Abstract
Aptamers are short, single stranded nucleic acids which bind a wide range of different ligands with extraordinary high binding affinity and specificity. The steadily increasing number of aptamers is accompanied by an expanding range of applications in biotechnology. We will describe new developments in the field including the use of aptamers for conditional gene regulation and as biosensors. In addition, we will discuss the potential of aptamers as tags to visualize RNA and protein distribution in living cells and as therapeutics. Furthermore, we will consider biotechnological applications of riboswitches for gene regulation and as drug target.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Nucleic acids have long been thought to be a mere coding device for proteins and inferior to proteins with regard to their enzymatic and regulatory activity. Ribozymes or functional RNA-protein complexes like self-splicing introns, the ribosome or ribonuclease P (RNase P), were seen as an exception, but the discovery of more and more noncoding RNAs in every kingdom of life with inherent catalytic properties or with an active function in regulating gene expression led to a reassessment of the biological potential of RNA and promoted research on the capabilities and occurrence of nucleic acids not only involved in protein-coding.
The hypothesis that—in ancient times—RNAs were the first players in early life (“The RNA world theory”, Chen et al. 2007) inspired the development of in vitro evolution techniques. The so-called systematic evolution of ligands by exponential enrichment (SELEX) technology allowed the identification of RNAs with the capability to bind to a plethora of different ligands, so called aptamers, but also the selection of catalytically active RNAs, like ligases (Levy et al. 2005) or polymerases (Zaher and Unrau 2007). The identification of RNAs capability to bind with high affinity and selectivity to a variety of target molecules and to catalyze complex chemical reactions is an impressive demonstration that RNA molecules can rival their protein companions in terms of their biological versatility.
Aptamers—versatile “RNA antibodies”
Aptamers are single stranded nucleic acids which bind their cognate ligands with high affinity and specificity. They can be isolated in an in vitro selection process called SELEX (Fig. 1; Ellington and Szostak 1990; Tuerk and Gold 1990). Thereby, a large combinatorial library of single stranded RNA or DNA molecules with random sequences is subjected to a selection process. Nonbinding molecules are removed during washing steps, whereas nucleic acids with binding affinity to the target of interest are eluted. These are amplified and subjected to further rounds of selection. During these iterative cycles, the selection pressure can be increased resulting in molecules, so-called aptamers, with remarkable binding affinities and specificities. In the last years, aptamers, which specifically bind to a plethora of different ligands including ions, small organic molecules like antibiotics, proteins, and even whole cells, have been selected. Beyond this, the SELEX process has been advanced to semi- and fully automated selections (Cox and Ellington 2001; Eulberg et al. 2005; Wochner et al. 2007), and aptamers can be commercially purchased.
SELEX: Systematic evolution of ligands by exponential enrichment. A library of single stranded DNA or RNA molecules (various symbols) is incubated with an immobilized target. Nonbinding molecules are separated from binding aptamers by washing steps. Aptamers are eluted either specifically by an excess of ligand or unspecifically by denaturation (e.g., pH change) and subsequently amplified using PCR and RT-PCR techniques resulting in a pool enriched for sequences binding to the target. The pool is then subjected to further rounds of selection. Enriched sequences are cloned and analyzed in detail
The steadily increasing number of aptamers is accompanied by an expanding range of applications. In this review, we will discuss new developments including the use of aptamers (a) for conditional gene regulation, (b) to visualize RNA and protein distribution, (c) as biosensors, and (d) in medicine.
Aptamers as regulators for conditional gene expression
The ability of aptamers to regulate gene expression has been exploited even before their natural counterparts, the riboswitches (see section below), have been discovered. One important characteristic of aptamers is that they often form a preformed binding pocket but adopt their final structure only in the presence of the ligand (Hermann and Patel 2000). This ligand-dependent induced fit has been employed to develop conditional gene expression systems.
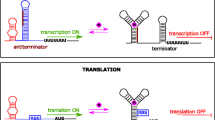
Translation initiation can be regulated by inserting aptamers into the 5′ untranslated region (UTR) of a messenger RNA (mRNA). In prokaryotes, the accessibility of the ribosomal binding site (Shine–Dalgarno (SD)) can be inhibited either by sterical hindrance due to a closely located stem loop structure or by base-pairing interactions (Fig. 2a, b). Control elements have been designed in a way that ligand-binding frees the SD by induction of aptamer-folding (Fig. 2a; Desai and Gallivan 2004; Fowler et al. 2008; Lynch et al. 2007; Lynch and Gallivan 2009) or by ligand-induced shifting of a stem-loop structure (Fig. 2b; Suess et al. 2004). In addition, off switches have been reported where the SD is masked in the presence of ligand leading to gene expression only in the nonbound state (Jo and Shin 2009; Topp and Gallivan 2008).
Aptamer-based control of gene expression in prokaryotes (a, b) and eukaryotes (c–e). Translation initiation in bacteria is controlled by the accessibility of the SD sequence. Aptamers can inhibit ribosome-binding either by base-pairing with the SD (a) or by sterical hinderance (b). In eukaryotes, aptamer-ligand complexes can interfere with the ribosomal scanning (c), pre-mRNA splicing (d), and siRNA processing (e)
In yeast, aptamers inserted into the 5′UTR are able to interfere with the binding of the small ribosomal subunit or with the scanning process only in the ligand-bound conformation (Fig. 2c; Hanson et al. 2003). Interestingly, only very few small molecule-binding aptamers can be used as molecular switch for conditional gene expression (Grate and Wilson 2001; Hanson et al. 2003; Harvey et al. 2002; Suess et al. 2003; Werstuck and Green 1998). Currently, we attempt to unravel the structural basis for the regulatory activity of these aptamers.
An aptamer which binds the drug tetracycline shows a remarkable dynamic range for gene regulation in yeast (Weigand et al. 2008). Three copies of the aptamer within a 5′UTR lead to a tight translational regulation which is sufficient even for the control of essential genes. This led to the development of an insertion cassette for conditional mutants by polymerase chain reaction (PCR)-based integration of the aptamers into the Saccharomyces cerevisiae chromosome (Kotter et al. 2009). Compared to protein-based conditional gene expression systems, this approach is independent of auxiliary factors like regulatory proteins or the strain background and offers easy and high efficient methods to produce conditional mutants in yeast.
One major step in mRNA processing in eukaryotes is pre-mRNA splicing. Alternative splicing is the main source producing protein diversity, and consequently, miss-splicing leads to severe diseases. We developed a system for conditional splicing in yeast using the tc aptamer (Weigand and Suess 2007). Inclusion of the 5′ splice site (SS) into the closing stem of the aptamer masks the 5′SS upon ligand-binding and subsequently abolished gene expression (Fig. 2d). Gaur and coworkers incorporated human splice sites into a theophylline aptamer thereby inhibiting pre-mRNA splicing in the presence of the ligand (Gusti et al. 2008; Kim et al. 2008; Kim et al. 2005).
Aptamers have also been used to control RNA interference. Small interfering RNAs (siRNAs) produced as double stranded precursors (short hairpin RNAs (shRNAs)) are processed by the RNase III-type endonuclease Dicer. The 21-nt long cleavage products are subsequently bound by the RNA-induced silencing complex and guided to their mRNA targets. Processing by Dicer can be controlled by aptamers inserted into the terminal loop of the shRNA precursor (Fig. 2e). Yokobayashi and coworkers showed that processing into siRNAs is only possible in the unbound form whereas the aptamer–ligand complex is not able to support cleavage (An et al. 2006; Tuleuova et al. 2008). In contrast, Smolke and coworkers designed off switches in which theophylline binding to the aptamer interferes with base-pairing of the shRNA stems and inhibits siRNA processing (Beisel et al. 2008).
Aptamers can also be used as binding domains for more complex regulatory RNA modules. Such modular RNA switches combine different functionalities, e.g., an aptamer as sensor and a ribozyme as catalytically active domain resulting in allosterically regulated self-cleaving ribozymes. Hartig and coworkers employed a theophylline-dependent hammerhead ribozyme to control gene expression in Escherichia coli (Fig. 3a; Wieland et al. 2009b; Wieland and Hartig 2008). The ribozyme is attached to the 5′UTR in a way that allows base-pairing with the SD, thereby preventing ribosome access. The ribozyme is inactive in the unbound state. Ligand-binding then induces proper folding of the active center. This promotes self-cleavage which frees the SD leading to gene expression. Smolke and coworkers inserted theophylline- or tetracycline-dependent hammerhead ribozymes into the 3′UTR of yeast mRNAs (Fig. 3b; Win and Smolke 2007). Self-cleavage of the ribozyme in the absence and presence of ligand, respectively (both is possible), promotes rapid degradation of the mRNA due to the missing poly(A) tail. In addition, they combined the two aptamers on one ribozyme leading to logical AND, NAND, OR, and NOR gates (Win and Smolke 2008).
Control of gene expression using allosterically regulated ribozymes. a Control of translation initiation in E. coli: The SD is sequestered in one of the three stems of the hammerhead ribozyme which interferes with binding of the small ribosomal subunit. Ligand binding to the aptamer either promotes self-cleavage of the ribozyme leading to unmasking of the SD due to dissociation of the 5′ part of the sequestration stem. b Control of mRNA stability in S. cerevisiae: The ribozyme is inserted into the 3′UTR. Ligand-dependent cleavage leads to a shortened UTR without poly(A) tail which is rapidly degraded. Ligand binding to the aptamer domain leads to a conformational shift either promoting or inhibiting the activity of the ribozyme
Reengineering natural riboswitches for the conditional control of gene expression
Riboswitches are regulatory elements which reside within the mRNA they control (Roth and Breaker 2009). They respond to small metabolites, like amino acids and cofactors, but also ion-dependent switches are known. Riboswitches are mainly present in the 5′UTRs and are composed of two domains, a binding domain (also called aptamer domain), which recognizes the specific ligand with high binding affinity and specificity and an expression platform. The expression platform interprets the binding status of the aptamer domain and mediates changes in gene expression either by transcription termination or the sequestration of the SD sequence. Reports about eukaryotic riboswitches are rare. A few examples are located within introns where they regulate alternative splicing (Cheah et al. 2007; Wachter et al. 2007).
Natural riboswitches combine both sensory and regulatory function and are independent of auxiliary regulatory proteins. The principle of direct RNA-ligand interaction has been exploited to develop aptamer-based conditional gene expression systems (see the first section of the review). Natural riboswitches depend on the metabolic status of the cell and are therefore not suited for conditionally control of heterologous genes. In addition, their binding pocket is highly specific for their cognate ligand. Thus, altering their ligand recognition mode towards nonmetabolizable ligands might be difficult.
However, Yokobayashi and coworkers showed that natural riboswitches can still be reengineered to acquire new functions. They reversed the ligand response of a thiamin pyrophosphate (TPP) riboswitch, which normally represses gene expression upon ligand-binding by turning it into an on switch (Muranaka et al. 2009; Nomura and Yokobayashi 2007). Furthermore, they exchanged the aptamer domain with a theophylline aptamer and created AND and NAND logic gates (Sharma et al. 2008). Hartig and coworkers used the aptamer domain of the TPP riboswitch to create allosterically regulatable ribozymes and used them for ligand-dependent control of translation in E. coli (see above and Fig. 2a; Wieland et al. 2009a). Here, both on and off switches were selected.
Aptamers as tags to visualize RNA and protein distribution
The imaging of intracellular RNA distribution and dynamics in living cells aids our understanding of biological processes, like nuclear export and localization to specific organelles or cytoplasmic regions. Most imaging techniques rely on targeting fluorescent proteins to a certain mRNA since no intrinsically fluorescent RNA motif has been discovered so far. To this end, green fluorescent protein (GFP) is fused to an RNA-binding protein, and the respective RNA recognition sequence is inserted (often in multiple repeats) into the untranslated region of the target mRNA. The phage-derived MS2-coat protein and its RNA recognition hairpin motif are widely used for this approach (Bertrand et al. 1998). An alternative approach employs an aptamer which binds the eukaryotic initiation factor 4A (eIF4A). eIF4A is comprised of two globular domains both of which are recognized by the aptamer. GFP is split in two halves, with each half fused to one domain of eIF4A (Valencia-Burton et al. 2007). Functional eIF4A and GFP get together only when both parts are bound and connected by the aptamer sequence. The advantage of this approach is in the lowered background fluorescence due to the splitting of GFP.
Tagging of RNAs with bulky proteins (usually 6–24 copies of the protein-binding motif are needed) might influence the integrity of an mRNA. Aptamers which directly bind a fluorescent dye would render auxiliary proteins unnecessary if the binding event is accompanied by a change in the fluorescence of the dye. Alternatively, ligands with intrinsic fluorescence can be modified in a way that they show fluorescence only upon aptamer-binding (Sando et al. 2007; Sparano and Koide 2007). A fluorescence increase has been reported for malachite green aptamers with a 2,000-fold-enhanced fluorescence upon binding (Babendure et al. 2003). However, application of this aptamer in vivo is restricted due to high toxicity of the ligand. Hahn and coworkers made use of an aptamer SRB2m which binds to the fluorophore sulforhodamine B (Holeman et al. 1998). The aptamer shows specific binding to its cognate ligand even in the presence of human total RNA extracts and in conjunction with long flanking sequences (Eydeler et al. 2009). Currently, the system might not be practical for studies in vivo due to nonspecific binding of sulforhodamine B to basic amino acids. However, the results promise that further studies will provide suitable tools for RNA detection in vivo (for a detailed review on the advantages and limitations of aptamers for in vivo imaging see Tyagi (2009)).
Aptamers have also been used to visualize proteins within cells. Commonly, GFP is fused to a protein of interest to track its spatial and temporal distribution but not all proteins can be expressed as GFP fusion. Liu and coworkers employed a fluorescently labeled aptamer to visualize protein internalization and its targeting to the cell nucleus in real time (Li et al. 2008). They attached the fluorophore Cy5 to the angiogenin-binding aptamer. Angiogenin is known to bind endothelial cells followed by a quick internalization through receptor-mediated endocytosis and subsequent nuclear localization. Using fluorescent microscopy, protein import into breast cancer cells has been monitored in real time.
Aptamers as biosensors
Biosensors are measuring heads equipped with a biological component. They rely on the direct spatial contact of a biodetector with a transducer and a signal processing unit (Fig. 4). Biosensors use biological systems like antibodies, enzymes, or cells to detect an analyte of choice. Several biophysical and biochemical detection systems have been developed which are based on electrochemical, optical, mass sensitive, and potentiometric principles and are reviewed in detail in Strehlitz et al. (2008). Here, we will focus on potential applications of aptamers as biosensors.
In many aspects, aptamers are comparable to monoclonal antibodies regarding their binding affinity and specificity (Liss et al. 2002). Beyond that, aptamers have some striking advantages. The selection process is in vitro and therefore can be performed against targets that are toxic or not immunogenic. In addition, the selection can be customized to later assay conditions and modifications for the immobilization of biochips, such as biotinylation, can easily be introduced. In spite of their reputation of being fragile (especially RNA), aptamers proved to be more resistant to denaturation and degradation than antibodies (Stadtherr et al. 2005). Furthermore, they have been shown to be active also in the presence of organic solvents (Ehrentreich-Forster et al. 2008). Aptamers can be produced in large scale through PCR and in vitro transcription, which is cheap, highly reproducible, and ensures the same quality for different batches compared to the animal-dependent production of antibodies. These advantages led to the development of biosensors based on DNA or RNA aptamers which recognize ions, small molecules, proteins, and whole cells.
Ions like potassium (K+), lead(II) (Pb2+), and mercury(II) (Hg2+) can be sensed indirectly through formation of the aptamer structure. Chang and coworkers detected K+ ions through G-quadruplex formation of an ATP-binding aptamer (Huang and Chang 2008). G-quadruplexes consist of a square arrangement of guanines stabilized by hydrogen-bonding and cations, especially K+. Therefore, folding of the aptamer is directly dependent on the K+ concentration. The biosensor was used to determine K+ levels in human urine which is an indicator for certain kidney diseases (Huang and Chang 2008). The same group employed ion-dependent aptamer-folding for a biosensor which detects both Pb2+ and Hg2+ in samples of soil and pond. Thereby, Hg2+ lead to a hairpin-like structure whereas Pb2+ induced G-quadruplex formation within an aptamer originally selected to bind thrombin (Liu et al. 2009).
Aptamer-based biosensors have also been developed for the detection of small molecules within biological samples. Aptamers were used for the detection of the antibiotic neomycin in milk (de-los Santos-Alvarez et al. 2007) or cocaine in blood serum (Swensen et al. 2009). Rimmele and coworkers developed an RNA aptamer-based biosensor for the fast (within minutes) detection of trinitrotoluene (TNT) in water and soil samples (Ehrentreich-Forster et al. 2008). They were able to determine the TNT concentrations directly in the field since the biosensor does not rely on complex instrumentation, and no elaborate extractions and concentration steps are needed.
A plethora of biosensors recognizing proteins like thrombin or IgE have also been developed and are reviewed in detail elsewhere (Strehlitz et al. 2008).
Aptamers can also be employed for the detection of pathogens. Cao and coworkers used a set of five DNA aptamers to detect Staphylococcus aureus in samples from patients with severe burn wound infection. In addition, the aptamers were able to distinguish S. aureus from evolutionary closely related bacteria. Using an aptamer mixture targeting different cell surface features further improved the applicability since the pathogen often changes its cell surface identity during the growth cycle (Cao et al. 2009). Oh and coworker used an aptamer for the detection of E. coli. E. coli cells were captured on antibody-conjugated magnetic beads and then decorated with aptamers. After separation, aptamers were released by heating and detected by quantitative real-time RT-PCR. The detection limit in pure E. coli samples was 10 cells/ml, in samples mixed with Listeria monocytogenes around 100 cells/ml (Lee et al. 2009b).
Göringer and coworker selected aptamers which target the cell surface of Trypanosoma brucei, the causative agent of sleeping sickness. Trypanosomes withstand the host immune response by the sequential expression of variable surface glycoproteins. Antigen-conjugated aptamers selected to recognize cell surface proteins have been shown to redirect the immune system to the pathogens (Homann et al. 2006). In addition, they developed aptamers which recognize the main endocytosis site of trypanosomes and become rapidly internalized. This opens up the possibility of toxin transport into the cell (Adler et al. 2008). Within these studies, aptamers have not only been used for detection but additionally as therapeutics implicating the potential of aptamers in drug development.
Aptamers in medical applications
Aptamers which bind specific proteins often inhibit their activity. This led to the development of the first aptamer-based drug. Macugen (or pegaptanib) is implicated in the treatment of the wet form of age-related macular degeneration (AMD; Apte 2008). In AMD, a splice isoform of the vascular-endothelial growth factor (VEGF) is overexpressed, promoting growth of new blood vessels. Macugen specifically binds and inhibits one splice isoform of VEGF leading to reduced pathologic vessel growth. Aptamers can also be employed as delivery vehicle for cytotoxic chemotoxins or siRNAs. Aptamers which specifically recognize cancer cells have been used leading to significant tumor reduction. Currently, many more aptamer-based drugs are in clinical trials. Recent developments and perspectives within this area are extensively reviewed in Mayer (2009).
The use of aptamers as drug is often limited due to low serum stability and delivery problems. Therefore, a method called aptamer displacement screening has been developed which makes use of aptamers to identify small molecule ligands of a given protein target with the same binding specificity (Hafner et al. 2008). Thereby, a small molecule library is screened for compounds which are able to displace a functional aptamer from the protein, assuming that they occupy the same binding site. Small molecules identified by this method often inherit the inhibition capacity of the aptamer. The method was successfully applied for the identification of new inhibitors for human immunodeficiency virus reverse transcriptase (Yamazaki et al. 2007).
Riboswitches as drug targets
Riboswitches control essential metabolic pathways within bacterial cells. They are highly specific toward their cognate ligand but to some extent also to close derivatives. It was recently shown that roseoflavin, an antibiotic produced by Streptomyces davawensis, binds to flavinmononucleotid (FMN) riboswitches and represses gene expression to the same extent as the natural ligand (Lee et al. 2009a; Ott et al. 2009; Serganov et al. 2009). However, resistant bacteria emerged which harbor mutations within the riboswitch sequence to escape this unnatural down regulation. Interestingly, S. davawensis also harbors an FMN riboswitch which binds to roseoflavin. Therefore, self-resistance has to be mediated by a different mechanism which is still undiscovered.
Since riboswitches are also present in pathogenic bacteria, they comprise novel targets for drug development. Specific inhibition of a riboswitch will be without any side effects on the eukaryotic host. The proof of principle has already been delivered by the Breaker group. They showed that the TPP riboswitches can also be recognized by pyrithiamine pyrophosphate (PTPP). PTPP binds the riboswitch with nearly the same efficiency and leads to repression of gene expression (Sudarsan et al. 2005; Thore et al. 2008). Similarly, lysine riboswitches can be recognized by lysine analogs like the antibacterial compound L-aminoethylcysteine (AEC; Blount et al. 2007; Serganov et al. 2008). However, unfortunately, resistance-conferring mutations within the respective riboswitches were rapidly identified.
High resolution crystal structures of riboswitches bound to their cognate ligand or a toxic derivative promise a structure guided design of novel drugs (Sudarsan et al. 2005; Thore et al. 2008). Time will tell if riboswitches really provide suitable targets for the development of new antibiotics.
References
Adler A, Forster N, Homann M, Goringer HU (2008) Post-SELEX chemical optimization of a trypanosome-specific RNA aptamer. Comb Chem High Throughput Screen 11:16–23
An CI, Trinh VB, Yokobayashi Y (2006) Artificial control of gene expression in mammalian cells by modulating RNA interference through aptamer-small molecule interaction. RNA 12:710–716
Apte RS (2008) Pegaptanib sodium for the treatment of age-related macular degeneration. Expert Opin Pharmacother 9:499–508
Babendure JR, Adams SR, Tsien RY (2003) Aptamers switch on fluorescence of triphenylmethane dyes. J Am Chem Soc 125:14716–14717
Beisel CL, Bayer TS, Hoff KG, Smolke CD (2008) Model-guided design of ligand-regulated RNAi for programmable control of gene expression. Mol Syst Biol 4:224
Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM (1998) Localization of ASH1 mRNA particles in living yeast. Mol Cell 2:437–445
Blount KF, Wang JX, Lim J, Sudarsan N, Breaker RR (2007) Antibacterial lysine analogs that target lysine riboswitches. Nat Chem Biol 3:44–49
Cao X, Li S, Chen L, Ding H, Xu H, Huang Y, Li J, Liu N, Cao W, Zhu Y, Shen B, Shao N (2009) Combining use of a panel of ssDNA aptamers in the detection of Staphylococcus aureus. Nucleic Acids Res 37(14):4621–4628
Cheah MT, Wachter A, Sudarsan N, Breaker RR (2007) Control of alternative RNA splicing and gene expression by eukaryotic riboswitches. Nature 447:497–500
Chen X, Li N, Ellington AD (2007) Ribozyme catalysis of metabolism in the RNA world. Chem Biodivers 4:633–655
Cox JC, Ellington AD (2001) Automated selection of anti-protein aptamers. Bioorg Med Chem 9:2525–2531
de-los Santos-Alvarez N, Lobo-Castanon MJ, Miranda-Ordieres AJ, Tunon-Blanco P (2007) Modified-RNA aptamer-based sensor for competitive impedimetric assay of neomycin B. J Am Chem Soc 129:3808–3809
Desai SK, Gallivan JP (2004) Genetic screens and selections for small molecules based on a synthetic riboswitch that activates protein translation. J Am Chem Soc 126:13247–13254
Ehrentreich-Forster E, Orgel D, Krause-Griep A, Cech B, Erdmann VA, Bier F, Scheller FW, Rimmele M (2008) Biosensor-based on-site explosives detection using aptamers as recognition elements. Anal Bioanal Chem 391:1793–1800
Ellington AD, Szostak JW (1990) In vitro selection of RNA molecules that bind specific ligands. Nature 346:818–822
Eulberg D, Buchner K, Maasch C, Klussmann S (2005) Development of an automated in vitro selection protocol to obtain RNA-based aptamers: identification of a biostable substance P antagonist. Nucleic Acids Res 33:e45
Eydeler K, Magbanua E, Werner A, Ziegelmuller P, Hahn U (2009) Fluorophore binding aptamers as a tool for RNA visualization. Biophys J 96:3703–3707
Fowler CC, Brown ED, Li Y (2008) A FACS-based approach to engineering artificial riboswitches. ChemBioChem 9:1906–1911
Grate D, Wilson C (2001) Inducible regulation of the S. cerevisiae cell cycle mediated by an RNA aptamer-ligand complex. Bioorg Med Chem 9:2565–2570
Gusti V, Kim DS, Gaur RK (2008) Sequestering of the 3′ splice site in a theophylline-responsive riboswitch allows ligand-dependent control of alternative splicing. Oligonucleotides 18:93–99
Hafner M, Vianini E, Albertoni B, Marchetti L, Grune I, Gloeckner C, Famulok M (2008) Displacement of protein-bound aptamers with small molecules screened by fluorescence polarization. Nat Protoc 3:579–587
Hanson S, Berthelot K, Fink B, McCarthy JE, Suess B (2003) Tetracycline-aptamer-mediated translational regulation in yeast. Mol Microbiol 49:1627–1637
Harvey I, Garneau P, Pelletier J (2002) Inhibition of translation by RNA-small molecule interactions. RNA 8:452–463
Hermann T, Patel DJ (2000) Adaptive recognition by nucleic acid aptamers. Science 287:820–825
Holeman LA, Robinson SL, Szostak JW, Wilson C (1998) Isolation and characterization of fluorophore-binding RNA aptamers. Fold Des 3:423–431
Homann M, Lorger M, Engstler M, Zacharias M, Goringer HU (2006) Serum-stable RNA aptamers to an invariant surface domain of live African trypanosomes. Comb Chem High Throughput Screen 9:491–499
Huang CC, Chang HT (2008) Aptamer-based fluorescence sensor for rapid detection of potassium ions in urine. Chem Commun (Camb) 12:1461–1463
Jo JJ, Shin JS (2009) Construction of intragenic synthetic riboswitches for detection of a small molecule. Biotechnol Lett. doi:https://doi.org/10.1007/s10529-009-0058-6
Kim DS, Gusti V, Pillai SG, Gaur RK (2005) An artificial riboswitch for controlling pre-mRNA splicing. RNA 11:1667–1677
Kim DS, Gusti V, Dery KJ, Gaur RK (2008) Ligand-induced sequestering of branchpoint sequence allows conditional control of splicing. BMC Mol Biol 9:23
Kotter P, Weigand JE, Meyer B, Entian KD, Suess B (2009) A fast and efficient translational control system for conditional expression of yeast genes. Nucleic Acids Res. doi:https://doi.org/10.1093/nar/gkp578
Lee ER, Blount KF, Breaker RR (2009a) Roseoflavin is a natural antibacterial compound that binds to FMN riboswitches and regulates gene expression. RNA Biol 6(2)
Lee HJ, Kim BC, Kim KW, Kim YK, Kim J, Oh MK (2009b) A sensitive method to detect Escherichia coli based on immunomagnetic separation and real-time PCR amplification of aptamers. Biosens Bioelectron 24(12):3550–3555
Levy M, Griswold KE, Ellington AD (2005) Direct selection of trans-acting ligase ribozymes by in vitro compartmentalization. RNA 11:1555–1562
Li W, Yang X, Wang K, Tan W, He Y, Guo Q, Tang H, Liu J (2008) Real-time imaging of protein internalization using aptamer conjugates. Anal Chem 80:5002–5008
Liss M, Petersen B, Wolf H, Prohaska E (2002) An aptamer-based quartz crystal protein biosensor. Anal Chem 74:4488–4495
Liu CW, Huang CC, Chang HT (2009) Highly selective DNA-based sensor for lead(II) and mercury(II) ions. Anal Chem 81:2383–2387
Lynch SA, Gallivan JP (2009) A flow cytometry-based screen for synthetic riboswitches. Nucleic Acids Res 37:184–192
Lynch SA, Desai SK, Sajja HK, Gallivan JP (2007) A high-throughput screen for synthetic riboswitches reveals mechanistic insights into their function. Chem Biol 14:173–184
Mayer G (2009) The chemical biology of aptamers. Angew Chem Int Ed Engl 48:2672–2689
Muranaka N, Sharma V, Nomura Y, Yokobayashi Y (2009) An efficient platform for genetic selection and screening of gene switches in Escherichia coli. Nucleic Acids Res 37:e39
Nomura Y, Yokobayashi Y (2007) Reengineering a natural riboswitch by dual genetic selection. J Am Chem Soc 129:13814–13815
Ott E, Stolz J, Lehmann M, Mack M (2009) The RFN riboswitch of Bacillus subtilis is a target for the antibiotic roseoflavin produced by Streptomyces davawensis. RNA Biol 6
Roth A, Breaker RR (2009) The structural and functional diversity of metabolite-binding riboswitches. Annu Rev Biochem 78:305–334
Sando S, Narita A, Aoyama Y (2007) Light-up Hoechst-DNA aptamer pair: generation of an aptamer-selective fluorophore from a conventional DNA-staining dye. ChemBioChem 8:1795–1803
Serganov A, Huang L, Patel DJ (2008) Structural insights into amino acid binding and gene control by a lysine riboswitch. Nature 455:1263–1267
Serganov A, Huang L, Patel DJ (2009) Coenzyme recognition and gene regulation by a flavin mononucleotide riboswitch. Nature 458:233–237
Sharma V, Nomura Y, Yokobayashi Y (2008) Engineering complex riboswitch regulation by dual genetic selection. J Am Chem Soc 130:16310–16315
Sparano BA, Koide K (2007) Fluorescent sensors for specific RNA: a general paradigm using chemistry and combinatorial biology. J Am Chem Soc 129:4785–4794
Stadtherr K, Wolf H, Lindner P (2005) An aptamer-based protein biochip. Anal Chem 77:3437–3443
Strehlitz B, Nikolaus N, Stoltenburg R (2008) Protein detection with aptamer biosensors. Sensors 8:4296–4307
Sudarsan N, Cohen-Chalamish S, Nakamura S, Emilsson GM, Breaker RR (2005) Thiamine pyrophosphate riboswitches are targets for the antimicrobial compound pyrithiamine. Chem Biol 12:1325–1335
Suess B, Hanson S, Berens C, Fink B, Schroeder R, Hillen W (2003) Conditional gene expression by controlling translation with tetracycline-binding aptamers. Nucleic Acids Res 31:1853–1858
Suess B, Fink B, Berens C, Stentz R, Hillen W (2004) A theophylline responsive riboswitch based on helix slipping controls gene expression in vivo. Nucleic Acids Res 32:1610–1614
Swensen JS, Xiao Y, Ferguson BS, Lubin AA, Lai RY, Heeger AJ, Plaxco KW, Soh HT (2009) Continuous, real-time monitoring of cocaine in undiluted blood serum via a microfluidic, electrochemical aptamer-based sensor. J Am Chem Soc 131:4262–4266
Thore S, Frick C, Ban N (2008) Structural basis of thiamine pyrophosphate analogues binding to the eukaryotic riboswitch. J Am Chem Soc 130:8116–8117
Topp S, Gallivan JP (2008) Riboswitches in unexpected places–a synthetic riboswitch in a protein coding region. RNA 14:2498–2503
Tuerk C, Gold L (1990) Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249:505–510
Tuleuova N, An CI, Ramanculov E, Revzin A, Yokobayashi Y (2008) Modulating endogenous gene expression of mammalian cells via RNA-small molecule interaction. Biochem Biophys Res Commun 376:169–173
Tyagi S (2009) Imaging intracellular RNA distribution and dynamics in living cells. Nat Methods 6:331–338
Valencia-Burton M, McCullough RM, Cantor CR, Broude NE (2007) RNA visualization in live bacterial cells using fluorescent protein complementation. Nat Methods 4:421–427
Wachter A, Tunc-Ozdemir M, Grove BC, Green PJ, Shintani DK, Breaker RR (2007) Riboswitch control of gene expression in plants by splicing and alternative 3′ end processing of mRNAs. Plant Cell 19:3437–3450
Weigand JE, Suess B (2007) Tetracycline aptamer-controlled regulation of pre-mRNA splicing in yeast. Nucleic Acids Res 35:4179–4185
Weigand JE, Sanchez M, Gunnesch EB, Zeiher S, Schroeder R, Suess B (2008) Screening for engineered neomycin riboswitches that control translation initiation. RNA 14:89–97
Werstuck G, Green MR (1998) Controlling gene expression in living cells through small molecule-RNA interactions. Science 282:296–298
Wieland M, Hartig JS (2008) Improved aptazyme design and in vivo screening enable riboswitching in bacteria. Angew Chem Int Ed Engl 47:2604–2607
Wieland M, Benz A, Klauser B, Hartig JS (2009a) Artificial ribozyme switches containing natural riboswitch aptamer domains. Angew Chem Int Ed Engl 48:2715–2718
Wieland M, Gfell M, Hartig JS (2009b) Expanded hammerhead ribozymes containing addressable three-way junctions. RNA 15:968–976
Win MN, Smolke CD (2007) A modular and extensible RNA-based gene-regulatory platform for engineering cellular function. Proc Natl Acad Sci U S A 104:14283–14288
Win MN, Smolke CD (2008) Higher-order cellular information processing with synthetic RNA devices. Science 322:456–460
Wochner A, Cech B, Menger M, Erdmann VA, Glokler J (2007) Semi-automated selection of DNA aptamers using magnetic particle handling. Biotechniques 43:344–344, 346, 348 passim
Yamazaki S, Tan L, Mayer G, Hartig JS, Song JN, Reuter S, Restle T, Laufer SD, Grohmann D, Krausslich HG, Bajorath J, Famulok M (2007) Aptamer displacement identifies alternative small-molecule target sites that escape viral resistance. Chem Biol 14:804–812
Zaher HS, Unrau PJ (2007) Selection of an improved RNA polymerase ribozyme with superior extension and fidelity. RNA 13:1017–1026
Acknowledgment
We thank Jens Wöhnert for the critical reading of the manuscript. We are grateful to the Aventis Foundation and the Deutsche Forschungsgemeinschaft for their financial support.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Weigand, J.E., Suess, B. Aptamers and riboswitches: perspectives in biotechnology. Appl Microbiol Biotechnol 85, 229–236 (2009). https://doi.org/10.1007/s00253-009-2194-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-009-2194-2