Abstract
A comprehensive approach of bioprocess design at various levels was used to optimize microbial production of extracellular fructofuranosidase, important as biocatalyst to derive fructooligosaccharides with broad application in food or pharmaceutical industry. For production, the recombinant strain Aspergillus niger SKAn1015 was used, which expresses the fructofuranosidase encoding gene suc1 under control of a strong constitutive promoter. In a first screening towards an optimized medium, glucose, nitrate, Fe2+, and Mn2+ were identified as beneficial for production. A minimal medium with optimized concentration of these key nutrients, obtained by central composite design experiments and quadratic modelling, provided a threefold increased fructofuranosidase activity in the culture supernatant (400 U/mL) as compared to the originally described medium. Utilizing the optimized medium, the process was then transferred from shake flask into a fed-batch-operated bioreactor. Hereby, the intended addition of talc microparticles allowed engineering the morphology of A. niger into a highly active mycelial form, which strongly boosted production. Fructofuranosidase production was highly specific as confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis. The secreted enzyme activity of 2,800 U/mL, corresponding to about 3 g/L of fructofuranosidase, achieved by the microparticle-enhanced fed-batch process, is tenfold higher than that of any other process reported so far, so that the presented bioprocess strategy appears as a milestone towards future industrial fructofuranosidase production.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Fructofuranosidases (EC 3.2.1.26) catalyze the conversion of the disaccharide sucrose into fructooligosaccharides, which receive high interest for new prebiotic products commercially available (Maiorano et al. 2008). Due to their interesting functional properties, the market potential of fructooligosaccharides further includes applications in the pharmaceutical or the diagnostic sector underling their great industrial value (Rubio and Maldonado 1995; Fernandez et al. 2004; Maiorano et al. 2008; Zuccaro et al. 2008). This potential has strongly stimulated the development of bioprocesses for production of fructofuranosidases (Mussatto et al. 2009a). Fructofuranosidases are mainly products of filamentous fungi including Aspergillus, Penicillium, or Aureobasidium, whereby only a few species are considered for industrial production (Sangeetha et al. 2005). Especially the genus Aspergillus, frequently applied in enzyme production due to the GRAS status, has received particular attention. Clearly, the economic feasibility of such an industrial process strongly depends on the enzyme quantity obtained in secreted form. As summarized in an excellent review, extensive studies in recent years, mainly using shake flask cultures, have aimed at the improvement of fructofuranosidase production (Maiorano et al. 2008). This comprised the screening for suitable production strains, new enzyme variants (Cuervo et al. 2007) and process conditions, such as temperature or pH value (Fernandez et al. 2007). Moreover, numerous studies investigated the effect of sources of carbon or nitrogen, of vitamins, inorganic salts, or additives such as polymers or surfactants (Hayashi et al. 1993; Balasubramaniem et al. 2001). Admittedly, secreted enzyme activities reached are typically below 100 U/mL, meaning that these processes appear not yet optimized for an industrial process (Maiorano et al. 2008). The high industrial relevance of fructofuranosidases and the still suboptimal production, demand for strong efforts in metabolic and biochemical engineering focusing on superior strains, as well as the design of appropriate media and process conditions towards improved production.
In this regard, the present work deals with the optimization of fructofuranosidase production by Aspergillus niger SKAn1015. In recent studies, this recombinant strain, expressing the fructofuranosidase gene under control of a strong constitutive promoter, showed encouraging production properties (Zuccaro et al. 2008; Driouch et al. 2010). The bioprocess design performed here combined optimization at various levels. First, we carried out a thorough design of the production medium using a statistical approach that included first the identification of nutrients important for production, followed by optimization of their concentration using central composite design. Utilizing the optimized medium, the process was then transferred from shake flask into bioreactor. This included the development and testing of suitable fed-batch strategies. Moreover, for the first time in fed-batch culture, microparticles were applied as novel approach to engineer the morphology of A. niger. This has been recently shown to be beneficial for enzyme production in A. niger (Driouch et al. 2010) and other fungal species (Kaup et al. 2007).
Materials and methods
Strains, maintenance, and incolulum preparation
In the present work, the fructofuranosidase overproducing strain A. niger SKAn1015 was used. This strain has been derived from A. niger N400 (ATCC 9029) as described previously (Zuccaro et al. 2008). It is protease deficient and expresses the fructofuranosidase gene (suc1) under the control of the constitutive pkiA promoter. In addition, the protease-deficient strain A. niger AB 1.13, derived from A. niger N400 (ATCC 9029) as described previously (Mattern et al. 1992), was used as host for construction of the gfp-expressing mutant ARAn701 to spatially resolve enzyme production in fungal aggregates. All strains were maintained in 50% glycerol as frozen spore suspension at −80 °C. A spore inoculum of A. niger was prepared by growing thawn spores from the maintenance culture at 30 °C for 3 days on potato dextrose agar. Spores were then harvested as spore suspension from the plate into 20 mL 0.9% NaCI solution, which was spread onto the plate. After filtration (Miracloth, 25 μm pore size, CalBioChem, Darmstadt, Germany), the spore concentration was determined photometrically at 600 nm. Cultivations were inoculated to an initial spore concentration of 106/mL.
Genetic construction of a GFP-expressing mutant
For construction of A. niger strain ARAn701, expressing gfp under control of the glaA promoter, the plasmid pARAn701 (PglaA-gfp) was integrated into the protease-deficient (prt) strain AB1.13 (Mattern et al. 1992) by protoblast transformation as described previously (Roth and Dersch 2009). The expression vector pARAn701 was derived from vector ANEp7 (Storms et al. 2005). First, the AMA1 sequence supporting autonomous replication was removed from ANEp7 by NotI digest and religation. Oligonucleotides MCS-NcoI-for (3′-GCC AGG CCA TGG GCT AGC TTA AGT TTA AAC GAT ATC GAT GGC GCC GGG CCC AGG CCG GCC CCA TGG AGA CGG-5′) and MCS-NcoI-rev (3′-CCG TCT CCA TGG GGC CGG CCT GGG CCC GGC GCC ATC GTT TAA ACT TAA GCT AGC CCA TGG CCT GGC-5′) including a multiple cloning site were annealed. The generated DNA fragment was inserted into the NheI/FseI sites of ANIp7 to obtain pARAn8 (P glaA -MCS). Subsequently, a polymerase chain reaction (PCR)-generated DNA fragment amplified from vector pAN56-gfp (Gordon et al. 2000) with primers PCR-gfp–AflII-for (3′-GGG AGC TTA AGC ATG GTG AGC AAG GGC GAG G-5′) and PCR-gfp–ClaI-rev (3′-CGC AGG ATC GAT TTA CTT GTA CAG CTC GTC-5′) was inserted into the AflII/ClaI sites of pARAn7.
Cultivation media
A solid medium, containing 30 g/L potato dextrose agar, was used to prepare the spore inoculum for all strains. The basic medium for batch culture of A. niger SKAn1015, as described previously (Driouch et al. 2010), contained per liter 20 g glucose, 6 g NaNO3, 0.5 g KCI, 1.5 g KH2PO4, 0.5 g MgSO4 ·7H2O (50 mL added from a 20× stock solution), and 10 mg EDTA, 4.4 mg ZnSO4 ·7H2O, 1.01 mg MnCl2 ·4H2O, 0.32 mg CuSO4 ·5H2O, 1 mg FeSO4 ·7H2O, 0.32 mg CoCl2 ·6H2O, 1.47 mg CaCl2 ·2H2O, and 0.22 mg (NH4)6Mo7O24 ·4H2O (1 mL added from a 1,000× stock solution). This medium formulation was varied as described in the “Results” section to achieve improved production performance. This included the replacement of glucose and nitrate by other carbon and nitrogen sources, respectively. Additionally, the level of nutrients was altered to investigate the effect on production. The optimal medium finally discovered differed from the basic composition by elevated levels of glucose (30 g/L), NaNO3 (9 g/L), FeSO4 ·7H2O (7.5 mg/L), and MnCl2 ·4H2O (1.5 mg/L). In fed-batch processes with A. niger SKAn1015 in the bioreactor, the above medium was used for the batch phase. The medium for the feeding phase contained per liter 200 g glucose, 40 g NaNO3, 0.5 g KCI, 1.5 g KH2PO4, 0.5 g MgSO4 ·7H2O, 10 mg EDTA, 4.4 mg ZnSO4 ·7H2O, 30 mg MnCl2 ·4H2O, 0.32 mg CuSO4 ·5H2O, 2.5 g FeSO4 ·7H2O, 0.32 mg CoCl2 ·6H2O, 1.47 mg CaCl2 ·2H2O, and 0.22 mg (NH4)6Mo7O24 ·4H2O. The batch medium for A. niger ARAn701 contained per liter 10 g xylose, 6.6 g/L (NH4)2SO4, 2.5 g/L KH2PO4, 0.2 g/L MgSO4 ·7H2O, 0.1 g/L CaCl2 ·H2O, and 5 mg/L citric acid H2O, 50 g/L ZnSO4 ·7H2O, 10 g/L Fe(NH4)2(SO4)·6H2O, 16 g/L CuSO4, 0.5 g/L H3BO3, 0.5 g/L Na2MoO4 ·H2O, and 0.37 g/L MnSO4 ·H2O. In fed-batch experiments with this strain, 200 g L−1 maltose was used as feeding solution. In all cases the solutions containing the carbon source, the inorganic salts and the trace element were sterilized separately. The maltose solution was sterilized by filtration. All other solutions were sterilized by autoclaving at 121 °C for 20 min and cooled down to room temperature prior to mixing.
Cultivation
Shake flask studies were carried out in triplicate in 250 mL shake flasks containing 50 mL medium on a rotary shaker at 37 °C at 120 min−1. For bioprocess optimization, a 3-L stirred tank bioreactor (Applikon, Schiedam, The Netherlands) with two six-bladed disc turbine impellers (equal to a volumetric power input of 44 W m−3) was employed. All bioreactor cultivations were carried out in duplicate. Aeration rate (1.0 L min−1), temperature (37 °C for strain SKAn1015 and 30 °C for strain ARAn701), and pH value (pH 5.0) were automatically kept constant. Antifoam (Ucolub N115) was added manually, when required. The concentration of oxygen and carbon dioxide in the exhaust gas was monitored online (BC perFerm, BlueSens, Germany). In batch processes (100 h), the working volume was 2.2 L, and the agitation speed was kept constant at 200 min−1. Fed-batch processes exhibited an initial batch phase with 1.5 L starting volume. An intermittent feeding was started immediately when the substrate level had decreased to about 5 g/L using a peristaltic pump (Applikon, Schiedam, The Netherlands). The feed was controlled such that the substrate level remained above 1 g/L, which was monitored by measurement in the broth. The total feed volume added was 1 L. The agitation speed was maintained at 200 min−1 during the first 24 h. After that, the agitation speed was increased stepwise every 30 min to 300, 400, 500, and 550 min−1, meaning that the latter value was then maintained for the rest of the process. In selected cultivations, microparticle of hydrous magnesium silicate (3 MgO·4SiO2 ·H2O, 6 µm, 5 g/L, talc powder) was added. Prior to use, the microparticles were re-suspended in 50 mM Na-acetate buffer (pH 6.5), autoclaved at 121 °C for 20 min and added to the sterile growth medium prior to inoculation.
Analysis of substrates and products
Concentration of dry biomass was determined in triplicate. For this purpose, 10-mL samples were filtered on a pre-weighted cellulose acetate filter (pore size 20 μm, Sartorius, Göttingen, Germany). Subsequently, the filter was rinsed twice with water and then dried at 100 °C until weight constancy. After cooling in a desiccator, the cell dry weight was determined by re-weighing. The quantification of sugars and of gluconate in cultivation supernatant, obtained by filtration (Minisart 0.2 μm, Sartorius, Göttingen, Deutschland) was carried out using HPLC (Elite Lachrome HITACHI Ltd., Japan) with a Metacarb 67H column (250 × 4.6 mm, 5 μm, VWR-Hitachi, Darmstadt, Germany) and 1 mM H2SO4 as mobile phase at a flow rate of 0.8 mL min−1 and 70 °C. Detection was performed with a UV detector (210 nm) or by refractive index. In addition, glucose concentration was determined using a glucose analyzer (Biochemistry Analyzer, YSI, Dayton, OH, USA).
Protein concentration in culture supernatant
The protein concentration in the culture supernatants (cell-free filtrate) was determined by the bicinchoninic acid method using bovine serum albumin as external standard (BSA protein assay kit, Pierce, Rockford, IL, USA) following the instruction given by the distributor. For this purpose, 10-mL samples were filtered (cellulose acetate filter, pore size 20 μm, Sartorius, Germany) and clarified by centrifugation (13,000×g, 10 min, 4 °C) prior to analysis.
Fructofuranosidase activity
The specific activity of fructofuranosidase was quantified in culture supernatant obtained by filtration of 10 mL culture broth through a cellulose acetate filter (pore size 20 μm, Sartorius, Göttingen, Germany). The reaction mixture (220 μL) consisted of 200 μL 1.65 M sucrose dissolved in 0.05 M phosphate buffer (pH 5.4) and 20 μL sample. Reaction was started by addition of 200 μL sucrose to a 20-μL sample and incubated at 55 °C for 20 min. The reaction was stopped by heating at 95 °C for 10 min. After cooling, the reaction mixture was centrifuged at 13,000×g for 10 min at 4 °C. Glucose formed from cleavage of sucrose by the enzyme was then quantified as described previously (Driouch et al. 2010). To account for residual glucose in the culture broth, negative controls were carried out by using samples in which fructofuranosidase was inactivated by heating at 95 °C for 10 min prior to incubation. The measured enzyme activity was on average 1.8-fold higher as compared to values determined at 40 °C. This ratio, estimated from the analysis of ten different culture samples at both temperatures, was used to compare the obtained results with previous studies which had employed 40 °C (Zuccaro et al. 2008; Driouch et al. 2010) or 55 °C (other studies listed in Table 4) for the fructofuranosidase assay.
Extracellular protein pattern
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was used to determine the secreted proteins in culture supernatant samples. First, 170 µL culture supernatant was mixed with 830 µL acetone for protein precipitation. The obtained pellet was dried and re-suspended in 10 µL buffer following the instruction given by the distributor. A volume of 3 µL (corresponding to 50 µL culture supernatant) was then loaded onto a 4–12% Bis-Tris-Mini Gel (Novex Mini-Cell 1 mm, 15-well, Invitrogen, Karlsruhe, Germany) for protein separation. The applied molecular mass marker was the PageRuler Unstained Protein Ladder (SM0661, Fermentas, St. Leon-Rot, Germany). The highly glycosylated fructofuranosidase has a molecular weight of about 110–120 kDa (Zuccaro et al. 2008). For quantification of the relative amount of fructofuranosidase in the supernatant, the gel was scanned using the Molecular Imager® PharosFX System (BioRad, Munich, Germany). The volume of the corresponding protein bands was determined by the 1-D Analysis Software (Version 4.6.9, BioRad, Munich, Germany).
Microscopy analysis
Culture morphology was analyzed by 3D photographs using a stereo-microscope (Stemi 2000-C, ZEISS, Germany) with an AxioCamMRc5 camera (Stemi 2000-C, ZEISS, Germany). Confocal laser scanning microscopy was applied to analyze the spatial distribution of green fluorescent protein (GFP) fluorescence within thin sections through cellular aggregates as described previously (Driouch et al. 2010).
Theoretical background of medium design
The effect of medium constituents on fructofuranosidase production was first investigated in shake flasks by the one-variable-at-a-time approach. Starting from the formulation of the basic medium, the concentration of each investigated nutrient was decreased to 50% (low level) or increased to 200% (high level). From these screening experiments, the key nutrients that significantly affected fructofuranosidase production were identified and optimized further using central composite design. As is described in the “Results” section, k = 4 nutrients were identified. From this, N = 30 nutrient combinations were designed for further shake flask studies, meaning that this included 16 combinations from a full 24 factorial composite rotary design, six at the center point (CP) and eight experiments, where one factor was set to an extreme level Eq. 1:
This methodology allows the modelling of the data by a second-order equation. The quadratic model for estimating the concentration of the investigated nutrients for optimum fructofuranosidase activity is given by Eq. 2:
Here, Y is the response variable, i.e., the fructofuranosidase activity; X i and X j are the independent input variables, i.e., the nutrient levels. The values for β 0, β ii , and β ij are the resulting regression variables for intercept, linear, quadratic, and interaction terms, respectively. The parameter estimation from the data by second-order multiple regression models was carried out using a statistical software (Design-Expert, Version 7.0, Stat-Ease, Minneapolis, MN, USA). This included a thorough statistical treatment of the data, involving a quality check of the fitted polynomial models by the coefficient of determination (R 2) and the correlation coefficient (R), as well as the analysis of variance combined with an F test to evaluate, if a given term had a significant effect (p < 0.05).
Results
Medium design
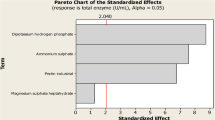
In a first step towards an optimized medium, the influence of carbon source, nitrogen source, and trace elements on production was studied. For this purpose, screening experiments in shake flasks were set up in triplicate each (Fig. 1). This included the test of alternative carbon and nitrogen sources as well as the variation of the level of the investigated nutrients. On the basic medium (control), A. niger secreted 160 U/mL fructofuranosidase. Among all studies, four conditions allowed a production of fructofuranosidase, which was higher than in the control. From a variety of carbon sources tested, the medium with 30 g/L glucose resulted in the highest enzyme activity. NaNO3 at a level of 12 g/L was found to be the most suited nitrogen source for production. The screening experiments further revealed a beneficial effect of elevated levels of the trace elements Mn2+ and Fe2+ on enzyme production. The four nutrients glucose, NaNO3, MnCI2 ·4H2O, and FeSO4 ·7H2O, were considered for further medium optimization. The strategy based on central composite design experiments with estimation of optimal nutrient levels using a quadratic model. In total, 30 different nutrient combinations were designed for this purpose and then tested in shake flask studies with three replicates each. The results from all experiments with the information on the corresponding medium composition are summarized in Table 1. Depending on the medium, the observed enzyme activity ranged from about 40 to almost 400 U/mL, underlining that the investigated nutrients obviously strongly influenced production. The experimental data were then fitted with a second-order polynomial expression. The fructofuranosidase activity (Y FFase) could be expressed as function of the concentration of glucose (x 1), NaNO3 (x 2), MnCI2 ·4H2O (x 3), and FeSO4 ·7H2O (x 4) by the following equation:
Identification of key nutrients for fructofuranosidase production by Aspergillus niger SKAn1015 using one-variable-at-a-time (OVAT) experiments in shake flasks. The fructofuranosidase activity, given as average with corresponding deviation from three replicate incubations, was determined after 100 h of cultivation. The control experiment was carried out with the basic medium containing 20 g/L glucose and 6 g/L sodium nitrate and corresponding trace elements. For the carbon sources tested, the low- and high-level conditions refer to 10 and 30 g/L. The low- and high-level concentrations for the nitrogen source were 3 and 12 g/L. Similarly, the low and high level for the trace elements corresponded to 50% and 200% of the concentration in the basic medium
The comparison of the experimental enzyme activity with the corresponding values predicted by the model revealed a good correlation (Fig. 2). The high coefficient of regression (r = 0.90) indicated a high goodness of fit. Statistical treatment of the data further underlined a highly consistent data set and high confidence for the obtained parameters (Table 2). Exemplified for glucose and sodium nitrate, the 3D response surface plot, determined from the obtained regression model, illustrates the strong effect of the key nutrients on production as well as the prediction of an optimal combination (Fig. 3). The optimal medium identified by the model contained 30 g/L glucose, 9 g/L NaNO3, 1.5 mg/L MnCI2 ·4H2O, and 7.5 mg/L FeSO4 ·7H2O, respectively. For these conditions, a maximum enzyme activity of 400 U/mL in shake flask culture was predicted, almost threefold more than in the original medium.
Batch production on optimized medium
The optimal medium was first compared in shake flasks to the original basic medium. On the original medium, A. niger SKAn1015 grew only to a maximum dry weight of 8.2 g/L. The enzyme activity reached 160 U/mL after 100 h of cultivation. Cultured on the new medium formulation, the strain revealed significantly improved performance. It reached a maximum dry weight of 12.8 g/L. The achieved enzyme activity of 400 U/mL corresponded nicely to the predicted value. The production performance of A. niger SKAn1015 on both media was then evaluated in bioreactor (Fig. 4a, b). On the newly designed medium, the strain grew much faster and reached a higher biomass concentration. Most impressively, the fructofuranosidase activity was more than sevenfold higher (750 U/mL) as compared to the control (100 U/mL).
Fructofuranosidase production by Aspergillus niger SKAn1015 in batch culture using 3 L stirred tank bioreactors on the original basic medium (a) and the optimized medium (b). The data comprise the time profile for glucose, cell dry weight, and enzyme activity over the cultivation time of 100 h as mean values from two replicate cultivations with corresponding deviations
Development of a fed-batch bioprocess
Compared with traditional batch operation, fed-batch operation mode often offers an improved efficiency. This was now exploited by transferring the production into a fed-batch process (Fig. 5a). After an initial batch phase of 30 h, the feed, which contained a concentrated form of the optimal nutrient mixture, was started. The intermittent feed with a total volume of 1 L was adjusted such that the glucose level in the culture was maintained above 1 g/L ensuring sufficient supply of the culture with the carbon source. The process was characterized by efficient growth of A. niger SKAn1015, meaning that 14 g/L of dry biomass were observed after about 80 h of cultivation time. The cells formed the characteristic pellet structure of filamentous fungi in submerged culture (Fig. 6a). During the first 50 h, the consumed glucose was partly oxidized into gluconate. Both substrates were then concurrently metabolized for the remaining process. The maximum fructofuranosidase activity (600 U/mL) was observed after about 100 h. The product level then dropped to a final value of 350 U/mL corresponding to a specific activity of 200 U/mgprotein. Overall, the performance of the fed-batch process, at least at this stage, was not better than that of the batch operation.
Fructofuranosidase production by Aspergillus niger SKAn1015 in fed-batch culture on the optimized medium without (a) and with addition of talc microparticles (6 µm, 5 g/L; b) using 3 L stirred tank bioreactors. The data comprise the time profile for glucose, gluconate, cell dry weight, and enzyme activity over 180 h cultivation time as mean values from two replicate cultivations with corresponding deviations
Microparticle-enhanced fed-batch process
For a further optimization of enzyme production in fed-batch mode, the batch medium was now additionally supplemented with inorganic talc microparticles (6 µm, 5 g/L), previously proven in shake flasks studies to stimulate production. All other process parameters remained unchanged. The cultivation profile of this microparticle-enhanced process is shown in Fig. 5b. It differed dramatically from the normal fed-batch process. Most strikingly, the fructofuranosidase activity reached a final value of 2,800 U/mL, meaning that the addition of the microparticles increased production dramatically. This was also observed for the specific enzyme activity (820 U/mgprotein). Cells grew as freely dispersed mycelium for the whole cultivation process (Fig. 6b). In addition to the morphology, the talc material obviously influenced the metabolic properties of A. niger SKAn1015. Compared to the normal fed-batch, the formation of gluconate was clearly stimulated at the expense of growth. This was especially pronounced in the first 80 h. Here, gluconate reached a maximum level of 24 g/L. Together with the added glucose, gluconate was completely re-used towards the end of the process. Interestingly, growth and enzyme production were maintained until the end of the process, which seems one of the major benefits as compared to the control fed-batch.
SDS-PAGE analysis of the supernatant at the end of the microparticle-enhanced fed-batch confirmed strong of fructofuranosidase, represented by the protein band at 110–120 kDa, by the recombinant strain (Fig. 7). The target enzyme was secreted almost exclusively, underlining the high specificity of production. In addition, only one band was visible at about 80 kDa. This coincided with low but significant activity of glucoamylase (90 U/mL) observed in the supernatant at the end of the process. A rough estimation via the volume of the observed bands revealed that fructofuranosidase accounted for about 85% of the totally excreted protein in the culture supernatant.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of culture supernatant (180 h) from Aspergillus niger SKAn1015 in fed-batch production with optimized medium and addition of talc microparticles. The left lane 1 (M) shows a molecular weight marker (Fermentas SM0661). The strong band at about 110–120 kDa corresponds to fructofuranosidase (Zuccaro et al. 2008). The total protein secreted was attributed to fructofuranosidase and to the other visible protein band at 80 kDa. The relative amount of fructofuranosidase was estimated as volume of the corresponding band related to that of all protein bands
Impact of morphology on enzyme production
Obviously, the microparticles specifically enhanced production of the target enzyme by A. niger in the fed-batch culture. A. niger, grown in the presence of the microparticles, exhibited a specifically higher production level, especially during the second half of the fermentation. To unravel the link between the production behavior and the obviously affected morphology, enzyme production was spatially resolved during the fed-batch process within the different fungal aggregates, mycelium, and pellet, employing a GFP-expressing variant of A. niger. Production was hereby localized in 70-µm thin cross-sections through biomass aggregates obtained from cultures with and without microparticles (Fig. 8). In both cases, GFP expression was immediately initiated after the induction by maltose after 40 h. Protein production in the pellet-based process, however, occurred only within a thin layer at the pellet surface (Fig. 8a–d). The inner pellet did not contribute to production, probably due to diffusion limitation of oxygen or of other nutrients. Despite glucose was still present, production in the surface layer markedly decreased after about 120 h, so that almost completely inactive pellets remained for a large time period of the process. For the microparticle-enhanced process, intensive fluorescence was present across the entire mycelium (Fig. 8e, f). Protein production remained high until 180 h. This indicates that the interaction with microparticles created a productive biocatalyst remaining highly active during the entire process.
Spatial resolution of fluorescent protein production (GFP2) in recombinant Aspergillus niger ARAn701 pellets (a–f, normal fed-batch cultivation) and freely dispersed mycelium (g–l, fed-batch cultivation with addition of 5 g/L talc microparticles of 6 µm diameter). GFP2 expression was induced by start of the maltose feed after 40 h of cultivation. Samples were taken before induction (39 h) and after 48, 72, 120, 144, and 172 h of cultivation time. The pictures were obtained from 70 µm cross-sections through fungal aggregates by confocal laser scanning microscopy
Discussion
Fructofuranosidases receive increasing interest as industrial biocatalysts for the synthesis of prebiotic fructooligosaccharides. This high potential demands for efficient strains and bioprocesses enabling industrially feasible fructofuranosidase production. In this regard, a number of studies have previously aimed at the isolation of fructofuranosidase-producing strains from nature (Cuervo et al. 2007) or the screening for suitable production media (Balasubramaniem et al. 2001). Most of these studies, however, resulted in relatively low production efficiency, which might be attributed to the fact that wild-type strains were employed and processes were performed in shake flask cultures. Towards an efficient production of fructofuranosidase, it appears important to develop efficient bioprocesses based on efficient media formulations and process conditions for tailor-made production strains. In this regard, we developed an optimized bioprocess for production of fructofuranosidase by the recombinant strain A. niger SKAn1015.
As first step, the production medium was systematically optimized using a modelling-based statistical approach. Among various carbon sources tested, glucose allowed fastest growth and also resulted in the highest enzyme activity. Previous fructofuranosidase production processes are almost exclusively based on sucrose and require high substrate concentration of typically more than 200 g/L. This is obviously due to the fact that these high sucrose levels are needed to induce fructofuranosidase expression in wild-type strains (Chen and Liu 1996). Accordingly, other sugars including glucose typically enable fast growth, but only low enzyme production (Zuccaro et al. 2008). This was not observed for the recombinant strain A. niger SKAn1015, which expresses the fructofuranosidase encoding gene under control of the strong constitutive promoter pkiA and is thus released from undesired endogenous control mechanisms. Due to this, the superior efficiency of glucose regarding growth as compared to sucrose (Fig. 1) could now be fully exploited for fructofuranosidase production which seems a major advantage. As further benefit, efficient enzyme production was possible even at a low glucose level below 5 g/L (Fig. 5b). This facilitates the set-up of efficient fed-batch processes with limited feeding and complete consumption of the carbon source.
Concerning the nitrogen source, recent studies revealed interesting effects of the chosen ingredient on production properties and the morphology of the cells (Balasubramaniem et al. 2001). It appeared that the higher enzyme activity on corn steep liquor, as compared to inorganic salts containing nitrate or ammonium, was indirectly related to the fact that the complex nitrogen source resulted in loose pellets or hyphae, whereas the salts led to pelleted growth. These medium-dependent morphology effects were no more required in the present work, since the intentional addition of microparticles allowed a direct and precise engineering of the morphology into the desired hyphal form. Accordingly, sodium nitrate could be efficiently used. It should be noticed that also complex nitrogen sources, such as peptone or yeast extract, enabled efficient production (data not shown), but were not considered since we aimed at a fully defined medium towards a robust, reproducible production process avoiding batch-to-batch variation, typically faced on complex nutrients. Among the microelements, Fe2+ and Mn2+ were identified as key nutrients. Their importance for fructofuranosidase production has not been observed before (Maiorano et al. 2008), which might be explained by the altered control of enzyme expression in A. niger SKAn1015 as compared to wild-type strains. It is interesting to note that in contrast to the present work, both trace elements have to be limiting for efficient production of citric acid (Papagianni 2007). The production of this organic acid demands for a downregulation of the tricarboxylic acid (TCA) cycle at the level of citric acid. This can be achieved by the lack of Fe2+ and Mn2+ required as cofactors of downstream TCA cycle enzymes. The stimulating role of the two metals here might indicate that an actively operating TCA cycle, supplying energy and building blocks, is needed to achieve high enzyme titers in A. niger SKAn1015. The subsequent optimization of the level of glucose, nitrate, Fe2+, and Mn2+ using central composite design allowed a threefold increase of fructofuranosidase in shake flask. This underlines the high value of a systematic, modelling-based medium design (Babu et al. 2008). Subsequently, the process was transferred from shake flask into bioreactor for further optimization. This step appears especially important, since for the production of fructofuranosidase, such investigations are quite rare (Maiorano et al. 2008). Already in batch operation, this allowed a higher enzyme production (Table 3). The increased enzyme level might be due to better aeration or mixing patterns provided in the bioreactor. When transferred into a fed-batch environment with intermittent feeding of glucose and the addition of microparticles, a fructofuranosidase activity of 2,800 U/mL could be finally achieved. Starting from the original medium in batch-operated shake flasks (140 U/mL), the product level could thus be optimized by a factor of 20. Among all fructofuranosidase-producing studies, providing data on the extracellular enzyme activity, values up to about 280 U/mL could be achieved so far (Table 4). The enzyme activity by the microparticle-enhanced fed-batch process on minimal medium is more than tenfold higher.
Moreover, the process was characterized by a high specificity of production. Fructofuranosidase accounted to about 85% of the totally secretion protein as verified by SDS-PAGE gel analysis. Taking the measured protein concentration in the culture supernatant into account (Table 3), this roughly corresponds to about 3 g/L of fructofuranosidase. It seems likely that glucoamylase formed as endogenous enzyme of the applied strain on glucose represents the weak protein band at around 80 kDa (Venkataraman et al. 1975). The presence of glucoamylase was, however, rather low. Its enzymatic activity (90 U/mL) was only 3% as compared to that of fructofuranosidase (2,800 U/mL). Future metabolic engineering of A. niger SKAn1015 could aim at disruption of the encoding gene to completely prevent formation of glucoamylase and achieve an even more pure culture supernatant.
Summarizing, the presented bioprocess strategy appears as a milestone towards future industrial fructofuranosidase production. A key to success seems the highly active mycelial biocatalyst present during the whole fed-batch process, exhibiting superior production properties as compared to the pelleted form, which only provided a small fraction of actively producing cells.
References
Babu SI, Ramappa S, Guru MD, Sunanda KK, Sita KK, Subba RG (2008) Optimization of medium constituents for the production of fructosyltransferase (FTase) by Bacillus subtilis using response surface methodology. Res Microbiol 3:114–121
Balasubramaniem AK, Nagarajan KV, Paramasamy G (2001) Optimization of media for fructofuranosidase production by Aspergillus niger in submerged and solid state. Process Biochem 36:1241–1247
Chen W, Liu C (1996) Production of β-fructofuranosidase by Aspergillus japonicus. Enzyme Microb Technol 18:153–160
Cruz R, Cruz VD, Belini MZ, Belote JG, Vieira CR (1998) Production of fructooligosaccharides by mycelia of Aspergillus japonicus immobilized in calcium alginate. Bioresour Technol 65:139–143
Cuervo RF, Ottoni CA, Silva ES, Matsubara RS, Carter JM, Magossi LR, Wada MAA, Rodrigues MFA, Guilarte BM, Maiorano AE (2007) Screening of β-fructofuranosidase producing microorganisms and effect of pH and temperature on enzymatic rate. Appl Microbiol Biotechnol 75:87–93
Dhake AB, Patil MB (2007) Effect of substrate feeding on production of fructosyltransferase by Penicillium purpurogenum. Braz J Microbiol 38:194–199
Driouch H, Sommer B, Wittmann C (2010) Morphology engineering of Aspergillus niger for improved enzyme production. Biotechnol Bioeng 105:1058–1068
Fernandez RC, Maresma BG, Juarez A, Martinez J (2004) Production of fructooligosaccharides by β-fructofuranosidase from Aspergillus sp. 27H. J Chem Techol Biotechnol 79:268–272
Fernandez RC, Ottoni CA, da Silva ES, Matsubara RM, Carter JM, Magossi LR, Wada MA, de Andrade Rodrigues MF, Maresma BG, Maiorano AE (2007) Screening of β-fructofuranosidase-producing microorganisms and effect of pH and temperature on enzymatic rate. Appl Microbiol Biotechnol 75:87–93
Gordon CL, Khalaj V, Ram AF, Archer DB, Brookman JL, Trinci AP, Jeenes DJ, Doonan JH, Wells B, Punt PJ, van den Hondel CA, Robson GD (2000) Glucoamylase::green fluorescent protein fusions to monitor protein secretion in Aspergillus niger. Microbiology 146(Pt 2):415–426
Guimaraes LHS, Somera AF, Terenzi HF, Polizeli MLTM, Jorge JA (2009) Production of fructofuranosidases by Aspergillus niveus using agroindustrial residues as carbon sources: characterization of an intracellular enzyme accumulated in the presence of glucose. Process Biochem 44:237–241
Hayashi S, Matsuzaki K, Takasaki Y, Ueno H, Imada K (1992) Production of fructofuranosidase by Aspergillus japonicus. World J Microbiol Biotechnol 8:155–159
Hayashi S, Hinotani T, Hayashi Y, Takasahaki Y, Imada K (1993) Development of medium composition for the production of glucosyltransferring enzyme by Aureobasidium. World J Microbiol Biotechnol 9:248–250
Hidaka H, Hirayama M, Sumi N (1988) A fructooligosaccharide-producing enzyme from Aspergillus niger ATCC20611. Agric Biol Chem 52:1181–1187
Kaup BA, Ehrich K, Pescheck M, Schrader J (2007) Microparticle-enhanced cultivation of filamentous microorganisms: increased chloroperoxidase formation by Caldariomyces fumago as an example. Biotechnol Bioeng 99:491–198
Maiorano AE, Piccoli RM, Sabino da Silva E, Rodrigues MFA (2008) Microbial production of fructosyltransferases for synthesis of pre-biotics. Biotechnol Lett 30:1867–1877
Mattern IE, van Noort JM, van den Berg P, Archer DB, Roberts IN, van den Hondel CA (1992) Isolation and characterization of mutants of Aspergillus niger deficient in extracellular proteases. Mol Gen Genet 234:332–336
Mussatto SI, Dragone G, Rodrigues LRM, Teixeira JA (2008) Fructooligosaccharides production using immobilized cells of Aspergillus japonicus. In: Ferreira EC, Mota M (eds) Chempor 2008: Proceedings of the International Chemical and Biological Engineering Conference, 10, Braga, 2008. Universidade do Minho, Departamento de Engenharia Biológica, Braga, Portugal, pp 1279–1284
Mussatto SI, Rodrigues LR, Teixeira JA (2009a) β-Fructofuranosidase production by repeated batch fermentation with immobilized Aspergillus japonicus. J Ind Microbiol Biotechnol 36:923–928
Mussatto SI, Rodrigues LR, Teixeira JA (2009b) Fructofuranosidase production by repeated batch fermentation with immobilized Aspergillus japonicus. J Microbiol Biotechnol 36:923–928
Papagianni M (2007) Advances in citric acid fermentation by Aspergillus niger: biochemical aspects, membrane transport and modelling. Biotechnol Adv 5:244–263
Roth AH, Dersch P (2009) A novel expression system for intracellular production and purification of recombinant affinity-tagged proteins in Aspergillus niger. Appl Microbiol Biotechnol 86:659–670
Rubio MC, Maldonado MC (1995) Purification and characterization of invertase from Aspergillus niger. Curr Microbiol 31:80–83
Sangeetha PT, Ramesh MN, Prapulla SG (2005) Recent trends in the microbial production, analysis and application of fructooligosaccharides. Trends Food Sci Technol 16:442–457
Shin HT, Baig SY, Lee SW, Suh DS, Kwon ST, Lim YB, Lee JH (2004) Production of fructo-oligosaccharides from molasses by Aureobasidium pullulans cells. Bioresour Technol 93:59–62
Storms R, Zheng Y, Li H, Sillaots S, Martinez-Perez A, Tsang A (2005) Plasmid vectors for protein production, gene expression and molecular manipulations in Aspergillus niger. Plasmid 53:191–204
Venkataraman K, Manjunath P, Raghavandra MRR (1975) Glucoamylases of Aspergillus niger NRRL 330. Ind J Biochem Biophys 12:107–114
Wang LM, Zhou HM (2006) Isolation and identification of a novel Aspergillus japonicus JN19 producing fructofuranosidase and characterization of the enzyme. J Food Biochem 30:641–658
Yoshikawa J, Amachi S, Shinoyama H, Fujii T (2006) Multiple fructofuranosidases by Aureobasidium pullulans DSM2404 and their roles infructooligosaccharide production. FEMS Microbiol Lett 265:159–163
Yoshikawa J, Amachi S, Shinoyama H, Fujii T (2007) Purifuction and some properties of β-fructofuranosudase I formed by Aureobasidium pullulans DSM 2404. J Biosci Bioeng 103:491–493
Zuccaro A, Götze S, Kneip S, Dersch P, Seibel J (2008) Tailor-made fructooligosaccharides by a combination of substrate and genetic engineering. Chem Bio Chem 9:143–149
Acknowledgements
The authors acknowledge financial support by the German Research Foundation (DFG) within the framework of the Collaborative Research Centre “From Gene to Product” (SFB 578).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Driouch, H., Roth, A., Dersch, P. et al. Optimized bioprocess for production of fructofuranosidase by recombinant Aspergillus niger . Appl Microbiol Biotechnol 87, 2011–2024 (2010). https://doi.org/10.1007/s00253-010-2661-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-010-2661-9