Abstract
Laccases are blue multicopper oxidases with potential applications in environmental and industrial biotechnology. In this study, a new bacterial laccase gene of 1.32 kb was obtained from a marine microbial metagenome of the South China Sea by using a sequence screening strategy. The protein (named as Lac15) of 439 amino acids encoded by the gene contains three conserved Cu2+-binding domains, but shares less than 40% of sequence identities with all of the bacterial multicopper oxidases characterized. Lac15, recombinantly expressed in Escherichia coli, showed high activity towards syringaldazine at pH 6.5–9.0 with an optimum pH of 7.5 and with the highest activity occurring at 45 °C. Lac15 was stable at pH ranging from 5.5 to 9.0 and at temperatures from 15 to 45 °C. Distinguished from fungal laccases, the activity of Lac15 was enhanced twofold by chloride at concentrations lower than 700 mM, and kept the original level even at 1,000 mM chloride. Furthermore, Lac15 showed an ability to decolorize several industrial dyes of reactive azo class under alkalescent conditions. The properties of alkalescence-dependent activity, high chloride tolerance, and dye decolorization ability make the new laccase Lac15 an alternative for specific industrial applications.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Laccases (benzenediol:oxygen oxidoreductases, EC1.10.3.2), a family of blue multicopper oxidases, are capable of oxidizing a wide range of aromatic compounds, with concomitant reduction of molecular oxygen to water (Hoegger et al. 2006). The substrate range for laccases is greatly broadened by low-molecular-weight (MW) redox mediators, such as 2,2′-azino-bis (3-ethylbenzothazoline-6-sulfonate) (ABTS) and 1-hydroxybenzotriazole (HBT). In the presence of appropriate mediators, laccases can degrade several hardly degraded compounds, e.g., nonphenolic compounds (Bourbonnais and Paice 1990), polycyclic aromatic hydrocarbons (Pickard et al. 1999), and dye pollutants (Arora and Sharma, 2010). Laccases are thus regarded as potential candidates for biotechnological applications in industrial effluent detoxification, herbicide or pesticide elimination (Murugesan 2003; Wesenberg et al. 2003), organic synthesis (Mikolasch and Schauer 2009; Plonka and Grabacka 2006), and wood-based material production (Rodríguez Couto and Toca Herrera 2006; Witayakran and Ragauskas 2009), etc.
Laccases exist widely in higher plants, fungi, bacteria, and arthropods (Hoegger et al. 2006), and most of the industrial laccases, such as the DeniLite and the Zylite, are from fungi, especially basidiomycetes (Rodríguez Couto and Toca Herrera 2006). Unfortunately, nearly all fungal laccases lose their activities under alkaline conditions (Sharma et al. 2007), though they can recover to some extent under acidulous conditions after alkaline treatment (Xiao et al. 2003). Furthermore, fungal laccases are sensitive to chloride and lose most of the laccase activities at concentrations higher than 100 mM (Jimenez-Juarez et al. 2005). These disadvantages of fungal laccases make it an important work to screen for laccases bearing activity at high pH and high concentrations of chloride. Bacteria-derived laccases may be such ideal alternatives.
Analysis on prokaryotic genomes suggested that laccase-like enzymes may be widespread in bacteria (Alexandre and Zhulin 2000; Sharma et al. 2007). Laccase-like genes or activity have been found in several bacterial strains, including Bacillus halodurans (Ruijssenaars and Hartmans 2004), Bacillus licheniformis (Koschorreck et al. 2008), Bacillus subtilis (Martins et al. 2002), Escherichia coli (Roberts et al. 2001), γ-proteobacterium (Singh et al. 2007), Thermus thermophilus (Miyazaki 2005), and streptomycetes (Arias et al. 2003; Endo et al. 2003). However, few bacterial laccases have been biochemically characterized, and little is known about their applications in biotechnology industries (Arias et al. 2003; Koschorreck et al. 2009), albeit their applications in industrial processes are so potent. Identifying and characterizing new laccases from bacteria and evaluating their application potential will greatly help us better use them in industrial processes.
To obtain new laccases with ideal characteristics, a marine microbial metagenome of the South China Sea was constructed and screened using sequence strategy. A new bacterial laccase with alkalescence-dependent activity and excellent chloride tolerance was obtained and characterized. The potential of the enzyme in decolorization of industrial dyes was also evaluated.
Materials and methods
Microorganisms and agents
Escherichia coli EPI300 and pIndigoBAC-5 vector were obtained from Epicentre (Madison, WI, USA); E. coli DH5α and E. coli BL21(DE3) were from TransGen (Beijing, China); ABTS, 2,6-dimethoxyphenol (DMP) and syringaldazine were from Sigma-Aldrich (St. Louis, MO, USA). All chemicals and reagents were of analytical grade.
Construction of bacterial artificial chromosome (BAC) library and screening for laccase genes
Microbes were collected from the surface water of the South China Sea. High-MW DNA was prepared according to the protocol of Chu et al. (2008). DNA was partially digested with HindIII (NEB, Beijing, China) and separated by pulse field gel electrophoresis (Bio-Rad, Hercules, CA, USA). DNA fragments of 50–150 kb were recovered and ligated into pIndigoBAC-5 vector. The recombinant plasmids were transformed into E. coli EPI300 according to the manufacturer's protocol. The clones obtained were then placed in 384-well plates and stored at −70 °C.
A sequence screening strategy, based on the conserved region of laccases in copper-binding sites, was adopted to clone laccase genes as follows: Firstly, 384 clones in the same plate were cultured in a bottle of Luria–Bertani (LB) liquid medium, and mixed plasmids were extracted from the total culture and used as polymerase chain reaction (PCR) templates. Degenerate PCR primers of Cu1F (ACMWCKGTTCAYTGGCACGG) and Cu4R (TGNTCNAGNAWGTGRCARTG), which were designed based on the relatively conserved sequences coding for the copper binding sites I and IV in bacterial laccases (Fig. S1), were used to amplify DNA fragments of potential laccases (Hoegger et al. 2006). Plates with a fragment about 1,100 bp in PCR products were considered as positive plates. Subsequently, the same PCR reaction was performed for every clone of positive plates and those with a fragment of about 1,100 bp in PCR product were considered as positive clones.
Inverse PCR was used to obtain the full length laccase genes. Briefly, the positive plasmids in positive clones were fully digested with NdeI (TaKaRa, Dalian, China), self-ligated by T4 DNA ligase (TaKaRa) and then used as templates. The flanking sequences of the laccase DNA fragments were amplified using LA-Taq (TaKaRa) with inverse primers of D15IF (TACACTTGTGGATGCGGGTGAGAC) and D15IR (AGACCCTTCGCAACTTGTTCCCAT). The PCR products were ligated into pMD18-T vector (TaKaRa) and sequenced.
Sequence analysis of lac15
The ORF of lac15 was determined using the ORF Finder provided by the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). Similar sequence searching was performed using BlastP at NCBI. The module structure of the enzyme was analyzed with simple modular architecture research tool SMART (http://smart.embl-heidelberg.de/). The presence and location of the signal peptides in lac15 was estimated using the Neural Networks and Hidden Markov models trained on Gram-negative bacteria with SignalP3.0 (http://www.cbs.dtu.dk/services/SignalP/) (Bendtsen et al. 2004). Multiple sequence alignment of lac15 with other related laccase sequences was performed using Clustal X 2.0 and GENEDOC.
Heterologous expression of lac15 in E. coli
The open reading frame (ORF) of lac15 was amplified using the primer pair of AAACATATGAACAGGCGAGACTTCCTGG (NdeI site italicized) and AAACTCGAGTGCGACCTCCACCCAGGTCT (XhoI site italicized). Construction of plasmid pET22b–lac15 was performed according to Koschorreck et al. (2008). Escherichia coli BL21(DE3) cells carrying pET22b–lac15 were grown at 16 °C in 200 mL of LB medium containing 100 μg/mL ampicillin. Isopropyl-β-d-thiogalactoside (IPTG) at a final concentration of 1 mM was added into the culture of OD 600 ≈ 0.6 to induce enzyme expression. After an additional incubation for 16 h, the cells were collected by centrifugation. The pellets were resuspended in cold 20 mM Tris–HCl buffer (pH 7.9) containing 500 mM NaCl and 5 mM imidazole, disrupted by sonication, and then centrifuged at 30,000×g for 30 min. The supernatant was applied to Ni–NTA (Novagen, Darmstadt, Germany) affinity chromatography to purify the recombinant Lac15 (Lac15). The purified protein was stored at 4 °C.
Characterization of Lac15
The assay mixture consisted of 10 μL of appropriately diluted protein stock and 990 μL of 50 mM Na2HPO4–KH2PO4 buffer (pH 7.5) containing 100 μM syringaldazine (ε 525 = 65,000 M−1 cm−1) and 100 μM CuSO4. The reaction was initiated by adding 10 μL syringaldazine into the solution. The mixture was incubated at 45 °C for 5 min and then transferred into ice-water bath for 30 s. One activity unit (U) was defined as the amount of Lac15 required for oxidizing 1 μmol of syringaldazine per minute. Alternative substrates for measurement of laccase activity were ABTS (ε 420 = 36,000 M−1 cm−1) and DMP (ε 468 = 49,600 M−1 cm−1) at final concentrations of 0.5 and 2 mM, respectively. Reactions with heat-treated Lac15 were used as controls.
Protein concentration of Lac15 was determined following the Bradford method. The MW of Lac15 was estimated by sodium dodecyl sulfate (SDS) polyacrylamide gel electrophoresis (PAGE). The pH optimal for activity was evaluated at 45 °C in 50 mM sodium citrate buffer (4.5–5.5), Na2HPO4–KH2PO4 buffer (5.5–8.0), and Tris–HCl buffer (8.0–9.0). The stability against pH was determined by measuring the residual activities of Lac15 after incubation at 4 °C for 1 h in the aforementioned buffer at different pH values. Effect of temperature on the enzyme activity was measured by incubating Lac15 at pH 7.5 and a temperature range from 15 to 55 °C. Thermostability was determined by incubating Lac15 at various temperatures (15 to 55 °C) at pH 7.5 for 15 min.
Effects of Mg2+, Co2+, Mn2+, Zn2+, Ca2+, K+, NaN3, SDS, and EDTA on Lac15 activity were investigated by incubating Lac15 with each effector for 15 min at 4 °C prior to substrate syringaldazine addition. The laccase assays were carried out under the aforementioned conditions. Chloride effect was determined at concentrations ranging from 1 to 2,000 mM. Control was carried out under the conditions with Na2SO4, KCl, or without NaCl in the normal manner.
Decolorization of dyes
Two anthraquinone dyes: Reactive Brilliant Blue X-BR (maximal absorbance at 600 nm) and K-GR (600 nm) and four azo dyes: Reactive Deep Blue M-2GE (620 nm), Reactive Brilliant Orange K-7R (490 nm), Reactive Red KM-8B (520 nm), and KD-8B (550 nm) were representatively selected to evaluate the ability of Lac15 to decolorize industrial dyes. Enzymatic treatment of the dyes was performed at 45 °C in the presence or absence of 100 μM mediator (ABTS, methyl-syringate, or syringaldehyde) in 1 mL Na2HPO4–KH2PO4 buffer (pH 7.5) containing 50 μM of dyes and 5 U/L or 10 U/L Lac15. The decolorization ability of Lac15 was determined spectrophotometrically as the relative decrease of absorbance at each maximal absorbance wavelength of the dyes.
Nucleotide sequence accession number
The nucleotide sequence data has been disposited in the GenBank database with accession number of HM623889. The corresponding protein ID is ADM87301.
Results
Cloning of a bacterial laccase gene
The marine microbial metagenome library obtained consisted of about 20,000 clones, harboring 1.4 Gb DNA. To search for new laccase genes, we screened for DNA fragments of laccase genes by PCR based on conserved sequences of copper binding sites I and IV in laccases (Fig. S1). A fragment (designated as lac15p) about 1.1 kb in length was amplified from one colony named as pSB46D15 using primers of Cu1F and Cu4R.
Inverse PCR was adopted to obtain the flanking sequences adjacent to lac15p. A fragment of about 5 kb (designated as lac15rp) was amplified from the self-ligated products of the NdeI-treated pSB46D15 BAC plasmids. The full-length laccase ORF of 1,320 bp (lac15) was thus obtained by integrating lac15p with lac15rp.
Analysis of lac15 sequence
The ORF of lac15 encodes a putative protein of 439 amino acids with a predicted MW of 47,860 Da. The deduced amino acid sequence of Lac15 is the most similar to an uncharacterized hypothetical multicopper oxidase deduced from the genome of Roseobacter sp. MED193 (ZP_01057124) with 84% identity. Further analysis indicated that Lac15 has low sequence identities of less than 40% to all of the bacterial multicopper oxidases already characterized (Table S1), with 22% identity to the typical bacterial laccase from B. subtilis (CAB12449), 24% to that from E. coli (BAB96698), 26% and 21% to those from marine bacteria Marinomonas mediterranea (AAF75831), and Oceanobacillus iheyensis (BAC13302), respectively. Lac15 also shares low identity to the bacterial laccases of RL5 from a bovine rumen metagenome (CAK32503, 7%) and Lac591 from a mangrove soil metagenome (ACV83921, 25%). Potential bacterial laccase relatives for Lac15 were also found by searching against the database of environmental samples at NCBI using BlastP, and all of the hits were uncharacterized hypothetical proteins from marine metagenomes (Yooseph et al. 2007).
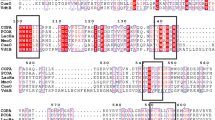
Module analysis suggested that Lac15 possesses three conserved copper-oxidase domains (Fig. 1), which are characteristics of laccases (Hoegger et al. 2006), with the Pfam database accession numbers of PF07732, PF00394, and PF07731. The first 22 amino acid residues (MNRRDFLVTTSAATLFPRIALA) at the N-terminus were recognized as a Tat (twin arginine translocation; Pfam: PF10518) signal sequence.
Multialignment of Lac15 with some other bacterial laccases. The protein sequences were retrieved from GenBank with the following accession numbers: uncultured bacterium of this study (ADM87301), B. subtilis (CAB12449), E. coli (BAB96698), M. mediterranea (AAF75831), O. iheyensis (BAC13302), RL5 from a bovine rumen metagenome (CAK32503), and Lac591 from a mangrove soil metagenome (ACV83921). Sequence alignment was performed using Clustal X 2.0 and GENEDOC. Four histidine-rich copper binding domains were indicated by full-length vertical boxes
Heterologous expression of Lac15
To analyze the biochemical properties of Lac15, its ORF absent of the 22-amino-acid-residue signal peptide was cloned into the plasmid pET-22b(+) and heterologously expressed in E. coli BL21(DE3). Lac15 was expressed mostly in inclusion body when induced at 30 °C, but partly soluble when induced at 16 °C. However, no soluble protein was obtained even at 16 °C when the signal peptide was retained. The pure protein showed an apparent MW of 49 kDa as determined by SDS-PAGE (Fig. 2), in accordance with the value of 47.9 kDa calculated based on the amino acid sequence with the His6 tag taken into account.
Characterization of Lac15
Lac15 was stable at pH values ranging from 5.5 to 9.0 and was the most stable at pH 7.0 (Fig. 3a). Specifically, it displayed the maximum activity at pH 7.5 (Fig. 3a) when using syringaldazine as substrate. On the other hand, the enzyme exhibited the highest activity at 45 °C and remained active even at 15 or 55 °C (Fig. 3b). The half-time of Lac15 was 72 min when incubated at pH 7.5 and 45 °C.
a Optimum pH (white circle) and pH stability (black circle) of Lac15 were investigated by measuring the enzyme activity at 45 °C with syringaldazine as substrate. b Optimum temperature (white circle) and themostability (black circle) for enzyme activity were measured in 50 mM Na2HPO4–KH2PO4 at pH 7.5, with syringaldazine as substrate. The results presented in the figures are average values calculated from triplicate technical repeats of measurements. Error bars represent ± 5 standard deviations
Effects of several potential inhibitors on Lac15 activity were also investigated. Under the test conditions, Lac15 was inactive in the presence of 10 μM EDTA, but retained 50% of the original activity in 2.5 mM NaN3. In contrast, SDS at a low concentration of 50 μM stimulated the enzyme activity up to 127%. The activity was gradually decreased as SDS concentration further increased, and retained 50% of the original value in the presence of 0.5 mM SDS.
Effects of cations on Lac15 activity were also assayed (Fig. 4). In the presence of 1 mM Mg2+ or Mn2+, the activity was stimulated up to 146% and 130%, respectively, compared to controls without the corresponding metal ions. Ca2+ and K+ had no obvious effect on Lac15 activity, whereas addition of Zn2+ or Co2+ dramatically reduced enzyme activity to 60% and 51%, respectively.
Effects of metal ions on the activity of Lac15. Sulphates are the donators of the mental ions. The activity was measured in 50 mM Na2HPO4–KH2PO4 at pH 7.5, supplemented with 0.1 mM CuSO4, at 45 °C with syringaldazine as substrate. The results presented in the figure are average values calculated from triplicate technical repeats of measurements. Error bars represent ± 5 standard deviations
Since chloride affects fungal laccase activities dramatically (Xu 1996), the effect of chloride on Lac15 activity was determined using NaCl as the chloride donator. Interestingly, the activity was enhanced to about 200% by NaCl at concentrations from 100 to 700 mM. Furthermore, the activity retained the original level even in the presence of 1,000 mM NaCl. As the NaCl concentration further increased, Lac15 activity decreased slowly, with the I 50 being 1,500 mM (Fig. 5).
Effect of chloride on the activity of Lac15. The activity was measured in 50 mM Na2HPO4–KH2PO4 at pH 7.5, supplemented with 0.1 mM CuSO4, at 45 °C with syringaldazine as substrate. The results presented in the figure are average values calculated from triplicate technical repeats of measurements. Error bars represent ± 5 standard deviations
The specificities of Lac15 towards syringaldazine and ABTS, under the corresponding optimum conditions, were 1.0 and 0.5 U mg−1, respectively. The K m and V max of Lac15 were 4.5 μM and 18.5 μmol min−1 mg−1, 123 μM, and 6.7 μmol min−1 mg−1, for syringaldazine and ABTS, respectively. However, no activity was detected for guaiacol.
Decolorization of artificial dyes
Six industrial dyes, including two of anthraquinone class and four of azo class, were used to evaluate the dye decolorization ability of Lac15. Under the test conditions, Lac15 was unable to decolorize two anthraquinone dyes either with or without the low MW mediators. In contrast, decolorization of the azo dyes was enhanced by addition of the mediators (except for K-7R and KM-8B, with ABTS as the mediator). Methyl-syringate was the best mediator for Lac15 to decolorize azo dyes. In the presence of methyl-syringate, 5 U/L decolorized 70% of K-7R and 60% of M-2GE after 1 h at 45 °C. However, less than 10% of K-7R was decolorized with ABTS as the mediator or without mediator (Fig. 6). K-7R and M-2GE in concentrations of 50 mM can be completely decolorized by 10 U/L Lac15 after 1 h at 45 °C.
Decolorization of different reactive azo dyes by Lac15. The assay was measured in 50 mM Na2HPO4–KH2PO4 at pH 7.5, supplemented with 0.1 mM CuSO4, at 45 °C for 1 h, with ABTS, methyl-syringate, or syringaldehyde as mediator. The results presented in the figure are average values calculated from triplicate technical repeats of measurements. Error bars represent ± 5 standard deviations
Discussion
Fungal laccases have been used commercially in textile industries (Rodríguez Couto and Toca Herrera 2006) and possess potential in more industrial applications. Compared to fungal laccases, bacteria-derived ones have some unique characteristics, such as activity and stability at high pH values (Sharma et al. 2007), which make bacterial laccases alternatives for some special fields where fungal laccases are inactive. Studies in recent years have suggested that laccases are widespread in the bacterial kingdom (Alexandre and Zhulin 2000; Sharma et al. 2007), but only a few bacterial laccases have been characterized so far. The diversity of marine microbes and the unique environment properties of the South China Sea potentially contribute to the diversity, novelty, and uniqueness of laccase genes. Laccases from marine microbes may exhibit some characteristics valuable for specific biotechnological application. Thus, we constructed and screened the microbial metagenomic library of the South China Sea to obtain new bacterial laccases.
A function-driven screening method based on enzymatic activity against chromogenic substrates is usually adopted to screen new laccases from microbial cultures. Recently, two bacterial laccases, RL5 and Lac591, were obtained by using this method from a bovine rumen microflora metagenome expression library (Beloqui et al. 2006) and a microbial metagenomic library from mangrove soil (Ye et al. 2010), respectively. Unfortunately, no laccase activity was detected from the microbial metagenomic library we constructed with syringaldazine or guaiacol as substrate (data not shown), which might be due to the failure of potential laccase genes in the library to express under the test conditions. Alternatively, a sequence screening strategy using conserved degenerate primers based on the copper-binding sites of laccases was adopted to amplify laccase genes in the library, and a new laccase gene (lac15) was screened out. Lac15 has low sequence identities (less than 40%) with all of the bacterial laccases characterized, though it harbors four conserved copper-binding domains characteristic of laccases. In addition, Lac15 showed excellent chloride tolerance but no guaiacol oxidizing activity compared to the two metagenome-derivered bacterial laccases mentioned above (Beloqui et al. 2006; Ye et al. 2010). The success of the sequence screening strategy demonstrated that it is an effective way to mine new bacterial laccase genes from microbial metagenomic libraries.
It is well known that fungal laccases catalyze the typical substrate syringaldazine at optimum pH around 5 (Jimenez-Juarez et al. 2005) and lose their activities at pH over 7. However, most bacterial laccases are active at pH 7.5–8.5. Recently, a bacterial laccase RL5 from a bovine rumen microflora metagenome expression library revealed a multipotent capacity to oxidize a wide range of substrates over an unusually broad range of pH values from 3.5 to 9.0, with the pH optimum at 4.0–5.0 for syringaldazine oxidation (Beloqui et al. 2006). Different from RL5, Lac15 showed excellent syringaldazine-catalyzing activity under alkalescent conditions and kept most of the highest activity even at pH 9. The alkalescence-dependent activity for Lac15 is similar to the bacterial laccase Lac591, which was stable in the pH range of 7.0–10.0, with the pH optimum at 8.0 for syringaldazine oxidation (Ye et al. 2010), and other bacterial laccases reported (Ruijssenaars and Hartmans 2004; Jimenez-Juarez et al. 2005; Singh et al. 2007). In addition, potential inhibitors exerted various effects on Lac15 activity. The chelating agent EDTA inhibited the activity, indicating that Lac15 is a metalloprotein, in accordance with the deduction that the metal ion Cu2+ is essential for bacterial laccase activity (Solano et al. 2001). In contrast, SDS at low concentrations enhanced Lac15 activity, similar to the laccases from Sinorhizobium meliloti (Castro-Sowinski et al. 2002) and Azospirillum lipoferum (Diamantidis et al. 2000). The activity enhancement might be attributed to a limited conformational change, exerted by SDS, capable of inducing a latent enzymatic form of more activity.
One of the major obstacles that have prevented rapid progress in the practical application of laccases in biotechnique industries is the requirement for activity under high concentrations of chloride (Jimenez-Juarez et al. 2005; Xu 1996). Recently, the activity of laccase Lbh1, found in B. halodurans C-125, was reported to be stimulated by NaCl at concentrations of 100–450 mM (Ruijssenaars and Hartmans 2004). The laccase PPO1 from M. mediterranea was also tolerant to NaCl at pH 5 with an I 50 value of 547 mM, and was entirely insensitive to 100 mM NaCl at pH 7 (Jimenez-Juarez et al. 2005). In the present study, Lac15 activity was enhanced twofold by NaCl at less than 700 mM and kept the original level even in the presence of 1,000 mM NaCl, with an I 50 of 1,500 mM. Thus, Lac15 showed excellent chloride tolerant ability compared to other bacterial laccases, not to mention fungal laccases. This advantage highlights the potential for Lac15 in several applications, such as biobleaching of paper pulp and dyestuffs processing, where most fungal laccases are unsuitable (Singh et al. 2009).
Laccases from fungi are well known to be capable of decolorizing synthetic dyes, but little is known about the decolorization potential of bacterial laccases. It has been reported that laccase-containing spores of B. subtilis SF and the corresponding immobilized spores could decolorize textile dyes such as Mordant Black 9 (Held et al. 2005). The laccase-positive bacterium Stenotrophomonas maltophilia AAP56 was also proven to be capable of decolorizing several synthetic dyes at some degrees (Galai et al. 2009). In this study, we demonstrated that the pure laccase Lac15, other than the laccase-containing strain, could completely decolorize reactive azo dyes K-7R and M-2GE, which unambiguously provided proof for the dye decorization ability of bacterial laccases. However, the laccase purified from a typical laccase-producing fungus, Ganoderma sp. 77002, could only decolorize 80% M-2GE and 63% K-7R in the optimal condition with HBT as mediator (Lin Zhu, personal communication).
In conclusion, we obtained a new bacterial laccase (Lac15) from a marine microbial metagenomic library and showed that Lac15 had an alkalescence-dependent activity, chloride tolerance, and an ability of dye decolorization.
References
Alexandre G, Zhulin LB (2000) Laccases are widespread in bacteria. Trends Biotechnol 18:41–42
Arias ME, Arenas M, Rodriguez J, Soliveri J, Ball AS, Hernandez M (2003) Kraft pulp biobleaching and mediated oxidation of a nonphenolic substrate by laccase from Streptomyces cyaneus CECT 3335. Appl Environ Microbiol 69:1953–1958
Arora DS, Sharma RK (2010) Ligninolytic fungal laccases and their biotechnological applications. Appl Biochem Biotechnol 160:1760–1788
Beloqui A, Pita M, Polaina J, Martinez-Arias A, Golyshina OV, Zumárraga M, Yakimov MM, García-Arellano H, Alcalde M, Fernández VM, Elborough K, Andreu JM, Ballesteros A, Plou FJ, Timmis KN, Ferrer M, Golyshin PN (2006) Novel polyphenol oxidase mined from a metagenome expression library of bovine rumen. J Biol Chem 281:22933–22942
Bendtsen JD, Nielsen H, von Heijne G, Brunak S (2004) Improved prediction of signal peptides — SignalP 3.0. J Mol Biol 340:783–795
Bourbonnais R, Paice MG (1990) Oxidation of non-phenolic substrates, an expanded role for laccase in lignin biodegradation. FEBS Lett 267:99–102
Castro-Sowinski S, Gloria M-D, Okon Y (2002) Laccase activity in melanin-producing strains of Sinorhizobium meliloti. FEMS Microbiol Lett 209:119–125
Chu XM, He HZ, Guo CQ, Sun BL (2008) Identification of two novel esterases from a marine metagenomic library derived from South China Sea. Appl Microbiol Biotechnol 80:615–625
Diamantidis G, Effosse A, Potier P, Bally R (2000) Purification and characterization of the first bacterial laccase in the rhizospheric bacterium Azospirillum lipoferum. Soil Biol Biochem 32:919–927
Endo K, Hayashi Y, Hibi T (2003) Enzymological characterization of EpoA, a laccase-like phenol oxidase produced by Streptomyces griseus. J Biochem 133:671–677
Galai S, Limam F, Marzouli MN (2009) A new Stenotrophomonas maltophilia strain producing laccase. Use in decolorization of synthetics dyes. Appl Biochem Biotechnol 158:416–431
Held C, Kandelbauer A, Schroeder M, Cavaco-Paulo A, Guebitz GM (2005) Biotransformation of phenolics with laccase containing bacterial spores. Environ Chem Lett 3:74–77
Hoegger PJ, Kilaru S, James TY, Thacker JR, Kües U (2006) Phylogenetic comparison and classification of laccase and related multicopper oxidase protein sequences. FEBS J 273:2308–2326
Jimenez-Juarez N, Roman-Miranda R, Baeza A, Sánchez-Amat A, Vazquez-Duhalt R, Valderrama B (2005) Alkali and halide-resistant catalysis by the multipotent oxidase from Marinomonas mediterranea. J Biotechnol 117:73–82
Koschorreck K, Richter SM, Ene AB, Roduner E, Schmid RD, Urlacher VB (2008) Cloning and characterization of a new laccase from Bacillus licheniformis catalyzing dimerization of phenolic acids. Appl Microbiol Biotechnol 79:217–224
Koschorreck K, Schmid RD, Urlacher VB (2009) Improving the functional expression of a Bacillus licheniformis laccase by random and site-directed mutagenesis. BMC Biotechnol 9:12. doi:10.1186/1472-6750-9-12
Martins LO, Soares CM, Pereira MM, Teixeira M, Costa T, Jones GH, Henriques AO (2002) Molecular and biochemical characterization of a highly stable bacterial laccase that occurs as a structural component of the Bacillus subtilis endospore coat. J Biol Chem 277:18849–18859
Mikolasch A, Schauer F (2009) Fungal laccases as tools for the synthesis of new hybrid molecules and biomaterials. Appl Microbiol Biotechnol 82:605–624
Miyazaki K (2005) A hyperthermophilic laccase from Thermus thermophilus HB27. Extremophiles 9:415–425
Murugesan K (2003) Bioremediation of paper and pulp mill effluents. Indian J Exp Biol 41:1239–1248
Pickard MA, Roman R, Tinoco R, Vazquez-Duhalt R (1999) Polycyclic aromatic hydrocarbon metabolism by white rot fungi and oxidation by Coriolopsis gallica UAMH 8260 laccase. Appl Environ Microbiol 65:3805–3809
Plonka PM, Grabacka M (2006) Melanin synthesis in microorganisms — biotechnological and medical aspects. Acta Biochim Pol 53:429–443
Roberts SA, Weichsel A, Grass G, Thakali K, Harrard JT, Tollin G, Rensing C, Montfort WR (2001) Crystal structure and electron transfer kinetics of CueO, a multicopper oxidase required for copper homeostasis in Escherichia coli. Proc Natl Acad Sci USA 99:2766–2771
Rodríguez Couto S, Toca Herrera JL (2006) Industrial and biotechnological applications of laccases: A review. Biotechnol Adv 24:500–513
Ruijssenaars HJ, Hartmans S (2004) A cloned Bacillus halodurans multicopper oxidase exhibiting alkaline laccase activity. Appl Microbiol Biotechnol 65:177–182
Sharma P, Goel R, Capalash N (2007) Bacterial laccases. World J Microbiol Biotechnol 23:823–832
Singh G, Capalash N, Goal R, Sharma P (2007) A pH-stable laccase from alkali-tolerant γ-proteobacterium JB: purification, characterization and indigo carmine degradation. Enzyme Microb Technol 41:794–799
Singh G, Sharma P, Capalash N (2009) Performance of an alkalophilic and halotolerant laccase from γ-proteobacterium JB in the presence of industrial pollutants. J Gen Appl Microbiol 55:283–289
Solano F, Lucas-Elio P, Lopez-Serrano D, Fernandez E, Sanchez-Amat A (2001) Dimethoxyphenol oxidase activity of different microbial blue multicopper proteins. FEMS Microbiol Lett 204:175–181
Wesenberg D, Kyriakides I, Agathos SN (2003) White-rot fungi and their enzymes for the treatment of industrial dye effluents. Biotechnol Adv 22:161–187
Witayakran S, Ragauskas AJ (2009) Synthetic applications of laccase in green chemistry. Adv Synth Catal 351:1187–1209
Xiao YZ, Tu XM, Wang J, Zhang M, Cheng Q, Zeng WY, Shi YY (2003) Purification, molecular characterization and reactivity with aromatic compounds of a laccase from basidiomycete Trametes sp. strain AH28-2. Appl Microbiol Biotechnol 60:700–707
Xu F (1996) Oxidation of phenols, anilines, and benzenethiols by fungal laccases: correlation between activity and redox potentials as well as halide inhibition. Biochem 35:7608–7614
Ye M, Li G, Liang WQ, Liu YH (2010) Molecular cloning and characterization of a novel metagenome-derived multicopper oxidase with alkaline laccase activity and highly soluble expression. Appl Microbiol Biotechnol 87:1023–1031
Yooseph S, Sutton G, Rusch DB, Halpern AL, Williamson SJ, Remington K, Eisen JA, Heidelberg KB, Manning G, Li W, Jaroszewski L, Cieplak P, Miller CS, Li H, Mashiyama ST, Joachimiak MP, van Belle C, Chandonia JM, Soergel DA, Zhai Y, Natarajan K, Lee S, Raphael BJ, Bafna V, Friedman R, Brenner SE, Godzik A, Eisenberg D, Dixon JE, Taylor SS, Strausberg RL, Frazier M, Venter JC (2007) The Sorcerer II Global Ocean Sampling expedition: expanding the universe of protein families. PLoS Biol 5:432–466
Acknowledgements
The authors are grateful to Prof. Baolin Sun for providing the metagenome library and to Prof. Ursula Kües for his constructive suggestions. This work was funded by the National High Technology Research and Development Program of China (863 Program; No. 2007AA09Z421), Science and Technology Foundation of Distinguished Young Scholars of Anhui Province (No. 08040106908).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Table S1
Comparison of sequence identities of Lac15 with other bacterial multicopper oxidases (DOC 47 kb)
Fig. S1
Multialignment of bacterial laccases for primer designing. The protein sequences were retrieved from GenBank with following accession numbers: Bacillus sp. (ACM46021), Bradyrhizobium japonicum (BAC47475), Brucella abortus (AAX75945), Cyanothece sp. (EAZ88980), E. coli (BAB96698), Lyngbya sp. (EAW36583), Marine actinobacterium (EAR25244), Pseudoalteromonas haloplanktis (CAI89062), Streptomyces clavuligerus (EDY51894), Streptomyces lavendulae (BAC16804), Synechococcus sp. (EAQ67893), T. thermophilus (AAS81712), and Vibrio harveyi (ABU69389). Sequence alignment was performed using Clustal X 2.0 and GENEDOC. Fragments used for primer designing were indicated by full-length vertical boxes (JPEG 1224 kb)
Rights and permissions
About this article
Cite this article
Fang, Z., Li, T., Wang, Q. et al. A bacterial laccase from marine microbial metagenome exhibiting chloride tolerance and dye decolorization ability. Appl Microbiol Biotechnol 89, 1103–1110 (2011). https://doi.org/10.1007/s00253-010-2934-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-010-2934-3