Abstract
The bacterium Actinoplanes sp. ATCC 53771 is known to perform drug metabolism of several xenobiotics similarly to humans. We identified a cytochrome P450 enzyme from this strain, CYP107E4, and expressed it in Escherichia coli using the pET101 vector. The purified enzyme showed the characteristic reduced-CO difference spectra with a peak at 450 nm, indicating the protein is produced in the active form with proper heme incorporation. The CYP107E4 enzyme was found to bind the drug diclofenac. Using redox enzymes from spinach, the reconstituted system is able to produce hydroxylated metabolites of diclofenac. Production of the human 4′-hydroxydiclofenac metabolite by CYP107E4 was confirmed, and a second hydroxylated metabolite was also produced.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cytochrome P450 enzymes (P450s) are an important superfamily of enzymes that are found in all domains of life and are responsible for a range of functions including production of antibiotics and steroids and detoxification of xenobiotics. In mammalian systems, the hepatic P450s are involved in the majority of drug metabolism (Guengerich 2005). Understanding the toxicology of drug metabolites is a critical issue in drug development and commercialization. Since current methods for producing these metabolites are slow and expensive, there have been many efforts to produce drug metabolites via biotransformation in bacteria (Chen et al. 1992a, b; Kuhnt et al. 1996, 1997; Li et al. 2008; Ohta et al. 2005; Osorio-Lozada et al. 2008; Otey et al. 2006; Vail et al. 2005; Zmijewski et al. 2006). Here, we describe the identification, cloning, expression, and characterization of a P450 enzyme from the bacterium Actinoplanes sp. ATCC 53771, which produces human-like metabolites of diclofenac.
Current methods for producing drug metabolites typically rely upon either liver microsomes, which have a limited half-life and are expensive, or chemical synthesis, which is not always feasible and is often costly. There have been some efforts towards expressing human P450s in bacteria for purification and/or biotransformation. In working with expression of mammalian P450s in Escherichia coli, there are several issues that must be overcome; these include expression of membrane-bound enzymes, the requirement for a FAD/FMN P450 reductase, and incorporation of the heme cofactor for proper folding and activity (Gillam 1998). Several human P450s have been successfully expressed in E. coli after some engineering to address these issues. The N-terminal membrane-binding domain can be altered to improve expression (Barnes et al. 1991; Yun et al. 2006), the human P450 reductase can be co-expressed (Blake et al. 1996; Dong and Porter 1996; Iwata et al. 1998; Parikh et al. 1997; Shimada et al. 1998; Vail et al. 2005), and addition of the heme precursor δ-aminolevulinic acid (ALA) increases heme incorporation (Guengerich et al. 1996). Despite these improvements, high-level expression and activity of the human enzymes for drug metabolism in heterologous hosts has been difficult to achieve (Dong and Porter 1996; Iwata et al. 1998; Parikh et al. 1997; Vail et al. 2005). Alternatively, microbial models of mammalian metabolism offer the ability to produce increased yields of human drug metabolites without the expense of chemical synthesis (Abourashed et al. 1999; Smith and Rosazza 1975a, b; Srisilam and Veeresham 2003). Many bacteria and fungi have the natural ability to perform mammalian-like metabolism. The challenge is that many of the strains used are uncharacterized and often exhibit poor growth characteristics. A potential solution is to clone P450 enzymes from these strains into platform host organisms that are more easily cultivated and then use such recombinant organisms to produce relevant drug metabolites via biotransformation. Unlike the mammalian hepatic P450s, the bacterial P450s are generally soluble enzymes. The system for electron transport is most typically via a soluble iron-sulfur ferredoxin and a soluble FAD-containing reductase (Munro and Lindsay 1996). There are other bacterial electron transport systems that are less common in bacteria including flavodoxins as with CYP107H1 (Lawson et al. 2004) and soluble fusion proteins containing a P450 domain and a reductase domain (De Mot and Parret 2002; Narhi and Fulco 1987). Several bacterial P450s that utilize a variety of redox systems have been cloned and expressed at high levels for both in vitro as well as in vivo activity in E. coli for production of many natural products and for biocatalysis (Budde et al. 2006; Hussain and Ward 2003; Jungmann et al. 2005; Peters et al. 2003; Sawada et al. 2004; Shrestha et al. 2008).
The bacterium Actinoplanes sp. has been identified as having the same metabolite profile as humans for several different drugs (Chen et al. 1992b; Hsu et al. 2002; Kuhnt et al. 1996, 1997; Ohta et al. 2005; Osorio-Lozada et al. 2008). One of these, diclofenac is a nonsteroidal anti-inflammatory drug that has been studied extensively with respect to drug metabolism (Bort et al. 1999; Shen et al. 1999; Stierlin and Faigle 1979; Stierlin et al. 1979; Tang et al. 1999a, b). Two major (4′-hydroxy and 5-hydroxy) and one minor (4′,5-dihydroxy) mammalian metabolites of diclofenac have been produced by biotransformation using Actinoplanes sp. cultures (Osorio-Lozada et al. 2008). In our efforts working towards improving bacterial drug metabolism, we have identified a P450 from Actinoplanes sp. that is capable of producing monohydroxylated diclofenac metabolites.
Materials and methods
Strains and culture conditions
Actinoplanes sp. ATCC 53771 was obtained from the ATCC library (Manassas, VA, USA). Culturing conditions for drug metabolism with Actinoplanes sp. were as described by Osorio-Lozada et al. (2008). Genomic DNA extraction was performed using the method of Pospiech and Neumann (1995). Cloning and expression vectors were purchased from Invitrogen (Carlsbad, CA, USA). E coli Top 10 (Invitrogen) was used as the host for cloning, and E. coli BL21(DE3) Codon Plus RP (Stratagene, La Jolla, CA, USA) was used for protein expression. The E. coli strains were grown in LB broth at 37°C with shaking at 225 rpm, and expression was performed at 30°C. Plasmid isolation was performed using the Qiaprep miniprep kit (Qiagen, Valencia, CA, USA). Primers for polymerase chain reaction (PCR) were synthesized by Operon (Huntsville, AL, USA). Diclofenac sodium salt, spinach ferredoxin, and spinach ferredoxin-NADP+ reductase were purchased from Sigma Aldrich (St. Louis, MO, USA). The 4′-hydroxydiclofenac standard was purchased from BD Biosciences (San Jose, CA, USA). All other chemicals and reagents were purchased from Sigma Aldrich except where noted and were of reagent grade or better. All high-performance liquid chromatography (HPLC) reagents were of HPLC grade.
Gene fishing
Gene fishing was performed using the Vectorette II system from SigmaGenosis (St. Louis, MO, USA). The Vectorette library was constructed according to the product manual using Actinoplanes sp. genomic DNA. The degenerate primer, O2-degen (5′-TSCTSCTSATCGCSGGSCACGAGAC-3′), was used with the Vectorette primer for PCR with Jumpstart Taq Polymerase (Sigma Aldrich) using a 10:1 O2-degen:vectorette primer molar ratio and the following thermal cycler program: 35 cycles of 94°C for 1 min, annealing at 68°C for 30 s, and extension at 72°C for 2 min. The PCR products were cloned into the pCR4-TOPO vector for sequencing (Macrogen, Seoul, Korea). Gene fishing was continued as described in the Vectorette manual to identify entire open reading frames (ORFs). P450 genes were confirmed by comparison to the Basic Local Alignment Search Tool (BLAST) database (Altschul et al. 1990), and the coding sequence was confirmed using the FrameD program (Schiex et al. 2003). A multiple sequence alignment was constructed using the Clustal W2 program (Larkin et al. 2007).
Construction of expression vectors
The cyp107E4 gene was isolated for directional cloning into the pET101/D-TOPO vector by PCR using Platinum Pfx DNA polymerase (Invitrogen) according to the manufacturer’s directions using the forward (5′-CACCATGACCACCACAGACACCGAA-3′) and reverse (5′-CCATCCGATCGGCATGGAGCG-3′) primers. The primers were designed for directional in-frame cloning into the TOPO vector, and to exclude the stop codon to allow expression of the C-terminal 6×-histidine tag for purification and detection purposes. The TOPO ligation product was transformed into Top 10 cells, a clone was isolated, and the plasmid DNA purified. The purified plasmid was transformed after confirmation of the insert by sequencing into BL21(DE3) Codon Plus RP cells according to the manufacturers protocol and confirmed by plasmid miniprep.
Gene expression and purification
Overnight cultures of BL21 Codon Plus were used to inoculate 200 mL LB in a 500-mL baffled flask and grown to an OD600 of 0.6. ALA was added to a final concentration of 1 mM and the culture incubated an additional 30 min. Expression was induced by adding isopropyl β-D-1-thiogalactopyranoside to a final concentration of 1 mM, and the culture was incubated 10 h at 30°C with shaking. Cells were harvested and stored at −80°C.
For purification of CYP107E4, the cell pellet was resuspended in 10 mL binding buffer (50 mM sodium phosphate buffer, 300 mM sodium chloride, and 10 mM imidazole, pH 8.0), and 10 mg lysozyme and 100 µL protease inhibitor cocktail (Sigma Aldrich) were added. The suspension was incubated on ice for 30 min and then lysed by three passages through a French pressure cell at 12,000 psi. The crude lysate was centrifuged at 30,000 × g for 30 min at 4°C and the supernatant filtered through a 0.45-μm nylon filter. The filtrate was added to a column containing 10 mL Ni-NTA agarose (Invitrogen). Using an AKTA fast protein liquid chromatography system (GE Healthcare, Piscataway, NJ, USA), the column was washed with buffer A (50 mM sodium phosphate, 300 mM sodium chloride, and 20 mM imidazole, pH 8.0) for eight column volumes, and bound protein was eluted by a linear gradient from 100% buffer A to 75% buffer B (50 mM sodium phosphate, 300 mM sodium chloride, and 400 mM imidazole, pH 8.0) over 300 min. Fractions were collected and analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) before being pooled. The pooled fractions were concentrated and desalted using a 10-kDa molecular weight cutoff Amicon Ultra centrifugal filter unit (Millipore, Billerica, MA, USA). Western blotting was performed using the WesternBreeze Chromogenic detection kit and the Anti-his(C-term) antibody (Invitrogen).
Spectral analyses
The purified P450 was diluted in 100 mM Tris–HCl, pH 7.4 and split between two microfuge tubes. CO gas was bubbled through one of the tubes for 1 min, and then a 20% volume of freshly prepared 300 mM sodium dithionite solution was added to both tubes and mixed well. The solutions were added to a 96-well plate, and the spectrum of each sample was read between 400 and 500 nm. The reduced-CO difference spectrum was found by subtracting the spectrum of the reduced sample from the reduced plus CO sample according to the method of Omura and Sato, and the P450 concentration was calculated using the difference extinction coefficient of 91 mM−1 cm−1 (Omura and Sato 1964).
The substrate-binding difference spectrum was found similarly. The purified P450 was diluted to 5 μM in 50 mM phosphate buffer, pH 7.3, and diclofenac dissolved in 1:1 DMSO:water was added at varying concentrations from 0 to 3.5 mM. The spectrum was read between 350 and 550 nm for each sample, and the difference spectrum between samples with diclofenac and with DMSO:water alone were calculated (Jefcoate 1978). The spectral binding constant, K s, was found by weighted nonlinear least squares regression, fit to Eq. 1, where ∆Abs is the difference in absorbance between 389 and 419 nm of the substrate-binding difference spectrum and [S] is the substrate concentration.
Diclofenac metabolism
Drug metabolism was performed in vitro with CYP107E4 in a reaction mixture containing 1 μM purified P450, 3.5 μM spinach ferredoxin, 0.1 U spinach ferredoxin reductase, 50 μL nicotinamide adenine dinucleotide phosphate (NADPH) regenerating system solution A, and 10 μL NADPH regenerating system solution B (BD Gentest, San Jose, CA, USA) in a final volume of 1 mL of 100 mM Tris–HCl, pH 7.5. Diclofenac was added at varying final concentrations of 25–500 μM and the reaction incubated 2 h at 30°C. Control incubations were performed without addition of the NADPH regenerating system as well as without addition of the P450. The reactions were stopped by adding 0.5 mL acetonitrile and centrifuged at 13,000 rpm for 10 min at 4°C. Supernatants were stored at −80°C for later analysis. Drug metabolism was also performed in vivo with the Actinoplanes sp. strain and the metabolites extracted as described by Osorio-Lozada et al. (2008) for comparison with the metabolites from the reconstituted system.
LC-MS analysis
Metabolites were analyzed by liquid chromatography-mass spectrometry (LC-MS). LC was carried out on an Agilent 1100 HPLC with a G1267A autosampler in combination with a G1330A thermostat, a G1312A binary pump, G1322A degasser, and a G1316A column thermostat, in-line with a G1315B diode array detector and a G1956A ion-trap detector. The system was controlled using the ChemStation software (Agilent Technologies, Santa Clara, CA, USA). Separation was performed using two columns in series (Eclipse XDB-C8, 4.6 × 250 mm, 5 μm particle size, Agilent Technologies) with the column temperature set to 65°C. The mobile phase was 0.02% formic acid (solvent A) and methanol (solvent B). The following elution profile was used, with a flow rate of 1.0 mL/min and an injection volume of 100 μL: 55% B from 0 to 1 min, 55% to 85% B from 1 to 4 min, 85% to 98% B from 4 to 12 min, 98% to 99% B from 12 to 14 min, 99% to 55% B from 14 to 14.1 min, and 55% B from 14.1 to 17 min. Absorbance was monitored at 282 nm. The ion-trap mass spectrometer was run in positive scan mode. The drying gas (nitrogen) flow was set to 9 L/min, the temperature to 360°C, the nebulizer pressure to 45 psi, and the capillary voltage to 4,000 V. The mass range was set to m/z = 100–1,100. The hydroxylated metabolites (m/z = 310) were identified by comparison with the 4′-hydroxydiclofenac reference material and the extracted metabolites from Actinoplanes sp.
Genbank submission
The gene and translated protein sequences for CYP107E4 have been submitted to Genbank and assigned the accession number FJ754024.
Results
Identification of a P450 gene in Actinoplanes sp.
P450 genes in Actinoplanes were identified by PCR using degenerate primers specific for Actinomycete P450s (Hyun et al. 1998). The gene fragments were then extended using the Vectorette II system, sequenced, and compared against the BLASTx database. New primers were designed to continue walking along the genome until the entire ORF was found for each P450 gene. One full ORF that was identified as a P450 was cyp107E4, which is a 1,191-bp gene that encodes a 43.9-kDa protein as determined by the deduced amino acid sequence. Comparison with known bacterial P450s indicates this gene belongs to the CYP107 subfamily due to the greater than 40% identity with other members of the same family. Based upon sequence identity, it is part of the 107E family, having as high as 61% identity with CYP107E2 from the erythromycin producing strain Saccharopolyspora erythrae NRRL 2338 (Oliynyk et al. 2007). The P450 from Actinoplanes sp. has been designated CYP107E4 according to the established P450 nomenclature (personal communication, D. R. Nelson). An alignment with other P450s from the 107 family was constructed and shows the presence of several highly conserved motifs associated with bacterial P450s (Fig. 1).
Clustal W2 alignment of deduced amino acid sequence of CYP107E4 with other P450s from the CYP107 family. CYP107E2 (GI 134098012) shares 61% identity, CYP107A1 (GI 152697) shares 39% identity, and CYP107L1 (GI 3800840) shares 42% identity with CYP107E4. The solid box indicates the Gx(D,E)T motif of the oxygen binding site, the dotted box indicates the ExxR charge pair motif of the K-helix, and the dashed box indicates the conserved F(G,S)xGx(H,R)xCxG motif of the heme binding region with the absolutely conserved cysteine (denoted by asterisk). Multiple sequence alignment imported to GeneDoc for display
Cloning and expression
The cyp107E4 gene was amplified from a genomic library of Actinoplanes sp. by PCR and TOPO cloned into the pET101 expression vector under control of the strong T7 promoter. The gene was cloned such that the protein was expressed as a fusion with a C-terminal 6× histidine tag for detection and purification. The resulting vector was transformed into BL21(DE3) Codon Plus RP cells for expression due to the 70% GC content of the P450 gene. Additionally, ALA was added prior to the start of expression to increase the heme available during protein production. Without the addition of ALA, we primarily produced the apo-form of the protein with lower overall expression levels (results not shown). The P450 was purified from crude extract by affinity chromatography using a Ni-NTA agarose column (Fig. 2). A second band appears on the SDS-PAGE gel of the purified fraction below the expected CYP107E4 protein. This is most likely a C-terminal truncated product; it contains a C-terminal histidine tag as shown by Western blot (Fig. 2b) and did not appear in cultures with the vector control.
Purification of CYP107E4. a Sodium dodecyl sulfate polyacrylamide gel electrophoresis gel stained with Coomassie blue. Lane M, molecular marker; lane 1, crude lysate; lane 2, purified fraction. Top band in lane 2 (arrow) corresponds to the full-length protein. b Western blot with anti-his(C-term) antibody. Top band corresponds to the full-length CYP107E4 protein; bottom band is truncated protein (C-terminal) that was co-purified due to the C-terminal histidine tag
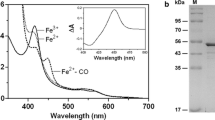
The reduced-CO difference spectrum was used to determine the active P450 content of the purified fraction. The difference spectrum of the purified enzyme from cultures grown in the presence of ALA show that the enzyme is primarily in the active form by the relatively large peak at 450 nm compared to the small peak at 420 nm (Fig. 3). Under the expression conditions used, we were able to purify 14.2 ± 0.3 mg holo-P450/L culture, 83% of the total purified protein fraction was P450. The substrate-binding difference spectrum was used to determine the ability of CYP107E4 to bind with diclofenac in the substrate-binding pocket. The resulting type I-binding difference spectrum indicated that CYP107E4 binds diclofenac (Fig. 4). The spectral shift from 415 nm to 387 nm is representative of the heme iron shifting from the low- to the high-spin state, which occurs upon substrate binding. The spectral binding constant of 0.48 ± 0.07 mM was found by plotting the substrate concentration versus the difference in absorbance at 387 and 415 nm and fitting the points to Eq. 1 (Fig. 4).
Substrate binding. a Substrate-binding difference spectrum. Increasing spectral shift from 416 to 387 nm with increasing diclofenac concentration, indicating a shift from the low-spin state to the high-spin state of CYP107E4 upon substrate binding. Arrow indicates increasing diclofenac concentrations, ranging from 0.1 to 3.5 mM. b Binding curve with diclofenac. The spectral binding constant, K s, was calculating by fitting the binding curve to Eq. 1
Drug metabolism
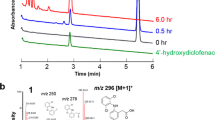
Drug metabolism was performed by reconstituting the activity of the P450 using ferredoxin and ferredoxin-NADP+ reductase from spinach. An NADPH regenerating system was used to increase conversion of diclofenac. The metabolites were extracted after 2 h of incubation at 30°C with the reconstituted system and analyzed by LC-MS (Fig. 5). The mass spectra of the compounds eluting at 11.4 min (metabolite M1) and 11.6 min (metabolite M2) were indicative of a single hydroxylation with the expected chlorine isotope pattern (not shown). The metabolites from the purified enzyme system with the available 4′-hydroxydiclofenac standard confirmed that M1 was the 4′-OH metabolite. There is no readily available standard for 5-hydroxydiclofenac for comparison with M2, and we were unable to obtain sufficient quantities for nuclear magnetic resonance (NMR). However, we were able to compare the metabolite profile from the reconstituted CYP107E4 system to the profile from Actinoplanes sp. that produces both the 4′-OH and 5-OH metabolites as major products. Comparison of the HPLC profiles was consistent with one another (Fig. 5a, b). Control incubations were performed for the system without P450 or without addition of NADPH (Fig 5c).
Liquid chromatography analysis of diclofenac metabolites. a Extracted metabolites from Actinoplanes sp. showing the main metabolites produced. M1 is 4′-hydroxydiclofenac as determined by comparison to authentic standard. M2 is a second hydroxylated metabolite, which is likely 5-hydroxydiclofenac. b Reconstituted reaction with 100 μM diclofenac. c Control sample without nicotinamide adenine dinucleotide phosphate. Control samples without CYP107E4 were identical
Discussion
The study of drug metabolism is a key step in the drug development process. The acquisition of sufficient amounts of metabolites for such studies is often costly. New methods for inexpensively and rapidly producing metabolites are needed. There has been much work focused on the ability of different microorganisms to perform the desired metabolism, but often with mixed results due to the difficulty in growing the strains. We have identified enzymes responsible for drug metabolism in Actinoplanes sp. In particular, we have reported a P450 enzyme that catalyzes the hydroxylation of diclofenac in a reconstituted system in vitro. This and other Actinoplanes sp. P450 enzymes could ultimately be used to develop a more efficient in vitro or in vivo biotransformation system for the production of additional drug metabolites.
Actinoplanes sp. has been found to perform human-like metabolism of several drugs via P450 reactions. This is the first study of a P450 from this strain. The CYP107E4 enzyme contains several highly conserved motifs that are characteristic of bacterial P450s (Fig. 1), including the ExxR charge pair and the conserved cysteine responsible for heme binding in the F(G,S)xGx(H,R)xCxG motif (Nelson 1995). The enzyme is part of the CYP107 family based on the high level of sequence identity. The CYP107 family is one of the largest bacterial P450 families, and many of the members have been shown to be involved in the tailoring reactions for the biosynthesis of several drugs. Most of these reactions involve hydroxylations as with rapamycin (CYP107G1) (Chung et al. 2001; Molnar et al. 1996), lankamycin (CYP107A2 and CYP107AB1; Arakawa et al. 2006), and erythromycin biosynthesis (CYP107A1) (Weber et al. 1991), but there are also examples of epoxidation as with CYP107D1 in oleandomycin synthesis (Shah et al. 2000) and CYP107E1 which performs both hydroxylation and epoxidation reactions in the synthesis of mycinamicin (Anzai et al. 2008). The broad range of substrates, as well as the range of reactions performed by members of a single P450 family makes it difficult to derive function from sequence alone. In order to identify the P450 responsible for diclofenac metabolism in Actinoplanes sp., the gene encoding CYP107E4 was expressed in E. coli for characterization.
Gene expression was performed in the BL21(DE3) Codon Plus RP strain, because initial attempts at expression in the BL21(DE3) strain resulted in poor expression as the cyp107E4 gene has a GC content much higher than that of wild type E. coli. Overall expression levels were improved in the codon optimized BL21 strain. The addition of ALA before induction has been shown to increase production of active P450 in E. coli (Guengerich et al. 1996; Taylor et al. 1999). We found that addition of the heme precursor in our system also increased yield of the correctly folded and active P450. After purification from either the BL21(DE3) or the codon optimized strain, we found an approximately 38-kDa soluble protein product in addition to our full CYP107E4 protein (Fig 2). This band also appears in Western blots using anti-his C-terminal antibody, which leads us to believe it is a truncated form of CYP107E4.
The ability of CYP107E4 to bind diclofenac was confirmed by a spectral shift associated with the transition from the low- to high-spin state of the heme iron upon addition of the substrate. The observed K s of 0.48 ± 0.07 mM is quite high, although not unexpected since diclofenac is not a natural substrate of this P450. Despite the high binding constant, we have been able to confirm through in vitro studies that CYP107E4 is able to produce the 4′-hydroxylated metabolite through LC-MS and comparison to an authentic standard. There is no standard commercially available for the 5-hydroxylated metabolite, and the fragmentation pattern by MS/MS is similar for both the 4′-OH and 5-OH metabolites (Osorio-Lozada et al. 2008), which has complicated our efforts to confirm the identity of the M2 metabolite produced by CYP107E4. We were able to compare the metabolite profile from Actinoplanes sp. incubations with the reconstituted system and see very good alignment between the two, which has led us to conclude that M2 is in fact 5-hydroxydiclofenac (Fig. 5). We are looking at producing larger quantities of the M2 metabolite in order to perform NMR to confirm that it is 5-hydroxydiclofenac.
We have not identified the 4′,5-dihydroxylated metabolite that has been reported with Actinoplanes sp. in vivo. The genome of Actinoplanes sp. has not been fully sequenced. Therefore, we do not have easy access to any electron transport genes present in the strain. The ability of the P450 to produce diclofenac metabolites is likely limited by electron transport from the spinach redox enzymes, which explains both the low conversion of diclofenac as well as the absence of the dihydroxylated metabolite. The spinach redox enzymes are commonly used for reconstitution of bacterial P450s when the native redox partners are unknown. There have been several reports of reconstituting activity of P450s with redox partners from other strains with suitable activity for biotransformation, especially with the putidaredoxin and putidaredoxin reductase from Pseudomonas putida (Celik et al. 2005; Fujii et al. 2006; Mouri et al. 2006; Shrestha et al. 2008), but this is not always successful and requires significant screening efforts to identify candidate redox systems. Our current efforts are aimed at identifying redox enzymes from Actinoplanes sp. or another source that improve electron transport to the P450; we expect that we will be able to produce the dihydroxylated metabolite using CYP107E4 with an improved system.
Biotransformation systems have been previously developed for the production of drug metabolites. Modern enzyme and metabolic engineering technologies now allows for the development of improved recombinant biotransformation systems (Kim et al. 2008; Landwehr et al. 2007; Otey et al. 2006; Stjernschantz et al. 2008). The identification of a P450 responsible for diclofenac metabolism in Actinoplanes sp. is the first step in developing a recombinant biotransformation system based on the breadth of drug modification activities previously reported in Actinoplanes sp. We have identified several other P450 sequences in Actinoplanes sp. and are currently characterizing their drug modification activities. We expect that the combination of the native activities of such enzymes, along with the tools of directed enzyme evolution and metabolic engineering, will enable the development of recombinant microbial strains capable of rapidly and efficiently producing a range of valuable drug metabolites.
References
Abourashed EA, Clark AM, Hufford CD (1999) Microbial models of mammalian metabolism of xenobiotics: an updated review. Curr Med Chem 6:359–374
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Anzai Y, Li S, Chaulagain MR, Kinoshita K, Kato F et al (2008) Functional analysis of MycCI and MycG, cytochrome P450 enzymes involved in biosynthesis of mycinamicin macrolide antibiotics. Chem Biol 15:950–959
Arakawa K, Kodama K, Tatsuno S, Ide S, Kinashi H (2006) Analysis of the loading and hydroxylation steps in lankamycin biosynthesis in Streptomyces rochei. Antimicrob Agents Chemother 50:1946–1952
Barnes HJ, Arlotto MP, Waterman MR (1991) Expression and enzymatic activity of recombinant cytochrome P450 17 alpha-hydroxylase in Escherichia coli. Proc Natl Acad Sci U S A 88:5597–5601
Blake JAR, Pritchard M, Ding SH, Smith GCM, Burchell B et al (1996) Coexpression of a human P450 (CYP3A4) and P450 reductase generates a highly functional monooxygenase system in Escherichia coli. FEBS Lett 397:210–214
Bort R, Mace K, Boobis A, Gomez-Lechon MJ, Pfeifer A et al (1999) Hepatic metabolism of diclofenac: role of human CYP in the minor oxidative pathways. Biochem Pharmacol 58:787–796
Budde M, Morr M, Schmid RD, Urlacher VB (2006) Selective hydroxylation of highly branched fatty acids and their derivatives by CYP102A1 from Bacillus megaterium. ChemBioChem 7:789–794
Celik A, Flitsch SL, Turner NJ (2005) Efficient terpene hydroxylation catalysts based upon P450 enzymes derived from actinomycetes. Org Biomol Chem 3:2930–2934
Chen TS, Arison BH, Wicker LS, Inamine ES (1992a) Microbial transformation of immunosuppressive compounds. 2. Specific desmethylation of 13-methoxy group of Fk-506 and Fr-900520 by Actinomycete sp. Atcc-53828. J Antibiot (Tokyo) 45:577–580
Chen TS, Arison BH, Wicker LS, Inamine ES, Monaghan RL (1992b) Microbial transformation of immunosuppressive compounds. 1. Desmethylation of Fk506 and Immunomycin (Fr-900520) by Actinoplanes sp. Atcc-53771. J Antibiot (Tokyo) 45:118–123
Chung L, Liu L, Patel S, Carney JR, Reeves CD (2001) Deletion of rapQONML from the rapamycin gene cluster of Streptomyces hygroscopicus gives production of the 16-O-desmethyl-27-desmethoxy analog. J Antibiot (Tokyo) 54:250–256
De Mot R, Parret AHA (2002) A novel class of self-sufficient cytochrome P450 monooxygenases in prokaryotes. Trends Microbiol 10:502–508
Dong JS, Porter TD (1996) Coexpression of mammalian cytochrome P450 and reductase in Escherichia coli. Arch Biochem Biophys 327:254–259
Fujii Y, Hirosue S, Fujii T, Matsumoto N, Agematu H et al (2006) Hydroxylation of oleanolic acid to queretaroic acid by cytochrome P450 from Nonomuraea recticatena. Biosci Biotechnol Biochem 70:2299–2302
Gillam EMJ (1998) Human cytochrome P450 enzymes expressed in bacteria: reagents to probe molecular interactions in toxicology. Clin Exp Pharmacol Physiol 25:877–886
Guengerich FP (2005) Human cytochrome P450 enzymes. In: Ortiz de Montellano PR (ed) Cytochrome P450: structure, mechanism, and biochemistry, 3rd edn. Plenum, New York, pp 377–396
Guengerich FP, Martin MV, Guo Z, Chun YJ (1996) Purification of functional recombinant P450s from bacteria. Methods Enzymol 272:35–44
Hsu FL, Hou CC, Yang LM, Cheng JT, Chi TC et al (2002) Microbial transformations of isosteviol. J Nat Prod 65:273–277
Hussain HA, Ward JM (2003) Enhanced heterologous expression of two Streptomyces griseolus cytochrome P450s and Streptomyces coelicolor ferredoxin reductase as potentially efficient hydroxylation catalysts. Appl Environ Microbiol 69:373–382
Hyun CG, Kim JM, Hong SK, Suh JW (1998) An efficient approach for cloning P450 hydroxylase genes from actinomycetes. J Microbiol Biotechnol 8:295–299
Iwata H, Fujita K, Kushida H, Suzuki A, Konno Y et al (1998) High catalytic activity of human cytochrome P450 co-expressed with human NADPH-cytochrome P450 reductase in Escherichia coli. Biochem Pharmacol 55:1315–1325
Jefcoate CR (1978) Measurement of substrate and inhibitor binding to microsomal cytochrome P-450 by optical-difference spectroscopy. Methods Enzymol 52:258–279
Jungmann V, Molnar I, Hammer PE, Hill DS, Zirkle R et al (2005) Biocatalytic conversion of avermectin to 4′-oxo-avermectin: characterization of biocatalytically active bacterial strains and of cytochrome p450 monooxygenase enzymes and their genes. Appl Environ Microbiol 71:6968–6976
Kim DH, Kim KH, Liu KH, Jung HC, Pan JG et al (2008) Generation of human metabolites of 7-ethoxycoumarin by bacterial cytochrome P450 BM3. Drug Metab Dispos 36:2166–2170
Kuhnt M, Bitsch F, France J, Hofmann H, Sanglier JJ et al (1996) Microbial biotransformation products of cyclosporin A. J Antibiot (Tokyo) 49:781–787
Kuhnt M, Bitsch F, Ponelle M, Fehr T, Sanglier JJ (1997) Microbial conversion of rapamycin. Enzyme Microb Technol 21:405–412
Landwehr M, Carbone M, Otey CR, Li Y, Arnold FH (2007) Diversification of catalytic function in a synthetic family of chimeric cytochrome p450s. Chem Biol 14:269–278
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA et al (2007) Clustal W and clustal X version 2.0. Bioinformatics 23:2947–2948
Lawson RJ, von Wachenfeldt C, Haq I, Perkins J, Munro AW (2004) Expression and characterization of the two flavodoxin proteins of Bacillus subtilis, YkuN and YkuP: biophysical properties and interactions with cytochrome P450 BioI. Biochemistry (Mosc) 43:12390–12409
Li W, Josephs JL, Skiles GL, Humphreys WG (2008) Metabolite generation via microbial biotransformations with Actinomycetes: rapid screening for active strains and biosynthesis of important human metabolites of two development-stage compounds, 5-[(5S,9R)-9-(4-cyanophenyl)-3-(3,5-dichlorophenyl)-1-methyl-2,4-dioxo-1,3,7-triazaspiro[4.4]non7-yl-methyl]-3-thiophenecarboxylic acid (BMS-587101) and dasatinib. Drug Metab Dispos 36:721–730
Molnar I, Aparicio JF, Haydock SF, Khaw LE, Schwecke T et al (1996) Organisation of the biosynthetic gene cluster for rapamycin in Streptomyces hygroscopicus: analysis of genes flanking the polyketide synthase. Gene 169:1–7
Mouri T, Michizoe J, Ichinose H, Kamiya N, Goto M (2006) A recombinant Escherichia coli whole cell biocatalyst harboring a cytochrome P450cam monooxygenase system coupled with enzymatic cofactor regeneration. Appl Microbiol Biotechnol 72:514–520
Munro AW, Lindsay JG (1996) Bacterial cytochromes P-450. Mol Microbiol 20:1115–1125
Narhi LO, Fulco AJ (1987) Identification and characterization of two functional domains in cytochrome P-450BM-3, a catalytically self-sufficient monooxygenase induced by barbiturates in Bacillus megaterium. J Biol Chem 262:6683–6690
Nelson DR (1995) Cytochrome P450 nomenclature and alignment of selected sequences. In: Ortiz de Montellano PR (ed) Cytochrome P450, 2nd edn. Plenum Press, New York, pp 575–606
Ohta K, Agematu H, Yamada T, Kaneko K, Tsuchida T (2005) Production of human metabolites of cyclosporin A, AM1, AM 4N and AM 9, by microbial conversion. J Biosci Bioeng 99:390–395
Oliynyk M, Samborskyy M, Lester JB, Mironenko T, Scott N et al (2007) Complete genome sequence of the erythromycin-producing bacterium Saccharopolyspora erythraea NRRL23338. Nat Biotechnol 25:447–453
Omura T, Sato R (1964) The carbon monoxide-binding pigment of liver microsomes. I. Evidence for its hemoprotein nature. J Biol Chem 239:2370–2378
Osorio-Lozada A, Surapaneni S, Skiles GL, Subramanian R (2008) Biosynthesis of drug metabolites using microbes in hollow fiber cartridge reactors: case study of diclofenac metabolism by Actinoplanes species. Drug Metab Dispos 36:234–240
Otey CR, Bandara G, Lalonde J, Takahashi K, Arnold FH (2006) Preparation of human metabolites of propranolol using laboratory-evolved bacterial cytochromes P450. Biotechnol Bioeng 93:494–499
Parikh A, Gillam EM, Guengerich FP (1997) Drug metabolism by Escherichia coli expressing human cytochromes P450. Nat Biotechnol 15:784–788
Peters MW, Meinhold P, Glieder A, Arnold FH (2003) Regio- and enantioselective alkane hydroxylation with engineered cytochromes P450 BM-3. J Am Chem Soc 125:13442–13450
Pospiech A, Neumann B (1995) A versatile quick-prep of genomic DNA from gram-positive bacteria. Trends Genet 11:217-218
Sawada N, Sakaki T, Yoneda S, Kusudo T, Shinkyo R et al (2004) Conversion of vitamin D3 to 1alpha, 25-dihydroxyvitamin D3 by Streptomyces griseolus cytochrome P450SU-1. Biochem Biophys Res Commun 320:156–164
Schiex T, Gouzy J, Moisan A, de Oliveira Y (2003) FrameD: a flexible program for quality check and gene prediction in prokaryotic genomes and noisy matured eukaryotic sequences. Nucleic Acids Res 31:3738–3741
Shah S, Xue Q, Tang L, Carney JR, Betlach M et al (2000) Cloning, characterization and heterologous expression of a polyketide synthase and P-450 oxidase involved in the biosynthesis of the antibiotic oleandomycin. J Antibiot (Tokyo) 53:502–508
Shen S, Marchick MR, Davis MR, Doss GA, Pohl LR (1999) Metabolic activation of diclofenac by human cytochrome P450 3A4: role of 5-hydroxydiclofenac. Chem Res Toxicol 12:214–222
Shimada T, Wunsch RM, Hanna IH, Sutter TR, Guengerich FP et al (1998) Recombinant human cytochrome P450 1B1 expression in Escherichia coli. Arch Biochem Biophys 357:111–120
Shrestha P, Oh TJ, Sohng JK (2008) Designing a whole-cell biotransformation system in Escherichia coli using cytochrome P450 from Streptomyces peucetius. Biotechnol Lett 30:1101–1106
Smith RV, Rosazza JP (1975a) Microbial models of mammalian metabolism. J Pharm Sci 64:1737–1759
Smith RV, Rosazza JP (1975b) Microbial systems for study of biotransformations of drugs. Biotechnol Bioeng 17:785–814
Srisilam K, Veeresham C (2003) Biotransformation of drugs by microbial cultures for predicting mammalian drug metabolism. Biotechnol Adv 21:3–39
Stierlin H, Faigle JW (1979) Biotransformation of diclofenac sodium (Voltaren) in animals and in man. II. Quantitative determination of the unchanged drug and principal phenolic metabolites, in urine and bile. Xenobiotica 9:611–621
Stierlin H, Faigle JW, Sallmann A, Kung W, Richter WJ et al (1979) Biotransformation of diclofenac sodium (Voltaren) in animals and in man. I. Isolation and identification of principal metabolites. Xenobiotica 9:601–610
Stjernschantz E, van Vugt-Lussenburg BM, Bonifacio A, de Beer SB, van der Zwan G et al (2008) Structural rationalization of novel drug metabolizing mutants of cytochrome P450 BM3. Proteins 71:336–352
Tang W, Stearns RA, Wang RW, Chiu SH, Baillie TA (1999a) Roles of human hepatic cytochrome P450s 2C9 and 3A4 in the metabolic activation of diclofenac. Chem Res Toxicol 12:192–199
Tang W, Stearns RA, Bandiera SM, Zhang Y, Raab C et al (1999b) Studies on cytochrome P-450-mediated bioactivation of diclofenac in rats and in human hepatocytes: identification of glutathione conjugated metabolites. Drug Metab Dispos 27:365–372
Taylor M, Lamb DC, Cannell R, Dawson M, Kelly SL (1999) Cytochrome P450105D1 (CYP105D1) from Streptomyces griseus: heterologous expression, activity, and activation effects of multiple xenobiotics. Biochem Biophys Res Commun 263:838–842
Vail RB, Homann MJ, Hanna I, Zaks A (2005) Preparative synthesis of drug metabolites using human cytochrome P450s 3A4, 2C9 and 1A2 with NADPH-P450 reductase expressed in Escherichia coli. J Ind Microbiol Biotechnol 32:67–74
Weber JM, Leung JO, Swanson SJ, Idler KB, McAlpine JB (1991) An erythromycin derivative produced by targeted gene disruption in Saccharopolyspora erythraea. Science 252:114–117
Yun CH, Yim SK, Kim DH, Ahn T (2006) Functional expression of human cytochrome P450 enzymes in Escherichia coli. Curr Drug Metab 7:411–429
Zmijewski M, Gillespie TA, Jackson DA, Schmidt DF, Yi P et al (2006) Application of biocatalysis to drug metabolism: preparation of mammalian metabolites of a biaryl-bis-sulfonamide AMPA (alpha-amino-3-hydroxy-5-methylisoxazole-4-propionic acid) receptor potentiator using Actinoplanes missouriensis. Drug Metab Dispos 34:925–931
Acknowledgements
We thank J. Warner for technical assistance and M. D. Lynch for thoughtful discussion. J. E. Prior received financial assistance through the Department of Education’s Graduate Assistance in Areas of National Need Program.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Prior, J.E., Shokati, T., Christians, U. et al. Identification and characterization of a bacterial cytochrome P450 for the metabolism of diclofenac. Appl Microbiol Biotechnol 85, 625–633 (2010). https://doi.org/10.1007/s00253-009-2135-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-009-2135-0