Abstract
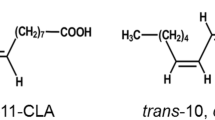
Lactobacillus plantarum AKU 1009a effectively transforms linoleic acid to conjugated linoleic acids of cis-9,trans-11-octadecadienoic acid (18:2) and trans-9,trans-11–18:2. The transformation of various polyunsaturated fatty acids by washed cells of L. plantarum AKU 1009a was investigated. Besides linoleic acid, α-linolenic acid [cis-9,cis-12,cis-15-octadecatrienoic acid (18:3)], γ-linolenic acid (cis-6,cis-9,cis-12–18:3), columbinic acid (trans-5,cis-9,cis-12–18:3), and stearidonic acid [cis-6,cis-9,cis-12,cis-15-octadecatetraenoic acid (18:4)] were found to be transformed. The fatty acids transformed by the strain had the common structure of a C18 fatty acid with the cis-9,cis-12 diene system. Three major fatty acids were produced from α-linolenic acid, which were identified as cis-9,trans-11,cis-15–18:3, trans-9,trans-11,cis-15–18:3, and trans-10,cis-15–18:2. Four major fatty acids were produced from γ-linolenic acid, which were identified as cis-6,cis-9,trans-11–18:3, cis-6,trans-9,trans-11–18:3, cis-6,trans-10–18:2, and trans-10-octadecenoic acid. The strain transformed the cis-9,cis-12 diene system of C18 fatty acids into conjugated diene systems of cis-9,trans-11 and trans-9,trans-11. These conjugated dienes were further saturated into the trans-10 monoene system by the strain. The results provide valuable information for understanding the pathway of biohydrogenation by anaerobic bacteria and for establishing microbial processes for the practical production of conjugated fatty acids, especially those produced from α-linolenic acid and γ-linolenic acid.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Interest in conjugated fatty acids as a novel class of functional lipids has increased in the last two decades, along with the discovery of unique biological/physiological effects of conjugated linoleic acid (CLA). Dietary CLA has been reported to reduce carcinogenesis (Ha et al. 1987, 1990; Pariza and Ha 1990; Ip et al. 1991), atherosclerosis (Lee et al. 1994), and body fat (Park et al. 1997). Recently, similar effects were found for conjugated trienoic acids.
The conjugated trienoic acid produced from α-linolenic acid [cis-9,cis-12,cis-15-octadecatrienoic acid (18:3)] through alkali isomerization showed cytotoxicity toward human tumor cells (Igarashi and Miyazawa 2000). There have been several reports on the occurrence of conjugated trienoic acids in plants, for example, α-eleostearic acid (cis-9,trans-11,trans-13–18:3) in Momordica charantia (Liu et al. 1997), β-eleostearic acid (trans-9,trans-11,trans-13–18:3) in paulownia oil, and punicic acid (cis-9,trans-11,cis-13–18:3) in Punica granatum and Cayaponia africana. These conjugated 9,11,13–18:3 isomers were also found to be cytotoxic toward mouse tumor and human monocytic leukemia cells (Suzuki et al. 2001). Secondary metabolism of fatty acids by marine algae also involves numerous polyunsaturated fatty acids (PUFAs) containing conjugated olefin systems, for example, cis-6,trans-8,trans-10,cis-12-octadecatetraenoic acid (18:4) produced from γ-linolenic acid (cis-6,cis-9,cis-12–18:3) by the coralline red alga Lithothamnion corallioides and bosseopentaenoic acid [cis-5,cis-8,trans-10,trans-12,cis-14-eicosapentaenoic acid (20:5)] produced from arachidonic acid [cis-5,cis-8,cis-11,cis-14-eicosatetraenoic acid (20:4)] by the marine red alga Bossiella orbigniana. On the other hand, except for the investigations on CLA production by anaerobic bacteria including our previous works (Ogawa et al. 2001; Kishino et al. 2002a, b, 2003; Ando et al. 2003, 2004), the microbial production of PUFA containing conjugated double bonds has not been extensively studied so far.
The production of conjugated 18:3 by rumen bacteria was reported in relation with analysis of fatty acids in dairy products (Destaillats et al. 2005; Wasowska et al. 2006; Jouany et al. 2007; Plourde et al. 2007). Conjugated 18:3 is regarded as an intermediate of PUFA biohydrogenation occurring in the rumen, which affects the fatty acid compositions of dairy products. Understanding of the PUFA biohydrogenation process in the rumen is important for improving the fatty acid profiles of diary products so as to be rich in n-3 polyunsaturated fatty acids and CLA (Scollan et al. 2001; Jenkins et al. 2008). With respect to this, α-linolenic acid transformation by rumen bacteria was investigated and it was found that cis-9,trans-11,cis-15–18:3 is an initial intermediate, which is further transformed into trans-11,cis-15-octadecadienoic acid (18:2) and then to trans-11-octadecenoic acid (18:1). However, the details of rumen biohydrogenation of PUFA remain unclear.
These situations led us to develop a novel microbial method for conjugated PUFA production and to understand the pathway of PUFA biohydrogenation by anaerobic bacteria. In this paper, we report that Lactobacillus plantarum AKU 1009a, which was selected as a potential strain producing CLA from linoleic acid (Kishino et al. 2002b), transformed various PUFAs into a variety of conjugated fatty acids. We also identified successive intermediates in α-linolenic acid and γ-linolenic acid transformation. Based on the results, novel pathways of PUFA biohydrogenation by L. plantarum involving isomerization of a cis-9,cis-12 double bond to cis-9,trans-11 and trans-9,trans-11 conjugated double bonds and further saturation to a trans-10 double bond are proposed. This paper also provides detailed structure information of these fatty acids involved in the novel biohydrogenation pathway. The information is valuable for further understandings of biohydrogenation by other anaerobic bacteria and for investigations of physiological activities of the conjugated fatty acids.
Materials and methods
Chemicals
γ-Linolenic acid and fatty acid-free (<0.02%) bovine serum albumin (BSA) were purchased from Sigma (St. Louis, MO, USA). The α-linolenic acid (Wako Pure Chemical, Osaka, Japan) used in this study was of 76% purity, and its fatty acid composition was 76% α-linolenic acid, 19% linoleic acid, and 5% oleic acid. Standard samples of CLA isomers, i.e., cis-9,trans-11–18:2 (CLA1) and trans-9,trans-11–18:2 (CLA2) and 10-hydroxy-12–18:1 were prepared as described previously (Ogawa et al. 2001). All other chemicals used were of analytical grade and were commercially available.
Microorganisms, cultivation, and preparation of washed cells
Washed cells of L. plantarum AKU 1009a (AKU culture collection, Faculty of Agriculture, Kyoto University, Kyoto, Japan) were used as the catalyst for fatty acid transformation (Kishino et al. 2002b). The strain was cultivated in de Man–Rogosa–Sharpe medium comprising 1.0% tryptone, 1.0% meat extract, 0.5% yeast extract, 2.0% glucose, 0.1% Tween 80, 0.2% K2HPO4, 0.5% sodium acetate, 0.2% diammonium citrate, 0.02% MgSO4·7H2O, 0.005% MnSO4·5H2O, and 0.06% linoleic acid (pH 6.5). The strain was inoculated into 550 ml of medium in 600-ml flasks and then incubated at 28°C with shaking (120 strokes/min) for 24 h. Growth was monitored as the optical density at 610 nm. Cells were harvested by centrifugation (12,000 × g, 10 min), washed twice with 0.85% NaCl, centrifuged again, and then used as the washed cells for the reactions.
Reaction conditions
The reaction mixture, 1 ml, in test tubes (16.5 × 125 mm) was composed of 0.4% (w/v) fatty acid complexed with BSA [0.08% (w/v)], 0.1 M potassium phosphate buffer (pH 6.5), and 22.5% (wet cells, w/v) washed cells [corresponding to 3.2% (dry cells, w/v)]. The fatty acids used as the substrates were cis-6–18:1, cis-9–18:1 (oleic acid), trans-9–18:1 (elaidic acid), cis-11–18:1 (cis-vaccenic acid), trans-11–18:1 (trans-vaccenic acid), cis-12–18:1, trans-12–18:1, cis-9,cis-12–18:2 (linoleic acid), cis-9,cis-12,cis-15–18:3 (α-linolenic acid), cis-6,cis-9,cis-12–18:3 (γ-linolenic acid), trans-9,trans-12,trans-15–18:3 (linolenelaidic acid), trans-5,cis-9,cis-12–18:3 (columbinic acid), cis-6,cis-9,cis-12,cis-15–18:4 (stearidonic acid), cis-8,cis-11,cis-14-eicosatrienoic acid (20:3) (dihomo-γ-linolenic acid), cis-5,cis-8,cis-11,cis-14–20:4 (arachidonic acid), cis-5,cis-8,cis-11,cis-14,cis-17–20:5 (EPA), cis-13-docosaenoic acid (22:1), and cis-15-tetracosaenoic acid (24:1). The reactions were carried out microaerobically under an O2-adsorbed atmosphere in a sealed chamber with O2-absorbent (AnaeroPack “Kenki”, Mitsubishi Gas Chemical Co., Inc., Tokyo, Japan), with gentle shaking (120 strokes/min) at 37°C for 24 to 72 h. Under the microaerobic conditions, the oxygen concentration, monitored with an oxygen indicator (Mitsubishi Gas Chemical Co., Inc., Tokyo, Japan), was kept under 1%. All experiments were performed in triplicate, and the averages of three separate experiments, which were reproducible within ±10%, are presented in the figures.
Lipid analyses
Lipids were extracted from the reaction mixture with chloroform–methanol (1:2, by vol.) according to the procedure of Bligh and Dyer (1959) and methylated with 10% methanolic HCl at 50°C for 20 min. The resultant fatty acid methyl esters were extracted with n-hexane and analyzed by gas–liquid chromatography (GC) using a Shimadzu (Kyoto, Japan) GC-1700 gas chromatograph equipped with a flame ionization detector and a split injection system and fitted with a capillary column (HR-SS-10, 50 m × 0.25 mm I.D., Shinwa Kako, Kyoto, Japan), as described previously (Ogawa et al. 2001).
Isolation, derivatization, and identification of reaction products
The fatty acid methyl esters of the reaction products were separated at 30°C by high-performance liquid chromatography (HPLC; monitored at 205 and 233 nm) using a Shimadzu LC-VP system fitted with a Cosmosil 5C18-AR-II-packed column (20 × 250 mm, Nacalai Tesque, Kyoto, Japan). The mobile phase was acetonitrile–H2O (8:2, by vol.) at the flow rate of 3.0 ml/min. The separated fatty acid methyl esters were purified with a HPLC fitted with a ChromSpher 5 Lipids-packed column (4.6 × 250 mm, Chrompack, Raritan, NJ, USA; Ostrowska et al. 2000). The mobile phase was hexane–acetonitorile (99.9:0.1, by vol.) at the flow rate of 1.0 ml/min. Free fatty acids and pyrrolidide derivatives were prepared by saponification with sodium hydroxide and direct treatment with pyrrolidine acetic acid, respectively, as described previously (Ogawa et al. 2001). The isolated fatty acid methyl esters were dissolved in CDCl3 and analyzed by proton nuclear magnetic resonance (1H-NMR), 1H–1H chemical shift correlation spectroscopy (DQF-COSY), two-dimensional nuclear Overhauser enhancement spectroscopy, and 1H clean-total correlation spectroscopy (clean-TOCSY; Kishino et al. 2003) with a Bruker Biospin CMX-750 (750 MHz for 1H). The chemical shifts were assigned relative to the solvent signal. The free fatty acids and pyrrolidide derivatives were subjected to mass spectroscopy (MS)–MS analysis and GC–MS analysis, respectively, as described previously (Ogawa et al. 2001).
MS and NMR spectra
CALA1
MS–MS analysis for the molecular ion peak of CALA1 (Li-complex, m/z 299 [M+Li]+); m/z (FAB+, 8.00 kV), 283(2), 269(2), 243(1), 230(15), 229(18), 215(2), 189(1), 171(4), 163(2), 156(8), 149(18), 135(12), 121(5), 107(40), 94(24), 93(100), 80(65). NMR δH (CDCl3); (the large alphabets indicate the proton signals on the carbons represented in Figs. 2A and 4) M, 6.31 (1H, dd, J = 15.1, 10.8 Hz, =CH–CH=); L, 5.94 (1H, dd, J = 11.0, 10.9 Hz, =CH–CH=); K, 5.66 (1H, dt, J = 15.1, 6.5 Hz, =CH–CH2–); J, 5.39 (1H, dt, J = 10.8, 7.0 Hz, =CH–CH2–); I, 5.33 (1H, dt, J = 10.8, 6.7 Hz, –CH2–CH=); H, 5.30 (1H, dt, J = 11.0, 7.0 Hz, –CH2–CH=); X, 3.67 (3H, s, –OCH 3); G, 2.30 (2H, t, J = 7.6 Hz, –COCH 2–); F, 2.15 (6H, m, –CH2–CH 2–CH=); E, 2.04 (2H, dq, J = 7.5, 7.4 Hz, =CH–CH 2CH3); D, 1.64 (2H, m, –CH2CH 2CH2–); C, 1.37 (2H, m, –CH2CH 2CH2–); B, 1.31 (6H, m, –CH2CH 2CH2–); A, 0.96 (3H, t, J = 7.5 Hz, –CH 3).
CALA2
MS–MS analysis for the molecular ion peak of CALA2 (Li-complex, m/z 299 [M+Li]+); m/z (FAB+, 8.00 kV), 283(5), 269(3), 243(2), 230(21), 229(26), 215(4), 189(2), 171(3), 163(4), 156(10), 149(29), 135(17), 121(9), 107(60), 94(46), 93(100), 80(99). NMR δH (CDCl3); (the large alphabets indicate the proton signals on the carbons represented in Figs. 2D and 4) L, 6.01 (2H, m, =CH–CH=); K, 5.58 (1H, dt, J = 14.3, 6.9 Hz, –CH2–CH=); J, 5.56 (1H, dt, J = 14.3, 7.0 Hz, =CH–CH2–); I, 5.38 (1H, dt, J = 10.7, 7.1 Hz, =CH–CH2–); H, 5.33 (1H, dt, J = 10.7, 6.6 Hz, –CH2–CH=); X, 3.67 (3H, s, –OCH 3); G, 2.30 (2H, t, J = 7.6 Hz, –COCH 2–); F, 2.11 (4H, dt, J = 15.1, 7.3 Hz, –CH2–CH 2–CH=); E, 2.04 (4H, m, –CH 2–CH=); D, 1.62 (2H, t, J = 7.2, –CH2CH 2CH2–); C, 1.36 (2H, tt, J = 7.2, 6.7, –CH2CH 2CH2–); B, 1.29 (6H, m, –CH2CH 2CH2–); A, 0.95 (3H, t, J = 7.5 Hz, –CH 3).
A1
MS–MS analysis for the molecular ion peak of A1 (Li-complex, m/z 301 [M+Li]+); m/z (FAB+, 8.00 kV), 285(4), 271(1), 245(1), 231(19), 218(7), 217(12), 203(3), 177(2), 163(23), 149(11), 135(18), 121(11), 107(36), 94(30), 93(100), 80(87). NMR δH (CDCl3); (the large alphabets indicate the proton signals on the carbons represented in Fig. S1 and Fig. 4) J, 5.40 (2H, m, –CH2–CH=); I, 5.37 (1H, dt, J = 10.8, 7.0 Hz, –CH2–CH=); H, 5.32 (1H, dt, J = 10.8, 7.2 Hz, =CH–CH2–); X, 3.67 (3H, s, –OCH 3); G, 2.30 (2H, t, J = 7.5 Hz, –COCH 2–); F, 2.03 (4H, m, –CH 2–CH=); E, 1.98 (4H, m, –CH 2–CH=); D, 1.61 (2H, tt, J = 7.5, 7.4 Hz, –CH2CH 2CH2–); C, 1.40 (2H, tt, J = 8.2, 7.3 Hz, –CH2CH 2CH2–); B, 1.29 (10H, m, –CH2CH 2CH2–); A, 0.97 (3H, t, J = 7.5 Hz, –CH 3).
CGLA1
MS–MS analysis for the molecular ion peak of CGLA1 ([M-H]+, 277); m/z (FAB−, 8.00 kv), 261(11), 247(6), 233(13), 219(12), 205(11), 191(25), 179(5), 177(10), 167(2), 165(4), 153(13), 139(13), 127(12), 125(11), 113(12), 100(7), 99(45), 86(32), 85(7), 72(23), 71(91), 58(100), 44(48). NMR δH (CDCl3); (the large alphabets indicate the proton signals on the carbons represented in Fig. S2 and Fig. 4) M, 6.31 (1H, dd, J = 15.0, 9.6 Hz, =CH–CH=); L, 5.96 (1H, dd, J = 11.0, 10.7 Hz, =CH–CH=); K, 5.69 (1H, dt, J = 15.0, 7.3 Hz, =CH–CH2–); J, 5.40 (1H, dt, J = 11.0, 5.9 Hz, –CH2–CH=); I, 5.37 (1H, dt, J = 11.0, 6.0 Hz, =CH–CH2–); H, 5.25 (1H, dt, J = 10.7, 7.4 Hz, –CH2–CH=); X, 3.67 (3H, s, –OCH 3); G, 2.90 (2H, dd, J = 6.0, 5.9 Hz, =CH–CH 2–CH=); F, 2.32 (2H, t, J = 7.5 Hz, –COCH 2–); E, 2.10 (4H, dt, J = 14.4, 7.2 Hz, –CH 2–CH=); D, 1.65 (2H, tt, J = 7.8, 7.5 Hz, –CH2CH 2CH2–); C, 1.39 (4H, m, –CH2CH 2CH2–); B, 1.28 (6H, m, –CH2CH2CH2–); A, 0.88 (3H, t, J = 6.9 Hz, –CH 3).
CGLA2
MS–MS analysis for the molecular ion peak of CGLA1 (Li-complex, m/z 299 [M+Li]+); m/z (FAB+, 8.00 kv), 283(4), 269(2), 255(5), 241(15), 228(13), 227(11), 213(4), 201(2), 187(1), 175(11), 162(8), 161(38), 147(9), 133(1), 121(1), 108(12), 107(27), 94(24), 93(50), 80(100). NMR δH (CDCl3); (the large alphabets indicate the proton signals on the carbons represented in Fig. S3 and Fig. 4) K, 6.01 (2H, m, =CH–CH=); J, 5.59 (1H, dt, J = 14.1, 7.0 Hz, =CH–CH2–); I, 5.52 (1H, dt, J = 14.2, 7.1 Hz, –CH2–CH=); H, 5.40 (2H, m, –CH2–CH=); X, 3.67 (3H, s, –OCH 3); G, 2.79 (2H, dd, J = 7.4, 5.3 Hz, =CH–CH 2–CH=); F, 2.32 (2H, t, J = 7.5 Hz, –COCH 2–); E, 2.05 (4H, dt, J = 15.4, 7.3 Hz, –CH2–CH 2–CH=) D, 1.64 (2H, tt, J = 7.7, 7.6 Hz, –CH2CH 2CH2–); C, 1.38 (4H, m, –CH2CH 2CH2–); B, 1.27 (6H, m, –CH2CH 2CH2–); A, 0.89 (3H, t, J = 7.0 Hz, –CH 3).
G2
MS–MS analysis for the molecular ion peak of G2 ([M-H]+, 279); m/z (FAB+, 8.00 kv), 263(9), 249(7), 235(13), 221(22), 207(15), 194(9), 193(10), 179(7), 153(3), 140(36), 139(36), 127(1), 125(1), 113(2), 100(1), 99(2), 86(29), 71(66), 58(100), 44(18). NMR δH (CDCl3); (the large alphabets indicate the proton signals on the carbons represented in Fig. S4 and Fig. 4) J, 5.40 (2H, dt, J = 14.6, 6.0 Hz, =CH–CH2–); I, 5.36 (2H, dt, J = 11.4, 7.0 Hz, –CH2–CH=); X, 3.67 (3H, s, –OCH 3); H, 2.31 (2H, t, J = 7.5 Hz, –COCH 2–); G, 2.08 (2H, m, –CH2–CH 2–CH=); F, 2.04 (4H, m, –CH2–CH 2–CH=); E, 1.97 (2H, m, =CH2–CH 2–CH–); D, 1.64 (2H, tt, J = 7.7, 7.6 Hz, –CH2CH 2CH2–); C, 1.38 (2H, m, –CH2CH 2CH2–); B, 1.28 (10H, m, –CH2CH 2CH2–); A, 0.88 (3H, t, J = 6.9 Hz, –CH 3).
G1
MS–MS analysis for the molecular ion peak of G1 (Li-complex, m/z 303 [M+Li]+); m/z (FAB+, 8.00 kv), 287(4), 273(2), 259(4), 245(9), 231(21), 218(8), 217(12), 203(3), 201(3), 177(4), 163(26), 149(15), 135(20), 121(15), 107(35), 94(31), 93(100), 80(83). NMR δH (CDCl3); (the large alphabets indicate the proton signals on the carbons represented in Fig. S5 and Fig. 4) F, 5.38 (2H, m, –CH2–CH=); X, 3.67 (3H, s, –OCH 3); E, 2.30 (2H, t, J = 7.6 Hz, –COCH 2–); D, 1.96 (4H, m, –CH2–CH 2–CH=); C, 1.62 (2H, m, –CH2CH 2CH2–); B, 1.30 (20H, m, –CH2CH 2CH2–); A, 0.88 (3H, t, J = 6.9 Hz, –CH 3).
Results
Transformation of polyunsaturated fatty acids by washed cells of L. plantarum AKU 1009a
Washed cells of L. plantarum AKU 1009a prepared under the optimum culture conditions for CLA production from linoleic acid were used as the catalyst for fatty acid transformation (Kishino et al. 2002b). Free fatty acids of cis-6–18:1, oleic acid, elaidic acid, cis-vaccenic acid, trans-vaccenic acid, cis-12–18:1, trans-12–18:1, linoleic acid, α-linolenic acid, γ-linolenic acid, linolenelaidic acid, columbinic acid, stearidonic acid, dihomo-γ-linolenic acid, arachidonic acid, EPA, cis-13–22:1, and cis-15–24:1 were used as the substrates. When linoleic acid, α-linolenic acid, γ-linolenic acid, columbinic acid, or stearidonic acid was used as the substrate, newly generated fatty acids were observed in the GC chromatograms of the methylated fatty acid products. The fatty acids recognized as substrates were C18 free fatty acids with a cis-9,cis-12-diene system. The other fatty acids tested were not transformed by the washed cells of L. plantarum.
Fatty acids produced from α-linolenic acid by washed cells of L. plantarum AKU 1009a
A GC chromatogram of the methylated fatty acids produced from α-linolenic acid by washed cells of L. plantarum AKU 1009a is shown in Fig. 1A. Three major newly generated fatty acids designated as A1, CALA1, and CALA2 were observed in the GC chromatogram of the methylated fatty acids. These materials were purified by HPLC from the mixture of fatty acid methyl esters and subjected to structure analysis.
Transformation of α-linolenic acid (A) and γ-linolenic acid (B) by L. plantarum AKU 1009a. The peaks indicated by big arrows are the newly generated fatty acids. 0 h and 24 h represent the reaction times. LA linoleic acid, ALA α-linolenic acid, CLA1 cis-9,trans-11-octadecadienoic acid, CLA2 trans-9,trans-11-octadecadienoic acid, GLA γ-linolenic acid. All chromatographs are presented in the same magnification
Identification of CALA1 and CALA2
Mass spectra of pyrrolidide derivatives of the isolated methyl esters of CALA1 and CALA2 both showed a molecular weight of m/z 331. These results suggested that CALA1 and CALA2 are C18 fatty acids containing three double bonds. The molecular ion peaks (Li-complex, m/z 299 [M+Li]+) obtained on FAB–MS analysis (FAB+) of the free fatty acids of CALA1 and CALA2 were fragmented again on MS–MS (the results are presented in “Materials and methods” section). The peaks derived on cleavage at single bonds 8–9, 10–11, 12–13, 13–14, 14–15, and 16–17, numbered from the carboxyl group, were detected. The peaks derived on the cleavage of the single bond between the α and β positions from the double bond (m/z 149, 229, and 283 peaks) were also clearly detected. On the basis of the results of MS analyses, CALA1 and CALA2 were identified as geometrical isomers of 9,11,15–18:3. These structures were further confirmed by the results of 1H-NMR analysis of the fatty acid methyl esters (the results are presented in “Materials and methods” section).
On the basis of the signal pattern of the interaction between adjacent protons observed on DQF-COSY (Fig. 2B), the sequence of the protons from the methyl end of CALA1 was deduced to be A–E–J–I–F–K–M–L–H–F–C–B–D–G or A–E–J–I–F–H–L–M–K–F–C–B–D–G. On clean-TOCSY analysis (Fig. 2C), the sequence was confirmed to be the former one by the appearance of an interaction signal between C and H, but not C and K, indicating that C was near to H, but that K was far from C. Coupling constants were obtained based on the decoupled 1H-NMR spectra of the methyl ester of CALA1. When the methyl ester was irradiated at 2.04 ppm (signal E), the coupling constant between I and J was 10.8 Hz, indicating that the double bond between I and J is of the cis-configuration. When it was irradiated at 2.15 ppm (signal F), the coupling constant between K and M was 15.1 Hz, indicating that the double bond between K and M is of the trans-configuration. With the same irradiation, the coupling constant between H and L was 11.0 Hz, indicating that the double bond between H and L is of the cis-configuration. On the basis of the results of the above spectral analyses, CALA1 was identified as cis-9,trans-11,cis-15–18:3 (Fig. 2A).
1H-NMR analysis of CALA1 and CALA2 and the structures of CALA1 and CALA2 identified. A Structure of CALA1; B1H–1H chemical shift correlation spectroscopic spectrum of the methyl ester of CALA1; C1H clean-total correlation spectroscopic spectrum of the methyl ester of CALA1; D Structure of CALA2; E1H–1H chemical shift correlation spectroscopic spectrum of the methyl ester of CALA2; F1H clean-total correlation spectroscopic spectrum of the methyl ester of CALA2
As shown in Fig. 2E, the DQF-COSY signal pattern of CALA2 indicated fragment proton sequences of A–E–I, K or C, I–H–F, K–L–J and E–C–B–D–G. As shown in Fig. 2F, the signal pattern on clean-TOCSY showed clear interaction between A and I, but not A and J or A and C. Decoupled 1H-NMR spectra with irradiation at 2.04 (signal E) and 2.11 ppm (signal F) indicated in the disappearance of signals K and J, respectively. These results confirmed that the proton sequence from the methyl end of CALA2 is A–E–I–H–F–J–L–K–E–C–B–D–G. The 1H-NMR coupling constant between J and L obtained on irradiation at 2.11 ppm (signal F) was 14.3 Hz, and those of H and I, and K and L obtained on irradiation at 2.04 ppm (signal E) were 10.7 and 14.3 Hz, respectively. These results indicated that the double bonds between J and L, H and I, and K and L are of the trans-, cis-, and trans-configurations, respectively. On the basis of the results of the above spectral analyses, CALA2 was identified as trans-9,trans-11,cis-15–18:3 (Fig. 2D).
Identification of A1
The mass spectrum of the pyrrolidide derivative of A1 revealed a molecular weight of m/z 333. This result suggested that compound A1 is a C18 fatty acid containing two double bonds. The molecular ion peak (Li-complex, m/z 301 [M+Li]+) obtained on FAB–MS analysis (FAB+) of the free fatty acid of A1 was fragmented again on MS–MS (the result is presented in “Materials and methods” section). The peaks derived on cleavage at single bonds 9–10, 11–12, 12–13, 13–14, 14–15, and 16–17, numbered from the carboxyl group, were detected. The peaks derived on the cleavage of the single bond between the α and β positions from the double bond (m/z 163, 217, 231, and 285 peaks) were also clearly detected. On the basis of the results of spectral analyses, A1 was identified as 10,15–18:2. This deduced structure was further confirmed by the result of 1H-NMR (the result is presented in “Materials and methods” section). On the basis of the signal pattern on DQF-COSY (Fig S1 and Fig. 4), the sequence of the protons from the methyl end of A1 was deduced to be A–F–H–I–F–C–E–J–E–B–D–G or A–F–I–H–F–C–E–J–E–B–D–G. The signal pattern on clean-TOCSY (Fig. S1) showed clear interaction between A and H, but not A and I. These results confirmed that the proton sequence from the methyl end of A1 is A–F–H–I–F–C–E–J–E–B–D–G (Fig. 4). The 1H-NMR coupling constant between H and I obtained on irradiation at 2.03 ppm (signal F) was 10.8 Hz and that of J and J obtained on irradiation at 1.98 ppm (signal E) were 15.3 Hz. These results indicated that the double bonds between H and I, and J and J are of the cis- and trans-configurations, respectively. On the basis of the results of the above spectral analyses, A1 was identified as trans-10,cis-15–18:2 (Fig. 4).
Time course of α-linolenic acid transformation by washed cells of L. plantarum AKU 1009a
The time course of changes in fatty acid composition during α-linolenic acid [0.3% (w/v)] transformation by washed cells [22.5% (wet cells, w/v)] of L. plantarum AKU 1009a was studied. CALA (sum of CALA1 and CALA2) reached 41.7% (wt.%) of total fatty acids after 48-h reaction (Fig. 3A). The amount of CALA produced after 48-h reaction was 1.59 mg/ml (CALA1, 0.27 mg/ml; CALA2, 1.32 mg/ml; molar conversion to α-linolenic acid, 47%). The proportion of A1 in total fatty acids was 3.8% (wt.%) after 24-h reaction, and it gradually increased with a decrease in the proportion of CALA. These results suggest that CALA was further converted to A1.
Time course of α-linolenic acid (A) and γ-linolenic acid (B) transformation by L. plantarum AKU 1009a. Cellular FA comprised myristic acid, palmitic acid, palmitoleic acid, oleic acid, trans-vaccenic acid, and 2-hexy-1-cyclopropane-octanoic acid. CALA1 cis-9,trans-11,cis-15-octadecatrienoic acid, CALA2 trans-9,trans-11,cis-15-octadecatrienoic acid, A1 trans-10,cis-15-octadecadienoic acid, LA linoleic acid, CLA1 cis-9,trans-11-octadecadienoic acid, CLA2 trans-9,trans-11-octadecadienoic acid, CGLA1 cis-6,cis-9,trans-11-octadecatrienoic acid, CGLA2 cis-6,trans-9,trans-11-octadecatrienoic acid, G2 cis-6,trans-10-octadecadienoic acid, G1 trans-10-octadecaenoic acid, Others possible hydroxylated fatty acids and other unknown fatty acids
Fatty acids produced from γ-linolenic acid by washed cells of L. plantarum AKU 1009a
A GC chromatogram of the methylated fatty acids produced from γ-linolenic acid by washed cells of L. plantarum AKU 1009a is shown in Fig. 1B. Four major newly generated fatty acids designated as G1, G2, CGLA1, and CGLA2 were observed in the GC chromatogram. They were purified by HPLC from the mixture of fatty acid methyl esters and subjected to structure analysis.
Identification of CGLA1 and CGLA2
The mass spectra of the pyrrolidide derivatives of CGLA1 and CGLA2 both showed a molecular weight of m/z 331. These results suggested that CGLA1 and CGLA2 are C18 fatty acids containing three double bonds. The molecular ion peak ([M-H]+, 277) obtained on FAB–MS analysis (FAB−) of the free fatty acid of CGLA1 was fragmented again on MS–MS (the result is presented in “Materials and methods” section). The peaks derived on cleavage at single bonds 5–6, 7–8, 8–9, 10–11, and 12–13, numbered from the carboxyl group, were detected. The peaks derived on the cleavage of the single bond between the α and β positions from the double bond (m/z 85/86 and 205 peaks) were also clearly detected. The molecular ion peak (Li-complex, m/z 299 [M+Li]+) obtained on FAB–MS analysis (FAB+) of the free acid of CGLA2 was fragmented again on MS–MS (the result is presented in “Materials and methods” section). The peaks derived on cleavage at single bonds 5–6, 7–8, 8–9, 10–11, and 12–13, numbered from the carboxyl group, were detected. The peaks derived on the cleavage of the single bond between the α and β positions from the double bond (m/z 107 and 227 peaks) were also clearly detected. On the basis of the results of MS analyses, CGLA1 and CGLA2 were identified as geometrical isomers of 6,9,11–18:3.
1H-NMR analysis also suggested that CGLA1 is an isomer of octadecatrienoic acid (the result is presented in “Materials and methods” section). On DQF-COSY analysis (Fig. S2 and Fig. 4), the sequence of the protons from the methyl end of the molecule was deduced to be A–B–C–E–K–M–L–H–G–I–J–E–C–D–F or A–B–C–E–J–I–G–H–L–M–K–E–C–D–F based on the signal pattern of interaction between adjacent protons. The sequence was confirmed to be the former one based on the results of MS analyses of the free fatty acid that clarified that the C8 carbon, numbered from the carboxyl group, is flanked by saturated bonds and the results of 1H-NMR analysis (Fig. S2 and Fig. 4) showing that signals H (5.25 ppm), I (5.37 ppm), J (5.40 ppm), K (5.69 ppm), L (5.96 ppm), and M (6.31 ppm) are on the double bonds. The 1H-NMR coupling constant between H and L obtained on irradiation at 2.90 ppm (signal G) was 10.7 Hz, and those of I and J, and K and M obtained on irradiation at 2.10 ppm (signal E) were 11.0 and 15.0 Hz, respectively. These results indicated that the double bonds between H and L, I and J, and K and M are of the cis-, cis-, and trans-configurations, respectively. On the basis of the results of the above spectral analyses, CGLA1 was identified as cis-6,trans-9,trans-11–18:3 (Fig. 4).
Putative pathway for α- and γ-linolenic acid transformation by L. plantarum AKU 1009a. The large alphabets on the carbons of each compounds indicate the corresponding proton signals observed on NMR analysis (see “Results” section for identification of each compounds)
1H-NMR analysis also suggested that CGLA2 is an isomer of octadecatrienoic acid (the result is presented in “Materials and methods” section). On DQF-COSY (Fig. S3 and Fig. 4), the sequence of the protons from the methyl end of CGLA2 was deduced to be A–B–C–E–J–K–I–G–H–E–C–D–F or A–B–C–E–H–G–I–K–J–E–C–D–F on the basis of the signal pattern of the interaction between adjacent protons. On clean-TOCSY analysis (Fig. S3 and Fig. 4), the sequence was confirmed to be the former one by the appearance of an interaction signal between D and H, but not D and J, indicating that D was near to H, but that D was far from J. The 1H-NMR coupling constant between I and K obtained on irradiation at 2.79 ppm (signal G) was 14.2 Hz, and those of H and H, and J and K obtained on irradiation at 2.05 ppm (signal E) were 10.9 and 14.1 Hz, respectively. These results indicated that the double bonds between I and K, H and H, and J and K are of the trans-, cis-, and trans-configurations, respectively. On the basis of the results of the above spectral analyses, CGLA2 was identified as cis-6,trans-9,trans-11–18:3 (Fig. 4).
Identification of G2
The mass spectrum of the pyrrolidide derivative of G2 showed a molecular weight of m/z 333. This result suggested that compound G2 is a C18 fatty acid containing two double bonds. The FAB–MS data for the free fatty acid of G2 revealed a molecular weight of m/z 280 ([M-H]+, 279). The molecular ion peak ([M-H]+, 279) obtained on FAB–MS analysis (FAB+) of the free fatty acid of G2 was fragmented again on MS–MS (the result is presented in “Materials and methods” section). The peaks derived on cleavage at single bonds 5–6, 7–8, 8–9, 9–10, and 11–12, numbered from the carboxyl group, were detected. The peaks derived on the cleavage of the single bond between the α and β positions from the double bond (m/z 86, 139 and 193 peaks) were also clearly detected. On the basis of the results of MS analyses, G2 was identified as 6,10–18:2.
This structure was further confirmed by the results of 1H-NMR analysis of the fatty acid methyl ester (the result is presented in “Materials and methods” section). On 1H-NMR analysis, signals around 2.9 ppm, which indicate the existence of protons of methylene flanked by double bonds, were not observed (Fig. S4). This result also supported that G2 is 6,10–18:2. On DQF-COSY (Fig. S4 and Fig. 4), the sequence of the protons from the methyl end of G2 was deduced to be A–B–E–J–G–F–I–F–C–D–H on the basis of the signal pattern of the interaction between adjacent protons. The 1H-NMR coupling constant between I and I was 11.4 Hz and that between J and J was 14.6 Hz. These results indicated that the double bonds between I and I, and J and J are of the cis- and trans-configuration, respectively. On clean-TOCSY analysis (Fig. S4 and Fig. 4), the absence of an interaction signal between G and E also indicated that the double bond between J and J is of the trans-configuration. On the basis of the results of the above spectral analyses, G2 was identified as cis-6,trans-10–18:2 (Fig. 4).
Identification of G1
The mass spectrum of the pyrrolidide derivative of G1 showed a molecular weight of m/z 335. This result suggested that G1 is a C18 fatty acid containing one double bond. The molecular ion peak (Li-complex, m/z 303 [M+Li]+) obtained on FAB–MS analysis (FAB+) of the free acid of G1 was fragmented again on MS–MS (the result is presented in “Materials and methods” section). The peaks derived on cleavage at single bonds 9–10 and 11–12, numbered from the carboxyl group, were detected. The peaks derived on the cleavage of the single bond between the α and β positions from the double bond (m/z 163 and 217 peaks) were also clearly detected. On the basis of the results of MS analyses, G1 was identified as 10–18:1.
This deduced structure was further confirmed by the results of 1H-NMR (the result is presented in “Materials and methods” section). On DQF-COSY (Fig. S5 and Fig. 4), the sequence of the protons from the methyl end of G1 was deduced to be A–B–D–F–D–B–C–E on the basis of the signal pattern of the interaction between adjacent protons. The results of chemical shifts were simulated with gNMR to determine the configurations of Δ10 double bonds. The simulation results supported that the structure of G1 is trans-10–18:1 (Fig. 4).
Time course of γ-linolenic acid transformation by washed cells of L. plantarum AKU 1009a
The time course of changes in fatty acid composition during γ-linolenic acid [0.4% (w/v)] transformation by washed cells [22.5% (wet cells, w/v)] of L. plantarum AKU 1009a was studied. CGLA (sum of CGLA1 and CGLA2) reached 58.8% (wt.%) of total fatty acids after 24-h reaction (Fig. 3B). The amount of CGLA produced after 24-h reaction was 1.94 mg/ml (CGLA1, 0.36 mg/ml; CGLA2, 1.58 mg/ml; molar conversion to γ-linolenic acid, 46%).The proportions of G2 and G1 in total fatty acids were 5.4% (wt.%) and 6.4% (wt.%) after 24-h reaction, respectively, and they gradually increased with a decrease in the proportion of CGLA. These results suggest that CGLA was converted to G2 and then further converted to G1.
Discussion
Transformation of PUFAs by anaerobic bacteria was studied in relation with ruminal PUFA biohydrogenation. The main aim of investigations on ruminal PUFA biohydrogenation is to create healthier ruminant products with low saturated fatty acid and high n-3 fatty acid contents (Scollan et al. 2001; Jenkins et al. 2008) and with potentially health-promoting CLA, mainly cis-9,trans-11-CLA. Considerable progress has been made in tracing intermediates of linoleic acid biohydrogenation. On the other hand, less is known about the identity of intermediates arising on biohydrogenation of 18:3 including α-linolenic acid and γ-linolenic acid.
Dawson and Kemp (1969) reported that incubation of 14C α-linolenic acid with rumen microorganisms resulted in a bewildering number of radioactive C18 acids exhibiting various degrees of unsaturation and positional isomerization. This finding indicated the existence of a variety of pathways for α-linolenic acid biohydrogenation. Destaillats et al. (2005) reported that two conjugated 18:3 isomers were present in milk fat, namely the cis-9,trans-11,cis-15–18:3 and cis-9,trans-13,cis-15–18:3 isomers. Based on their appearance in milk fat, they proposed that the two isomers are the initial intermediates of α-linolenic acid biohydrogenation, which are then subsequently reduced to two nonconjugated dienes (trans-11,cis-15–18:2 and cis-9,trans-13–18:2) and two conjugated dienes (cis-9,trans-11-CLA and trans-13,cis-15-CLA). Wasowska et al. (2006) presented an alternative view of α-linolenic acid biohydrogenation. α-Linolenic acid biohydrogenation in strained rumen fluid led to the accumulation of two 18:3 isomers (cis-9,trans-11,cis-15–18:3 and trans-9,trans-11,cis-15–18:3) and one nonconjugated 18:2 isomer (trans-11,cis-15–18:2), but accumulation of cis-9,trans-11-CLA was not seen. Jouany et al. also confirmed that the initial intermediate of α-linolenic acid biohydrogenation is cis-9,trans-11,cis-15–18:3, which is transformed into trans-11,cis-15–18:2 and then trans-11–18:1 (Jouany et al. 2007). However, it is difficult or impossible to determine the origin of a specific biohydrogenation intermediate in ruminal contents when the diet contains a multitude of fatty acids. Pure culture studies in which the media or reaction mixtures contain a single fatty acid as the substrate are required to determine the origin of biohydrogenation intermediates and the pathway of PUFA transformation in anaerobic bacteria.
There has been one earlier study involving a pure culture and defined substrates by Kepler and Tove (1967). They incubated Butyrivibrio fibrisolvens with linoleic acid or α-linolenic acid. They confirmed that linoleic acid was first isomerized to cis-9,trans-11-CLA followed by further hydrogenation to trans-11–18:1. When α-linolenic acid was used as the substrate, a conjugated fatty acid, which was tentatively assigned the structure of cis-9,trans-11,cis-15–18:3, was generated as an initial intermediate. This fatty acid was further hydrogenated to a nonconjugated 18:2 as the final product.
Our studies involving reaction mixtures with a pure culture of L. plantarum AKU 1009a and defined substrates revealed clear examples of biohydrogenation pathways catalyzed by anaerobic bacteria. The fatty acids recognized as the substrates by washed cells of L. plantarum AKU 1009a had the common structure of a C18 fatty acid with a cis-9,cis-12 diene system. The cis-9,cis-12 diene system is converted to the cis-9,trans-11- and trans-9,trans-11-diene systems and further saturated to the trans-10 monoene system by washed cells of L. plantarum AKU 1009a. Figure 4 shows the proposed pathways for α-linolenic acid and γ-linolenic acid transformation by washed cells of L. plantarum AKU 1009a. α-Linolenic acid is isomerized to CALA1 and CALA2 with cis-9,trans-11- and trans-9,trans-11-diene systems, respectively, and further saturated to trans-10,cis-15–18:2. Similarly, γ-linolenic acid is isomerized to CGLA1 and CGLA2 with cis-9,trans-11- and trans-9,trans-11-diene systems, respectively, and further saturated to trans-10–18:1 via cis-6,trans-10–18:2. Although the products derived from stearidonic acid and columbinic acid were not identified because of their insufficient amounts, on the basis of the above results, three major fatty acids produced from stearidonic acid are supposed to be cis-6,cis-9,trans-11,cis-15–18:4, cis-6,trans-9,trans-11,cis-15–18:4, and cis-6,trans-10,cis-15–18:3, and three major fatty acids produced from columbinic acid are supposed to be trans-5,cis-9,trans-11–18:3, trans-5,trans-9,trans-11–18:3, and trans-5,trans-10–18:2.
Our observations of the initial step of α-linolenic acid transformation resemble those in the previous works of Wasowska et al. (2006) and Kepler and Tove (1967) in that cis-9,trans-11,cis-15–18:3 and trans-9,trans-11,cis-15–18:3 were found to be the initial intermediates. But our finding was different from theirs in that the saturation of cis-9,trans-11 and trans-9,trans-11 double bonds to a trans-10 double bond was a successive step of α-linolenic acid biohydrogenation, while previous works reported that saturation to a trans-11 double bond was a successive step. Griinari et al. (1998) proposed that cis-9,trans-11-CLA originated from linoleic acid and that it was eventually converted into trans-10–18:1 in ruminal contents. Our results clearly indicate the involvement of trans-10-containing fatty acids as biohydrogenation intermediates.
The results presented here constitute the first example of detailed analysis of a biohydrogenation pathway catalyzed by anaerobic bacteria. However, besides the pathway presented here, various metabolic pathways should be investigated to understand the full diversity of the fatty acids existing in the rumen and in dairy products. This paper provides detailed structure information of these fatty acids involved in the biohydrogenation pathway. The information is valuable for further understandings of biohydrogenation by anaerobic bacteria. The results presented here are useful for establishing a microbial process for the practical production of conjugated fatty acids, especially those produced from α-linolenic acid and γ-linolenic acid, and helpful for investigating the physiological activities of the conjugated fatty acids. Further, exploitation of microbial conjugated fatty acid production and analysis of the enzymes involved in the biohydrogenation pathway are in progress.
References
Ando A, Ogawa J, Kishino S, Shimizu S (2003) CLA production from ricinoleic acid by lactic acid bacteria. J Am Oil Chem Soc 80:889–894
Ando A, Ogawa J, Kishino S, Shimizu S (2004) Conjugated linoleic acid production from castor oil by Lactobacillus plantarum JCM 1551. Enzyme Microb Technol 35:40–45
Bligh EG, Dyer WJ (1959) A rapid method of total lipid extraction and purification. Can J Biochem Physiol 37:911–917
Dawson RM, Kemp P (1969) The effect of defaunation on the phospholipids and on the hydrogenation of unsaturated fatty acids in the rumen. J Biochem 115:351–352
Destaillats F, Trottier JP, Galvez JM, Angers P (2005) Analysis of α-linolenic acid biohydrogenation intermediates in milk fat with emphasis on conjugated linolenic acids. J Dairy Sci 88:3231–3239
Griinari JM, Dwyer DA, McGuire MA, Bauman DE, Palmquist DL, Nurmela KV (1998) Trans-octadecenoic acids and milk fat depression in lactating dairy cows. J Dairy Sci 81:1251–1261
Ha YL, Grimm NK, Pariza MW (1987) Anticarcinogens from fried ground beef: heat-altered derivatives of linoleic acid. Carcinogenesis 8:1881–1887
Ha YL, Storkson J, Pariza MW (1990) Inhibition of benzo(a)pyrene-induced mouse forestomach neoplasia by conjugated dienoic derivatives of linoleic acid. Cancer Res 50:1097–1101
Igarashi M, Miyazawa T (2000) Newly recognized cytotoxic effect of conjugated trienoic fatty acids on cultured human tumor cells. Cancer Lett 148:173–179
Ip C, Chin SF, Scimeca JA, Pariza MW (1991) Mammary cancer prevention by conjugated dienoic derivative of linoleic acid. Cancer Res 51:6118–6124
Jenkins TC, Wallace RJ, Moate PJ, Mosley EE (2008) Recent advances in biohydrogenation of unsaturated fatty acids within the rumen microbial ecosystem. J Anim Sci 86:397–412
Jouany JP, Lassalas B, Doreau M, Glasser F (2007) Dynamic features of the rumen metabolism of linoleic acid, linolenic acid and linseed oil measured in vitro. Lipids 42:351–360
Kepler CR, Tove SB (1967) Biohydrogenation of unsaturated fatty acids. III. Purification and properties of a linoleate Δ12-cis, Δ11-trans-isomerase from Butyrivibrio fibrisolvens. J Biol Chem 242:5686–5692
Kishino S, Ogawa J, Ando A, Omura Y, Shimizu S (2002a) Ricinoleic acid and castor oil as substrates for conjugated linoleic acid production by washed cells of Lactobacillus plantarum. Biosci Biotechnol Biochem 66:2283–2286
Kishino S, Ogawa J, Omura Y, Matsumura K, Shimizu S (2002b) Conjugated linoleic acid production from linoleic acid by lactic acid bacteria. J Am Oil Chem Soc 79:159–163
Kishino S, Ogawa J, Ando A, Iwashita T, Fujita T, Kawashima H, Shimizu S (2003) Structural analysis of conjugated linoleic acid production by Lactobacillus plantarum, and factors affecting isomer production. Biosci Biotechnol Biochem 67:179–182
Lee KN, Kritchevsky D, Pariza MW (1994) Conjugated linoleic acid and atherosclerosis in rabbits. Atherosclerosis 108:19–25
Liu L, Hammond EG, Nikolau BJ (1997) In vivo studies of the biosynthesis of α-eleostearic acid in the seed of Momordica charantia. L Plant Physiol 113:1343–1349
Ogawa J, Matsumura K, Kishino S, Omura Y, Shimizu S (2001) Conjugated linoleic acid accumulation via 10-hydroxy-12-octadecaenoic acid during microaerobic transformation of linoleic acid by Lactobacillus acidophilus. Appl Environ Microbiol 67:1246–1252
Ostrowska E, Dunshea FR, Muralitharan M, Cross RF (2000) Comparison of silver-ion high-performance liquid chromatographic quantification of free and methylated conjugated linoleic acids. Lipids 35:1147–1153
Pariza MW, Ha YL (1990) Newly recognized anticarcinogenic fatty acids. In: Kuroda Y, Shankel D, Waters MD (eds) Antimutagenesis and anticarcinogenesis mechanism II. Plenum, New York, pp 167–170
Park Y, Albright KJ, Liu W, Storkson JM, Cook ME, Pariza MW (1997) Effect of conjugated linoleic acid on body composition in mice. Lipids 32:853–858
Plourde M, Destaillats F, Chouinard PY, Angers P (2007) Conjugated α-linolenic acid isomers in bovine milk and muscle. J Dairy Sci 90:5269–5275
Scollan ND, Choi NJ, Kurt E, Fisher AV, Enser M, Wood JD (2001) Manipulating the fatty acid composition of muscle and adipose tissue in beef cattle. Br J Nutr 85:115–124
Suzuki R, Noguchi R, Ota T, Abe M, Miyashita K, Kawada T (2001) Cytotoxic effect of conjugated trienoic fatty acids on mouse tumor and human monocytic leukemia cells. Lipids 36:477–482
Wasowska I, Maia MR, Niedzwiedzka KM, Czauderna M, Ribeiro JM, Devillard E, Shingfield KJ, Wallace RJ (2006) Influence of fish oil on ruminal biohydrogenation of C18 unsaturated fatty acids. Br J Nutr 95:1199–1211
Acknowledgment
This work was partially supported by the Industrial Technology Research Grant Program in 2007 (no. 07A08005a to S.K.) and the Project for Development of a Technological Infrastructure for Industrial Bioprocesses on R&D of New Industrial Science and Technology Frontiers (to S.S.) from New Energy and Industrial Technology Development Organization (NEDO) of Japan, Grants-in-Aid for Scientific Research (no. 19780056 to S.K., no. 16688004 to J.O., and no. 18208009 to S.S.) and COE for Microbial-Process Development Pioneering Future Production Systems from the Ministry of Education, Culture, Sports, Science and Technology, Japan. S.K. was a recipient of a Research Fellowship (no. 01985) from the Japan Society for the Promotion of Science for Yong Scientists.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Fig. S1
1H-NMR analysis of A1, and the structure of A1 identified (GIF 345 kb)
Fig. S1
1H-NMR analysis of A1, and the structure of A1 identified (GIF 345 kb)
Fig. S3
1H-NMR analysis of CGLA2, and the structure of CGLA2 identified (GIF 368 kb)
Fig. S4
1H-NMR analysis of G2, and the structure of G2 identified (GIF 433 kb)
Fig. S5
1H-NMR analysis of G1, and the structure of G1 identified (GIF 356 kb)
Rights and permissions
About this article
Cite this article
Kishino, S., Ogawa, J., Yokozeki, K. et al. Metabolic diversity in biohydrogenation of polyunsaturated fatty acids by lactic acid bacteria involving conjugated fatty acid production. Appl Microbiol Biotechnol 84, 87–97 (2009). https://doi.org/10.1007/s00253-009-1949-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-009-1949-0