Abstract
The aerobic sludge granules cultivated at high organic loading rates could effectively convert 100–700 mg l−1 nitrite to nitrogen gas with 400 or 1,200 mg l−1 dosed acetate. The denitrifying microbial community structure of the so-cultivated granules was investigated by 16S rRNA gene sequences and localized using fluorescence in situ hybridization (FISH). The 16S rRNA gene phylotypes in the clone library and FISH probes used exhibited high diversity among the bacteria and denitrifying communities, with the members of Betaproteobacteria predominant that were closely related to families Comamonadaceae, Nitrosomonadaceae, Alcaligenaceae, and Rhodocyclaceae. The confocal laser scanning microscope and staining test revealed that active microbial community principally distributed at 200–250 μm beneath the outer surface, embedded in extracellular polymeric substances.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Denitrifying bacteria converts nitrite into nitrogen gas by enzymatic pathway consisting three successive steps involving nitrite reductase, nitric oxide reductase, and nitrous oxide reductase (Mellor et al. 1992; Shapeligh et al. 1987).
The dissolved oxygen level affects the expression of nitrite reductase and nitrous oxide reductase, hence the denitrification efficiency of nitrite, too. High levels of nitrite inhibit denitrification process in wastewater treatment (Glass et al. 1997). A biological treatment process that can denitrify nitrite at high concentrations is desirable.
The aerobic sludge granulation technology had several advantages over conventional wastewater treatment systems, including possessing a dense and strong microbial structure, good settleability, high biomass retention, tolerance to high organic loading rate (OLR), and to high levels of toxic substances (Liu and Tay 2004; Adav et al. 2008). Aerobic granules were used to conduct simultaneous nitrification–denitrification process for wastewaters (Beun et al. 2001; Tsuneda et al. 2003; de Kreuk et al. 2005; Mosquera-Corral et al. 2005; Adav et al. 2009). Denitrifying bacteria thus present in excess quantity in cultivated aerobic granules. The microbial diversity in acetate fed aerobic sludge granules is seldom reported.
This work aims at cultivating stable aerobic granules that can degrade high concentrations of nitrite and at analyzing microbial community of acetate utilizing denitrifiers using 16S rRNA gene amplification and cloning. The spatial distributions of typical denitrifiers were probed using fluorescent in situ hybridization (FISH) probes.
Materials and methods
Reactor start-up
Aerobic-activated sludge was obtained from a local municipal wastewater treatment plant in Taipei, Taiwan. The sludge sample (1 l in each reactor) was seeded in column-type sequential batch reactors (120 × 6 cm). The column reactor was fed with synthetic wastewater containing acetate as the sole carbon source with the following media composition (in gl−1): sodium acetate, 0.4; (NH4)2SO4, 1.0; NaCl, 0.2; MgSO4•7H2O, 0.2; FeCl3, 0.02; CaCl2•2H2O, 0.01; K2HPO4, 1.65; KH2PO4, 1.35; pH 6.8 ± 0.2; and micronutrients, 1.0 ml l−1 (Moy et al. 2002). The chemical oxygen demand (COD) concentration of this initial feed was 292 ± 17 mgl−1 and was increased to attain higher OLR by proportionally adjusting the concentration of each chemical ingredient except buffer constitutes (K2HPO4 and KH2PO4). Fine air bubbles for aeration and mixing were fed through the reactor bottom at superficial velocity of 3.0 cm s−1. The reactors were operated sequentially in 4-h cycles with 5-min settling, 10-min effluent withdrawal, and 5-min filling, and the remaining time in a cycle was the reaction time. The volumetric exchange ratio of liquid was 50%.
PCR amplification of 16S rRNA gene sequences and library construction
The DNA from granules were extracted via enzymatic lysis using extraction buffer (100 mM Tris–HCl at pH 8.0, 100 mM EDTA, and 1.5 M NaCl) containing Proteinase K (10 mg ml−1; Amresco Inc., Solon, OH, USA), as described previously (Adav et al. 2007). The isolated DNA was purified by Gene-Spin™ (Protect Technology Co. Ltd, USA). PCR amplification of 16S rRNA gene sequences was carried out using the primers F27 (5′-CCA GAG TTT GAT CMT GGC TCA G-3′) and R1492 (5′-TAC CTT GTT ACG ACT T-3). PCR amplification was performed as recommended by Polz and Cavanaugh (1998) to reduce bias in amplification. Briefly, 50-μl reaction volumes contained PCR buffer with 1.5 mM MgCl2, 200 μM dNTPs, 100 ng genomic DNA, 0.5 U Taq DNA polymerase, and 1 μM of each primer. PCR amplification were performed in Eppendorf mastercycler (Eppendorf AG, Hamburg, Germany) under the following conditions: one cycle of 95°C for 3 min; 25 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 90 s; and one cycle of 10 min at 72°C. PCR products were purified, cloned using the Qiagen PCR cloning kit (Qiagen, Valencia, CA) as per the manufacturer’s instructions, and transformed into Escherichia coli TOP10F cells (Invitrogen, Carlsbad, CA).
In total, 102 clones were sequenced using the ABI Prism model 3730 (version 3.2). The 16S rRNA gene sequences were aligned by the multiple alignments ClustalW. Phylogenetic trees based on 16S rRNA gene sequences were constructed by the neighbor-joining method using the MEGA software. To estimate the confidence of the tree topologies, bootstrap re-sampling analysis for 1,000 replicates was performed. The gene sequences of the clones were deposited in GenBank under accession numbers GQ891759 to GQ891860.
FISH and CLSM imaging
Sludge granule samples were washed with deionized water and fixed with 4% paraformaldehyde. The fixed sludge granule samples were then dehydrated by successive passages through 50%, 80%, and 100% ethanol (10 min each) and were subsequently embedded in Shandon cryomatrix (Thermo Scientific, Pittsburg, PA, USA) and frozen at −20°C. The frozen sludge granules were then cut into 40-μm-thick sections with a cryotome (model CS-3306, Thermo Shandon, Runcorn, UK). The sections of sludge samples were immobilized on glass slides coated with polysine for FISH experiments. The hybridization was performed as described previously (Amann et al. 1990). The probes and formamide concentration used in this study were listed in Table 1. The samples were hybridized using hybridization buffer (0.9 M NaCl, 100 mM Tris–HCl at pH 7.4, 0.01% sodium dodecyl sulfate) containing 50 ng μl−1 of fluorescently labeled probes and incubated for 30 min. The samples were washed with washing buffer (0.9 M NaCl, 100 mM Tris–HCl at pH 7.4, 0.01% sodium dodecyl sulfate) and ringed with Milli-Q water. The cells hybridized with the probes were observed with a confocal laser scanning microscope (Leica TCS SP5 Confocal Spectral Microscope Imaging System, Germany).
Analytical methods
The dry weight of granules, volatile suspended solids (VSS), sludge volume index (SVI), and nitrate was measured according to Standard Methods (APHA 1998). The granules were sampled, washed with distilled water, and inoculated in 21 serum bottles (bottle volume, 200 ml) containing 100 ml synthetic wastewater supplemented with 400 mgl−1 acetate at different NO2–N concentrations. The test bottles were sealed with a butyl rubber stopper and incubated at room temperature (25 ± 2ºC). At fixed time interval, concentration of NO2–N in filtered samples was determined by high-performance liquid chromatography (Ecom LCP 4100 Pump, C18 column, LCD 2083) with 10 mM NaH2PO4 in acetonitrile/water (150:850 v/v) as described by Tsikas et al. (1999). Similarly, experiments were performed with different acetate concentrations. The size of sludge granules was measured by a laser particle size analysis system (Mastersizer Series 2600; Malvern, Instruments, Worcestershire, UK) or an image analysis system. Washed granules were prepared for scanning electron microscopic (SEM; Jeol JSM-5310, Tokyo, Japan) observation via fixing with 2.5% glutaraldehyde for 2 h and dehydration via successive passages through 30%, 50%, 75%, 85%, 90%, 95%, and 100% ethanol followed by critical drying in critical point dryer (HCP-2, Hitachi Co. Ltd., Tokyo, Japan).
Results
Reactor performance and granules characteristics
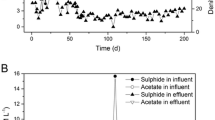
During the first week of reactor operation, most biomass was floc-like and small aggregates approximately 400–600 μm in size emerged in second week. With the beginning of sludge granulation, the COD removal rate increased from 60% to 75–80% in the test reactors. The granule size, SVI (Fig. 1) and settling velocity increased with reactor operation period and remain stable. Further reactor operation for 90 days, the COD removal rate ranged between 95.8% and 96.5%.The matured sludge granules had smooth surfaces without surface filaments and distinct-shaped various cell morphotypes, including rods and cocci (Fig. 2). At the end of first month of rector operation, COD removal rate was 95.5%. The settling velocity of the granule was 47.2 ± 7.3 m h−1.
The batch test of the granules showed nitrification–denitrification potential. At the initial concentration of 300 mg l−1 NO −2 –N and the supplement of 1,200 mg l−1 acetate, NO −2 –N removal rate was 2.35 ± 0.05 mg NO −2 –N g−1 VSS h−1. While the corresponding value for NO −2 –N removal at 400 mg l−1 acetate supplement was 1.18 ± 0.02 mg NO −2 –N g−1 VSS h−1 (Fig. 3). The granules could denitrify 600 mg NO −2 –N in 105 h when supplied with acetate 1,200 mgl−1. In denitrification test with the initial concentration of 100–700 mgl−1 NO −3 –N, complete denitrification without accumulation of nitrite was achieved (data not shown).
Bacterial diversity in aerobic sludge granule
The matured granules of size 3.5–5.0 mm from the SBR reactor that exhibited >95% COD removal rate at OLR 19.5 kg m−3 day−1 were used to analyze the microbial diversity. In total, 102 randomly selected bacterial clones from bacterial 16S rRNA gene clone libraries were analyzed. The compositions of the bacterial 16S rRNA gene libraries for the aerobic granules showed many sequences with high sequence similarity (97–99%) to sequences of uncultivated microorganisms. The microbial community composition in aerobic sludge granules demonstrated dominancy of Proteobacteria and exhibited 21 phylotypes. Of 102 clones, 66 clone sequences belong to phylum Proteobacteria (64.7%), 34 clones to Bacteroidetes (33.3%), and only two clones affiliated to Firmicutes phyla (2.0%). The Proteobacteria-related clone sequences were further grouped into Alphaproteobacteria, Betaproteobacteria, and Gammaproteobacteria.
Proteobcteria
Of the 53 clone sequences that related to the Betaproteobacteria, mostly belong to members of families that include denitrifying bacterial community, such as Rhodocyclaceae (one), Comamonadaceae (44), Nitrosomonadaceae (three), Alcaligenaceae (two), and Incertae sedis (two), while one sequence was unclassified Betaproteobacteria. The clone sequences (48) belonging to Burkholderiales showed dominancy of acetate-denitrifying clusters of members belonging to Comamonadaceae family (44), two clone sequences (G-23 and G-64) belong to genus Pelomonas; one each was affiliated to Alcaligenes (clone G-27) and Castellaniella genus (G-78). Phylotypes G-50 clustered within family Rhodocyclaceae (99% bootstrap) with Azoarcus sp. by 95% bootstrap. The clone sequences G-65, G-18, G-61, G-77, and G-58 clustered within Comamonadaceae family and were similar to Comamonas sp. The clone sequence G-14 affiliated with Delftia acidovorans (92%), while the other two clone sequences (G-64 and G-23) formed a separate sub-cluster with iron-reducing bacterium by 96% bootstrap value. Table 2 and Fig. 3 indicate the relative abundances of acetate utilizing members of the family Comamonadaceae in the acetate fed aerobic granule. Three clone sequences (G-10, G-67, and G-22) were closely related with members of family Nitrosomonadaceae and had 96% and 99% sequence similarity with Nitrosomonas sp. Phylotype G-78 clustered with the members of the family Alcaligenaceae.
The Alphaproteobacteria represents 2.9% of clone library. Three sequences belonged to Alphaproteobacteria with 97–99% similarity to strains affiliated to family Hyphomicrobiaceae. Clone G-90 had sequence similarity of 99% with strain Hyphomicrobium denitrificans strain DSM 1869, while clone G-21 and G-26 exhibited 97% sequence similarity with Devosia sp.
Ten sequences were affiliated with the Gammaproteobacteria. Seven sequences were related to family Pseudomonadaceae (Chryseomonas, 14.3%; Pseudomonas, 71.4%; and unclassified Pseudomonadaceae genus, 14.3%); six clones were related to strain Pseudomonas and one clone to Chryseomonas; and three belonged to Xanthomonadaceae family. The clone sequence G-24 and G-11clustered within Xanthomonadaceae family, G-24 clustered with Thermomonas sp. by 98% bootstrap while G-11 with Rhodonobacter sp.
Non-proteobacteria
Of the total 102 clone sequences, 34 grouped within Bacteroidetes and two under Firmicute. The Bacteroidetes were further categorized under class Flavobacteria (two), Bacteroidetes (four), Sphingobacteria (27), and unclassified Bacteroidetes (one). The two clone sequences (G-38 and G-54) clustered within Lactococcus with 100% bootstrap value. The three phylotypes grouped within Dysgonomonas gadei group. The clone sequence G-103 clustered with Cryomorphaceae bacterium with 92% bootstrap. The analysis of the clone sequences belonging to Sphingobacteriales (27) revealed 3.7% Crenotrichaceae, 3.7% Sphingobacteriaceae, 7.4% Flexibacteraceae, and 74.1% Saprospiraceae family members, while others (11.1%) were unclassified (Table 2 and Fig. 4).
Phylum Proteobacteria neighbor-joining phylogentic tree of 16S rRNA gene sequences. The sequences from the present study and close relatives were aligned by multiple alignments ClustalW. Bootstrap analyses were conducted on 1,000 samples, and percentage greater than 50% are indicated at the nodes. Scale bar, 0.02 changes per nucleotide position
Spatial distribution of microorganisms in granules
FISH, using EUB and Cy3-labeled DEN581 probes, revealed that the outer layer of 250 μm (Fig. 5a) was dominated by acetate-denitrifying bacterial community. The hybridization with probe DEB220 specific for acetate-denitrifying cluster and some members of the Comamonadaceae family demonstrated that members of Comamonadaceae family clustered within 100–200 μm granule depth (Fig. 5b). Some members of acetate-denitrifying community with Rhodocyclaceae and Comamonadaceae family members were clustered very close to each other at the granule interior within 100–200 μm granule depth (Fig. 5c, d). The layered structure of acetate-denitrifying cluster and members of Rhodocyclaceae and Comamonadaceae families was repeatedly observed in all the granular sections analyzed. The cells hybridized with probe AT1458, specific for Azoarcus–Thauera, were numerically important bacteria and were located in core region of the granule (Figs. 6 and 7).
Non-Proteobacteria neighbor-joining phylogentic tree of 16S rRNA gene sequences. The sequences from the present study and close relatives were aligned by multiple alignments ClustalW. Bootstrap analyses were conducted on 1,000 samples, and percentage greater than 50% are indicated at the nodes. Scale bar, 0.02 changes per nucleotide position
CLSM image of in situ functional analyses of denitrifying bacteria in the SBR sludge granule. a Hybridized with both the EUB (red) and DEN581 (yellow) probe targeting acetate-denitrifying cluster, bar = 200 μm; b hybridized with EUB (red) and DEN220 probe targeting Comamonadaceae family members(yellow), bar = 300 μm; c granule from denitrification batch test under anoxic condition, dual-hybridized for total bacteria (red) and Rhodocyclaceae family members (pink), bar = 75 μm; d granule from denitrification batch test under anoxic condition hybridized with EUB (red), probe DEN220 targeting Comamonadaceae family members (yellow), and probe DEN220a targeting Rhodocyclaceae family members (pink), bar = 75 μm
Discussion
The abundance and dominancy of the denitrifying bacterial community in cultivated aerobic granules was likely due to the denitrification activity within the granules at high dosed level of ammonium and sufficient electron donor for denitrification. Majority of non-proteobacterial clone sequences belong to Bacteroidetes, which are obligate anaerobes, indicating the presence of anoxic core in the granule (Adav et al. 2009). The majorities (64.7%) of the 16S rRNA gene sequences from clone analyzed were denitrifying microbial community within Proteobacteria phylum, and 51.9% were from the families Comamonadaceae. Denitrifying representatives of Alpha-, Beta-, and Gammaproteobacteria, Firmicutes, and Bacteroidetes were found in the sludge granules, and apart from genera known to harbor denitrifiers, such as Pseudomonas, Comamonas, and Acidovorax, genera less frequently observed in cultivation studies of denitrifiers were encountered prominently in this study. The major strains assigned for the clones in Comamonadaceae family were Comamonas, Alicycliphilus, Diaphorobacter, and Delftia. Similarly, Kim et al. (2006) reported members of Acidovorax, Alicycliphilus, Comamonas, and Diaphorobacter strain in denitrifying activated sludge, while Park et al. (2005) reported denitrifying Thauera sp. with dominancy of Hydrogenophaga sp. in autohydrogenotrophic biofilm reactor. The other clone sequences within protobacteria phylum were affiliated with the members of families Nitrosomonadaceae under Betaproteobacteria and Hyphomicrobiaceae, Pseudomonadaceae, and Xanthomonadaceae. A significant positive correlation between the SBR denitrification rates and the abundance of the denitrifiers was demonstrated. According to Otlanabo (1993), various species of Achromobacter, Agrobacterium, Alcaligenes, Bacillus, Chromobacterium, Flavobacterium, Hyphomicrobium, Pseudomonas, and Vibrio are responsible for denitrification.
Researchers attempted to identify denitrifiers in activated sludge (Drysdale et al. 2001; Etchebehere et al. 2001, 2002; Khan and Hiraishi 2002; Khan et al. 2002; Ginige et al. 2005). Our finding and that of Ginige et al. (2005) suggested that the members of the Comamonadaceae family play a major role in denitrification processes in the presence of acetate. The present study also identified members of Nitrosomonadaceae and Alcaligenaceae within Betaproteobacteria, which was not reported in related works. Members of denitrifying Fe(II) oxidizers that belong to Xanthomonadaceae and denitrifying Pseudomonas sp. of Pseudomonadaceae family were identified in the present study that may be attributable to the strong binding strength of EPS matrix of aerobic granules. The present study also identified members of families Flavobacteriaceae, Porphyromonadaceae, Flexibacteraceae, and Saprospiraceae within Bacteroidetes phylum that also involved in the processes of denitrification (Mills et al. 2008). The clone sequences belonging to Azoarcus were identified in aerobic sludge granule, corresponding to the works by Wagner and Loy (2002) on activated sludge.
The clone analysis from our acetate fed SBR differs from the reactor fed with methanol (Ginige et al. 2009). The denitrifying microbial community depends on the fed major carbon sources and electron donor. The acetate can be degraded by microbes using tricarboxylic acid cycle and is one of the best carbon sources for denitrification (Grabinskaloniewska 1991). The granules cultivated at high OLR are composed of diverse denitrifying microbial community and are able to degrade high-level nitrogenous compound from wastewaters.
References
Adav SS, Lee DJ, Lai JY (2009) Biological nitrification–denitrification with alternating oxic and anoxic operation using aerobic granules. Appl Microbiol Biotechnol. doi:10.1007/s00253-009-2129-y
Adav SS, Chen MY, Lee DJ, Ren NQ (2007) Degradation of phenol by aerobic granules and isolated yeast Candida tropicalis. Biotechnol Bioeng 96:844–852
Adav SS, Lee DJ, Show KY, Tay JH (2008) Aerobic granular sludge: Recent advances. Biotechnol Adv 26:411–423
Amann RI, Binder BJ, Olson RJ, Chisholm SW, Devereux R, Stahl DA (1990) Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol 56:1919–1925
APHA (1998) Standard methods for the examination of water and wastewater, 20th edn. American Public Health Association, Washington
Beun JJ, Van Loosdrecht MCM, Heijnen JJ (2001) N-removal in a granular sludge sequencing batch airlift reactor. Biotechnol Bioeng 75:82–92
de Kreuk MK, Heijnen JJ, van Loosdrecht MC (2005) Simultaneous COD, nitrogen, and phosphate removal by aerobic granular sludge. Biotechnol Bioeng 90:761–769
Drysdale GD, Kasan HC, Bux F (2001) Assessment of denitrification by the ordinary heterotrophic organisms in an NDBEPR activated sludge system. Water Sci Technol 43:147–154
Etchebehere C, Errazquin I, Barrandeguy E, Dabert P, Moletta R, Muxi L (2001) Evaluation of the denitrifying microbiota of anoxic reactors. FEMS Microbiol Ecol 35:259–265
Etchebehere C, Errazquin MI, Dabert P, Muxi L (2002) Community analysis of a denitrifying reactor treating landfill leachate. FEMS Microbiol Ecol 40:97–106
Ginige MP, Bowyer JC, Foley L, Keller J, Yuan Z (2009) A comparative study of methanol as a supplementary carbon source for enhancing denitrification in primary and secondary anoxic zones. Biodegradation 20:221–234
Ginige MP, Jurg K, Blackall LL (2005) Investigation of an acetate-fed denitrifying microbial community by stable isotope probing, full-cycle rRNA analysis, and fluorescent in situ hybridization-microautoradiography. Appl Environ Microbiol 71:8683–8691
Glass C, Silversterin JA, Oh J (1997) Inhibition of denitrification in activated sludge by nitrite. Water Environ Res 69:1086–1093
Grabinskaloniewska A (1991) Denitrification unit biocenosis. Water Res 25:1565–1573
Khan ST, Hiraishi A (2002) Diaphorobacter nitroreducens gen. nov., sp nov., a poly(3-hydroxybutyrate) degrading denitrifying bacterium isolated from activated sludge. J Gen Appl Microbiol 48:299–308
Khan ST, Horiba Y, Yamamoto M, Hiraishi A (2002) Members of the family Comamonadaceae as primary poly(3-hydroxybutyrate-co-3-hydroxyvalerate)- degrading denitrifiers in activated sludge as revealed by a polyphasic approach. Appl Environ Microbiol 68:3206–3214
Kim H, Vanparys B, Wittebolle L, Verstraete W, Boon N, Vos PD (2006) Cultivation of denitrifying bacteria: optimization of isolation conditions and diversity study. Appl Environ Microbiol 72:2637–2643
Liu Y, Tay JH (2004) State of the art of biogranulation technology for wastewater treatment. Biotechnol Adv 22:533–563
Mellor RB, Ronnennberg J, Campbell HW (1992) Reduction of nitrate and nitrite in water by immobilized enzymes. Nature 355:717–719
Mills HJ, Hunter E, Humphrys M, Kerkhof L, McGuinness L, Huettel M, Kostka JE (2008) Characterization of nitrifying, denitrifying, and overall bacterial communities in permeable marine sediments of the northeastern Gulf of Mexico. Appl Environ Microbiol 74:4440–4453
Mosquera-Corral A, de Kreuk MK, Heijnen JJ, van Loosdrecht MCM (2005) Effects of oxygen concentration on N-removal in an aerobic granular sludge reactor. Water Res 39:2676–2686
Moy BYP, Tay JH, Toh SK, Liu Y, Tay STL (2002) High organic loading influences the physical characteristics of aerobic sludge granules. Lett Appl Microbiol 34:407–412
Otlanabo NL (1993) Denitrification of ground water for potable purposes. WRC Report No. 403/1/93.
Park HI, Choi YJ, Pak D (2005) Autohydrogenotrophic denitrifying microbial community in a glass beads biofilm reactor. Biotechnol Lett 27:949–953
Polz MF, Cavanaugh CM (1998) Bias in template-to-product ratios in multitemplate PCR. Appl Environ Microbiol 64:3724–3730
Rabus R, Wilkes H, Schramm A, Harms G, Behrends A, Amann R, Widdel F (1999) Anaerobic utilization of alkylbenzenes and n-alkanes from crude oil in an enrichment culture of denitrifying bacteria affiliating with the beta-subclass of Proteobacteria. Environ Microbiol 1:145–157
Shapeligh JP, Davies KJP, Payne WJ (1987) Detergent inhibition of nitric oxide reductase. Biochim Biophys Acta 911:334–340
Tsikas D, Rossa S, Sandmann J, Frolich JC (1999) High-performance liquid chromatographic analysis of nitrite and nitrate in human plasma as S-nitroso-N-acetylcysteine with ultraviolet absorbance detection. J Chromatogr B Biomed Sci Appl 724:199–201
Tsuneda S, Nagano T, Hoshino T, Ejiri Y, Noda N, Hirata A (2003) Characterization of nitrifying granules produced in an aerobic upflow fluidized bed reactor. Water Res 37:4965–73
Wagner M, Loy A (2002) Bacterial community composition and function in sewage treatment systems. Current Opin Biotechnol 13:218–227
Acknowledgment
This project is financially supported by the State Key Laboratory of Water Resource and Environment (SKLWRE) and Harbin Institute of Technology (HIT), China
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Adav, S.S., Lee, DJ. & Lai, J.Y. Microbial community of acetate utilizing denitrifiers in aerobic granules. Appl Microbiol Biotechnol 85, 753–762 (2010). https://doi.org/10.1007/s00253-009-2263-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-009-2263-6