23:378–383
Finogenova TV, Shishkanova NV, Fausek EA, Eremia SS (1991) Biosynthesis of isocitric acid from ethanol by yeasts. Appl Microbiol Biotechnol 36:231–235
Finogenova TV, Kamzolova SV, Dedyukhina EG, Shishkanova NV, Il'chenko AP, Morgunov IG, Chernyavskaya OG, Sokolov AP (2002) Biosynthesis of citric and isocitric acids from ethanol by mutant Yarrowia lipolytica N 1 under continuous cultivation. Appl Microbiol Biotechnol 59(4–5):493–500
Finogenova TV, Morgunov IG, Kamzolova SV, Cherniavskaia OG (2005) Organic acid production by the yeast Yarrowia lipolytica: a review of prospects. Appl Biochem Microbiol 41:418–425
Flores CL, Gancedo C (2005) Yarrowia lipolytica mutants devoid of pyruvate carboxylase activity show an unusual growth phenotype. Eukaryot Cell 4(2):356–364
Förster A, Aurich A, Mauersberger S, Barth G (2007a) Citric acid production from sucrose using a recombinant strain of the yeast Yarrowia lipolytica. Appl Microbiol Biotechnol 75(6):1409–1417
Förster A, Jacobs K, Juretzek T, Mauersberger S, Barth G (2007b) Overexpression of the ICL1 gene changes the product ratio of citric acid production by Yarrowia lipolytica. Appl Microbiol Biotechnol 77(4):861–869
Gerber J (1999) Untersuchungen zur Optimierung des Elektronentransportsystems für die Cytochrom P450 katalysierte Biotransformation von Steroiden in Yarrowia lipolytica. Diplomarbeit, Institut für Mikrobiologie, TU Dresden
Hoffmann CS, Winston F (1987) A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 57(57):267–272
Holz M, Forster A, Mauersberger S, Barth G (2009) Aconitase overexpression changes the product ratio of citric acid production by Yarrowia lipolytica. Appl Microbiol Biotechnol 81(6):1087–1096
Hua Q, Yang C, Shimizu K (1999) Metabolic flux analysis for efficient pyruvate fermentation using vitamin-auxotrophic yeast of Torulopsis glabrata. J Biosci Bioeng 87(2):206–213
Huang HJ, Liu LM, Li Y, Du GC, Chen J (2006) Redirecting carbon flux in Torulopsis glabrata from pyruvate to alpha-ketoglutaric acid by changing metabolic co-factors. Biotechnol Lett 28(2):95–98
Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK (2003) Global analysis of protein localization in budding yeast. Nature 425(6959):686–691
Il'chenko AP, Cherniavskaia OG, Shishkanova NV, Finogenova TV (2002) Metabolism of Yarrowia lipolytica grown on ethanol under conditions promoting the production of alpha-ketoglutaric and citric acids: a comparative study of the central metabolism enzymes. Mikrobiologiia 71(3):316–322
Juretzek T, Le Dall M, Mauersberger S, Gaillardin C, Barth G, Nicaud J (2001) Vectors for gene expression and amplification in the yeast Yarrowia lipolytica. Yeast 18(2):97–113
Kamzolova SV, Finogenova TV, Morgunov IG (2008) Microbial production of citric and isocitric acids from sunflower oil. Food Technol Biotechnol 46:51–59
Kim E (1999) Effect of thiamine on the by-products formation by Yarrowia lipolytica. Biotechnol Bioproc Eng 4:185–188
Li Y, Chen J, Lun SY (2001) Biotechnological production of pyruvic acid. Appl Microbiol Biotechnol 57(4):451–459
Liu L, Li Y, Zhu Y, Du G, Chen J (2007) Redistribution of carbon flux in Torulopsis glabrata by altering vitamin and calcium level. Metab Eng 9(1):21–29
Mauersberger S, Wang HJ, Gaillardin C, Barth G, Nicaud JM (2001) Insertional mutagenesis in the n-alkane-assimilating yeast Yarrowia lipolytica: generation of tagged mutations in genes involved in hydrophobic substrate utilization. J Bacteriol 183(17):5102–5109
Mauersberger S, Kruse K, Barth G (2003) Induction of citric acid/isocitric acid and α-ketoglutaric acid production in the yeast Yarrowia lipolytica. In: Wolf KH, Breunig K, Barth G (eds) Non-conventional yeasts in genetics, biochemistry and biotechnology. Practical protocols Springer, Berlin, pp 393–400
Miyata R, Yonehara T (1999) Breeding of high-pyruvate-producing Torulopsis glabrata with acquired reduced pyruvate decarboxylase. J Biosci Bioeng 88(2):173–177
Nicaud JM, Fabre E, Gaillardin C (1989) Expression of invertase activity in Yarrowia lipolytica and its use as a selective marker. Curr Genet 16(4):253–260
Pronk JT, Yde Steensma H, Van Dijken JP (1996) Pyruvate metabolism in Saccharomyces cerevisiae. Yeast 12(16):1607–1633
Reinders J, Wagner K, Zahedi RP, Stojanovski D, Eyrich B, van der Laan M, Rehling P, Sickmann A, Pfanner N, Meisinger C (2007) Profiling phosphoproteins of yeast mitochondria reveals a role of phosphorylation in assembly of the ATP synthase. Mol Cell Proteomics 6(11):1896–1906
Repetto B, Tzagoloff A (1989) Structure and regulation of KGD1, the structural gene for yeast alpha-ketoglutarate dehydrogenase. Mol Cell Biol 9(6):2695–2705
Repetto B, Tzagoloff A (1990) Structure and regulation of KGD2, the structural gene for yeast dihydrolipoyl transsuccinylase. Mol Cell Biol 10(8):4221–4232
Repetto B, Tzagoloff A (1991) In vivo assembly of yeast mitochondrial alpha-ketoglutarate dehydrogenase complex. Mol Cell Biol 11(8):3931–3939
Ross J, Reid GA, Dawes IW (1988) The nucleotide sequence of the LPD1 gene encoding lipoamide dehydrogenase in Saccharomyces cerevisiae: comparison between eukaryotic and prokaryotic sequences for related enzymes and identification of potential upstream control sites. J Gen Microbiol 134(5):1131–1139
Stottmeister U, Hoppe K (1991) In: Ruttloff He (ed) Organische Genußsäuren. Lebensmittelbiotechnologie Akademie-Verlag, Berlin, pp 516–547
Stottmeister U, Behrens U, Weissbrodt E, Barth G, Franke-Rinker D, Schulze E (1982) Utilization of paraffins and other noncarbohydrate carbon sources for microbial citric acid synthesis. Z Allg Mikrobiol 22(6):399–424
Weissbrodt E, Gey M, Barth G, Weber H, Stottmeister U, Düresch R, Richter HP (1989) Verfahren zur Herstellung von 2-Oxoglutarsäure durch Hefen. Patent DD 267999
Zhang D, Liang N, Shi Z, Liu L, Chen J, Du G (2009) Enhancement of α-ketoglutarate production in Torulopsis glabrata: Redistribution of carbon flux from pyruvate to α-ketoglutarate. Biotechnol Bioproc Eng 14(2):134–139
Acknowledgements
This project was supported by the Government of North Rhine-Westphalia and co-financed by the European Union.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Holz, M., Otto, C., Kretzschmar, A. et al. Overexpression of alpha-ketoglutarate dehydrogenase in Yarrowia lipolytica and its effect on production of organic acids. Appl Microbiol Biotechnol 89, 1519–1526 (2011). https://doi.org/10.1007/s00253-010-2957-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-010-2957-9