Abstract
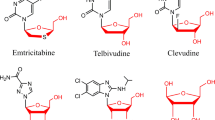
l-Ribose is a rare and expensive sugar that can be used as a precursor for the production of l-nucleoside analogues, which are used as antiviral drugs. In this work, we describe a novel way of producing l-ribose from the readily available raw material l-arabinose. This was achieved by introducing l-ribose isomerase activity into l-ribulokinase-deficient Escherichia coli UP1110 and Lactobacillus plantarum BPT197 strains. The process for l-ribose production by resting cells was investigated. The initial l-ribose production rates at 39°C and pH 8 were 0.46 ± 0.01 g g−1 h−1 (1.84 ± 0.03 g l−1 h−1) and 0.27 ± 0.01 g g−1 h−1 (1.91 ± 0.1 g l−1 h−1) for E. coli and for L. plantarum, respectively. Conversions were around 20% at their highest in the experiments. Also partially purified protein precipitates having both l-arabinose isomerase and l-ribose isomerase activity were successfully used for converting l-arabinose to l-ribose.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The l-enantiomers of nucleoside analogues have been widely used as antiviral drugs in treatments of severe viral diseases, such as those caused by the HIV or hepatitis virus (Wang et al. 1998). Since l-ribose can be used as a precursor for the synthesis of most of these compounds (Yun et al. 2005), there has been a growing interest in its production (Seo et al. 2003). Methods for the production of l-ribose by chemical synthesis from various sugar substrates have been described (Jung and Xu 1997; Seo et al. 2003; Shi et al. 2001; Takahashi et al. 2002). These methods are, however, not well suited for industrial production, since they include the use of expensive reagents, multiple steps, and/or several organic solvents, and the yields are in many cases low.
Biotechnological production of l-ribose has focused on converting l-ribulose to l-ribose. An l-ribose isomerase from Acinetobacter sp. DL-28 has been used for this purpose and the corresponding gene sequenced (Mizanur et al. 2001; Shimonishi and Izumori 1996). The use of toluene permeabilized cells of Acinetobacter sp. DL-28 for l-ribose production has also been described (Ahmed et al. 1999). A novel enzyme, d-lyxose isomerase, which also isomerizes l-ribulose to l-ribose as a side activity, has been isolated and sequenced recently (Cho et al. 2007). Furthermore, an isomerase having l-ribose isomerizing activity has been created by directed evolution. This was achieved by random mutagenesis of Escherichia colil-arabinose isomerase gene (De Muynck et al. 2007).
The production of the precursor of l-ribose, the rare sugar l-ribulose, has been studied mostly using acetic acid bacteria for the dehydrogenation of ribitol to l-ribulose (Ahmed et al. 1999; De Myunck et al. 2006; Kylmä et al. 2004). The enzyme catalyzing the oxidation of ribitol to l-ribulose in these bacteria has been shown to be a membrane-bound NAD(P)-independent dehydrogenase (Adachi et al. 2001). Unfortunately, ribitol is presently very expensive, and it is not available from natural sources in any significant amounts.
The l-form of ribulose is an intermediate in the pathway for l-arabinose utilization in many bacteria. The l-arabinose taken up by the cells is first isomerized to l-ribulose in an l-arabinose isomerase catalyzed reaction. The l-ribulose is then phosphorylated to l-ribulose-5-phosphate by an l-ribulokinase. l-Ribulokinase-deficient mutants of E. coli and Lactobacillus plantarum have been previously constructed by chemical mutagenesis (Englesberg 1961) and by targeted mutagenesis (Helanto et al. 2007), respectively. Unlike other l-sugars, l-arabinose is abundant in nature. It is a common component of polymers of lignocellulosic materials (Hayn et al. 1993). For example, sugar beet pulp, which is a by-product of the sugar industry, has been reported to contain significant amounts of l-arabinose (20% w w−1 of the deproteinated mass), which can be easily isolated from the pulp (Spagnuolo et al. 1999).
In the current work, we describe a novel way of producing l-ribose from the readily available raw material l-arabinose using metabolically engineered bacterial cells. For this purpose, we have introduced an l-ribose isomerase into l-ribulokinase-deficient mutants of E. coli and L. plantarum. We show how resting cells of these mutants can be used for the production of l-ribose from l-arabinose. We also describe the use of protein precipitates for converting l-arabinose to l-ribose.
Materials and methods
Bacterial strains and growth conditions
E. coli UP1110 and BPT234 strains were cultivated aerobically in Luria–Bertani (LB) medium (Pronadisa) at 37°C. Lactococcus lactis NZ9000 obtained from NIZO Laboratories (The Netherlands; Kuipers et al. 1998) was grown at 30°C in M17 medium (Difco) containing 5 g l−1 glucose. L. plantarum BPT197 and BPT232 strains were cultivated at 30°C in standard MRS growth medium (Lab M Limited) or in simplified MRS medium (Helanto et al. 2007) in the bioreactor cultivations. Ampicillin (100 mg l−1) and erythromycin (5 mg l−1) were used for selecting E. coli and L. plantarum transformants, respectively.
Plasmid constructions
A synthetic l-ribose isomerase gene (GenBank accession no. AB062121) purchased from GenScript Corporation (New Jersey, USA) was used as a template for the amplification of the l-ribose isomerase gene with the insertion of an NcoI site at the 5′ end and an XhoI site at the 3′ end. The primers used were l-RIFNcoIgly, 5′-ATA CCA TGG GTA CAA GGA CGT CGA TTA CTC GT-3′, and l-RIRXhoI, 5′-ATA CTC GAG CTA GCT GAT CGC GGT CTG AA-3′. The restriction sites are shown underlined. l-Ribose isomerase gene was cloned into NcoI and XhoI restriction sites of the pTrcHis 2B expression vector (Invitrogen), which then was transformed into E. coli UP1110 as described previously (Hanahan et al. 1991).
l-Ribose isomerase gene was cloned into an NcoI and XhoI restriction sites of the pSIP401 expression vector under the control of an SPPIP peptide inducible PsppA promoter (Sorvig et al. 2005). The resulting plasmid was transformed into L. lactis NZ9000 by electroporation (Holo and Nes 1989). Plasmid DNA was isolated from the positive transformants and transformed into L. plantarum BPT197 by electroporation as described previously (Aarnikunnas et al. 2003).
Expression of the l-ribose isomerase gene
E. coli BPT234 and L. plantarum BPT232 cells were grown overnight in standard LB medium and in standard MRS medium, respectively. Ten milliliters of the overnight cultures were used to inoculate 100 ml of LB medium supplemented with 50 g l−1 of l-arabinose or 50 ml of MRS medium containing 50 g l−1l-arabinose and 5 g l−1 glucose. Expression of the l-ribose isomerase gene in E. coli BPT234 was induced after 1 h by adding isopropyl-β-d-thiogalactopyranoside (IPTG) to the final concentration of 1 mM. Expression of the l-ribose isomerase gene in L. plantarum BPT 232 was induced after 1.5 h with 0 to 200 µg l−1 SPPIP (Innovagen AB, Lund, Sweden; Ejsink et al. 1996). E. coli BPT234 and L. plantarum BPT234 cells were grown for additional 3 and 2.5 h, respectively, and harvested by centrifugation at 5,000×g for 15 min.
Bioreactor cultivations
All bioreactor cultivations were carried out in a Biostat MD reactor (total volume, 2 l; B. Braun Biotech International). An overnight culture (200 ml) of E. coli BPT234 was used to inoculate 1.5 l of LB medium. The culture was aerated at a constant rate of 0.5 vvm. Dissolved oxygen was controlled at 30% by varying the stirring rate (minimum, 500 rpm). After 1 h of cultivation, the expression of the l-arabinose isomerase and l-ribose isomerase genes were induced by the addition of l-arabinose and IPTG to the final concentrations of 5 g l−1 and 1 mM, respectively. The cells were cultivated for additional 3 h after the induction.
A culture of L. plantarum BPT232 at late exponential growth phase (50 ml) was used to inoculate 2 l of simplified MRS medium supplemented with 5 g l−1l-arabinose. The cells were cultivated at a stirring rate of 200 rpm. pH was controlled at a minimum of 6.2 with 3 M NaOH. The expression of the l-ribose isomerase gene was induced by the addition of the peptide SPPIP to the final concentration of 100 µg l−1 after 3 h, and the cells were cultivated for further 5 h. Cell dry weights were determined in triplicate by centrifuging, washing, and drying the cells at 80°C from 4 ml samples taken from the cultivations.
l-Ribose production by resting cells
The E. coli BPT234 and L. plantarum BPT232 cells were cultivated in a bioreactor as described above, harvested by centrifugation at 5,000×g for 15 min, washed with saline, and suspended into saline at a volume of one tenth of the culture volume. The cell suspensions were distributed in 12-ml aliquots and 18 ml of production buffer was added to the aliquots, which resulted in final concentrations of 50 mM Tris–3-(N-morpholino) propanesulfonic acid (MOPS) (pH 8), 50 g l−1l-arabinose, and 2.5 mM MnCl2. E. coli BPT234 cells were incubated at 39°C for 24 h in 250-ml flasks with shaking at 200 rpm (amplitude, 2.5 cm). The L. plantarum BPT232 cells were incubated stationarily at 39°C for 22 h. The incubations were performed as triplicates. l-Arabinose, l-ribose, and l-ribulose concentrations were determined from the cell suspensions by high-performance liquid chromatography (HPLC) as described below.
Repetitive batch experiments with L. plantarum BPT232 cells
In order to test the reusability of L. plantarum BPT232 cells for l-ribose production, the cells from three individual experiments were collected by centrifugation at 4,500×g for 15 min and washed with saline. Cells were resuspended into the reaction medium as described above. After a 22-h incubation at 39°C, l-arabinose, l-ribose, and l-ribulose concentrations were determined from the cell suspensions by HPLC as described below.
Preparation of PEG6000 protein fractions for l-ribose production
L. plantarum BPT232 cells were cultivated in a bioreactor, harvested and suspended into buffer as described above. Dithiothreitol and MnCl2 were added to the cell suspension to the final concentrations of 1 and 10 mM, respectively, and the cells were disrupted by sonication. The cell lysate was centrifuged at 20,000×g for 20 min at 4°C, and the supernatants were collected. Polyethylenglycol (PEG6000) was added slowly to the stirred cell lysate to a final concentration of 5% (w/v). After an overnight precipitation at 4°C, the suspension was centrifuged (5,000×g, 20 min), and the precipitate and the supernatant were collected. The PEG concentration of the supernatant was subsequently increased to 15% (w/v) and finally to 20% (w/v). After each PEG additions, the precipitate and supernatant were collected. All three precipitate aliquots were suspended in 30 ml of buffer containing 50 mM Tris–HCl (pH 7.5) and 20% (w/v) PEG. The conversion studies were carried out in 50-ml test tubes in a buffer containing 50 mM Tris–MOPS (pH 7.5), 15% PEG (w/v) and 2.5 mM MnCl2 at 36°C with shaking.
Enzyme activity assays
Cell extracts were prepared by sonication and the l-arabinose isomerase activities determined at 30°C from the cell extracts as described previously (Helanto et al. 2007). The reaction mixture for l-ribose isomerase contained 50 mM Tris–glycine (pH 9.0), 10 mM MnCl2, 100 mM l-ribose, and cell extract in a volume of 950 μl. After incubation at 30°C, the reactions were stopped after 15 min by adding 50 µl of 0.5 M H2SO4. l-Ribulose and l-ribose concentrations were determined from the reaction mixtures by HPLC as described below. One unit of activity was defined as the amount of l-arabinose isomerase or l-ribose isomerase catalyzing the formation of 1 µmol min−1 of l-ribulose. Protein concentrations were determined using the Qubit™ fluorometer (Invitrogen) according to the instructions by the manufacturer.
HPLC analysis
l-Arabinose, l-ribose, and l-ribulose concentrations were determined using an Aminex HPX-87P column (Bio-Rad) at 70°C with distilled water as the mobile phase at an elution rate of 0.6 ml min−1. All components were analyzed with a refractive index detector.
Results
Expression of the l-ribose isomerase gene
To construct strains for l-ribose production, the l-ribose isomerase gene of Acinetobacter sp. Dl-28 was cloned into the ribulokinase-deficient strains E. coli UP1110 and L. plantarum BPT197. The growth medium of E. coli UP1110 was supplemented with l-arabinose to induce the expression of the endogenous l-arabinose isomerase gene of the strain. l-Ribose isomerase gene expression was induced by the addition of IPTG. The specific l-ribose isomerase activity determined from the cell extracts of E. coli UP1110 was 103 ± 1 U g−1 protein 3 h after the onset of the IPTG induction. The specific l-arabinose isomerase activity determined from the cell extracts was 610 ± 6 U g−1 protein at this point. Only negligible l-arabinose isomerase and l-ribose isomerase activity could be detected in non-induced E. coli cells.
The heterogenous expression of the l-ribose isomerase gene in L. plantarum BPT232 was studied using the peptide SPPIP as an inducer at concentrations ranging from 0 to 200 μg l−1. A specific l-ribose isomerase activity of 183 ± 28 U g−1 protein was determined from the cell extracts 1 h after the addition of 25 μg l−1 SPPIP, whereas no activity could be detected in the non-induced cells. The specific l-ribose isomerase activity reached a plateau above the SPPIP concentration of 25 μg l−1 (data not shown). At 100 µg l−1 SPPIP, the specific l-ribose isomerase activity determined was 155 ± 25 U g−1 protein, and this concentration was chosen for further studies. No l-arabinose isomerase activity was detected in the L. plantarum BPT232 cells during the expression experiment, most likely because the glucose concentration was above the repression limit (Helanto et al. 2007).
Production of l-ribose using resting cells
The production conditions used were chosen using the Modde 5.0 software (Umetrics; data not shown) as described earlier (Helanto et al. 2007). The cell concentration after the bioreactor cultivation was 0.99 ± 0.02 g l−1 for E. coli and 1.73 ± 0.05 g l−1 for L. plantarum. The cells were harvested as described in “Materials and methods” and resuspended in production buffer resulting in cell concentrations of 3.99 ± 0.06 and 6.91 ± 0.2 g l−1 for E. coli and L. plantarum, respectively. The process for l-ribose production by resting cells was investigated. The results are shown in Fig. 1. The initial l-ribose production rates (r i) determined between 0 and 3 h at 39°C and pH 8 were 0.46 ± 0.01 g g−1 h−1 (1.84 ± 0.03 g l−1 h−1) and 0.27 ± 0.01 g g−1 h−1 (1.91 ± 0.1 g l−1 h−1) for E. coli and for L. plantarum, respectively. Conversions of l-arabinose to l-ribose (x) were 19.7 ± 0.1% (mol mol−1) and 20 ± 1% (mol mol−1) for E. coli and for L. plantarum, respectively.
Repetitive batch experiments with L. plantarum BPT232
The reusability of L. plantarum BPT232 cells for l-ribose production was studied at 39°C and at pH 8.0 using three parallel samples of the first bioconversion cycle. The cells were washed and used for another batch under the same conditions. An r i of 0.22 ± 0.01 g g−1 h−1 (1.54 ± 0.02 g l−1 h−1) and an x of 21 ± 0.1% (mol mol−1) were achieved in this second production cycle. The l-ribose isomerase and l-arabinose isomerase activities determined from the cell lysates were 34% lower and 12% higher at the end of the second cycle than at the end of the first cycle, respectively. The results suggest that the cells can be used for several successive batches, even without addition of nutrients to the media.
Production of l-ribose by PEG6000 protein precipitates
It has previously been reported that the l-ribose isomerase of Acinetobacter sp. DL-28 is stable only for 10 min at 30°C (Shimonishi and Izumori 1996). In our studies, we were able to increase the stability of the enzyme by preparing a cell lysate and precipitating it with PEG. The l-ribose and l-arabinose isomerases containing cell extract was fractionated by PEG precipitation at three different concentrations. The protein precipitates with the highest activity were pooled and used for catalyzing the direct conversion of l-arabinose to l-ribose. The production of l-ribose and l-ribulose from l-arabinose by the protein precipitates is shown in Fig. 2. The results indicate that l-arabinose and l-ribose isomerase enzymes were both active in the PEG precipitate. The equilibrium between l-arabinose, l-ribulose, and l-ribose was not completely reached in 20-h experiments, but after 4 h, the reaction rates decreased significantly and continuation of the reaction was not reasonable. The conversions of l-arabinose to l-ribose were around 24% (mol mol−1), which corresponds to the level achieved by the resting cells. The production rates between 0 and 3 h were 18.1 ± 1.1 g g protein−1 h−1 (2.09 ± 0.13 g l−1 h−1) and 34.8 ± 4.4 g g protein−1 h−1 (2.71 ± 0.03 g l−1 h−1), for E. coli and Lb. plantarum, respectively.
Discussion
In this work, we have studied the possibility of developing a new and efficient process for l-ribose production from l-arabinose by metabolic engineering. For this purpose, we have introduced l-ribose isomerizing activity into l-ribulokinase-deficient mutants of E. coli and L. plantarum, which have an endogenous l-arabinose isomerase gene. The results indicate that the l-arabinose isomerase and l-ribose isomerase enzymes required for converting l-arabinose to l-ribose can be produced in active form in both strains
Resting cells have been widely used for the production of rare sugars and sugar alcohols (De Myunck et al. 2007; Doten and Mortlock 1985; Helanto et al. 2007; Nyyssölä et al. 2005). The advantages of this method include the simple purification of the product, since no major by-products are formed, and complex media components are omitted during the production phase. As shown in the present study, l-arabinose can be converted to l-ribose by resting cells in the two-step isomerization reaction with l-ribulose as the intermediate.
The initial l-ribose production rates were of the same order of magnitude: 0.46 and 0.27 g g−1 h−1 for E. coli and L. plantarum, respectively. Although the results suggest that E. coli BPT234 would produce l-ribose at a somewhat higher rate, it should be taken into account that the choice of production strain cannot be based only on the comparison of productivities. Concentration of the intermediate product, l-ribulose, was lower in the experiments with L. plantarum (Figs. 1 and 2), which makes the purifying process easier. If the l-ribose is produced for medical purposes, the L. plantarum process may be more favorable, as E. coli is known to produce endotoxins. Production of L. plantarum cell mass is simpler, since no aeration is required. Because of these considerations and because the productivity with the L. plantarum strain was comparable to the productivity with the E. coli strain, we chose L. plantarum BPT232 for the cell recycling experiments.
Cultivating the L. plantarum cells for production can be costly because of the price of the complex medium components. Therefore, an important consideration in using resting cells of these bacteria is the recyclability of the cells. As shown in the present study, the L. plantarum BPT232 cells could be used in two sequential batches with a 19% loss of productivity. However, further improvement in the viability of the cells could most likely be achieved by adding minor amounts of nutrients to the bioconversion medium.
An alternative for utilizing resting cells for l-ribose production is the use of enzyme preparates containing the two isomerases. It has previously been reported that the l-ribose isomerase of Acinetobacter sp. DL-28 is stable only for 10 min at 30°C, which makes its use unfeasible for industrial processes (Shimonishi and Izumori 1996). In our studies, we were able to increase the stability of the enzyme by preparing a cell lysate and precipitating it with PEG. The l-ribose isomerase was active in the precipitated protein aggregates at least for 24 h (results not shown). The precipitated protein aggregates were cross-linked using glutaraldehyde, and the experiment was repeated. l-ribose production was detected, but cross-linking decreased the conversion rate considerably (results not shown).
l-Arabinose isomerase activity of the cell lysates was increased substantially when l-ribose isomerase was active in the reaction mixture (results not shown). At the equilibrium of the isomerization reaction, the ratio of l-arabinose to l-ribulose has previously been determined to be 90:10 (Heath et al. 1958) and the ratio of l-ribulose to l-ribose 30:70 (Shimonishi and Izumori 1996). This would suggest that in a reaction mixture containing the substrate and the two isomerases, the maximum l-ribose yield would not exceed 26% (mol/mol). However, with whole cells, the situation is more complicated, since the reaction takes place in the cytoplasm and the produced l-ribose is excreted and/or transported into the medium. It may therefore be possible to reach even higher yields using whole cells. Be that as it may, it has been shown previously that l-ribose can be easily and efficiently separated from l-arabinose and l-ribulose by ion exclusion chromatography (Jumppanen et al. 2000). This would enable the recycling of the reagents back to the bioconversion.
The production of l-ribose from l-ribulose with a conversion of 70% using toluene-permeabilized cells of Acinetobacter sp. DL-28 has been described previously (Ahmed et al. 1999). The precursor, l-ribulose, can be produced by the quantitative dehydrogenation of ribitol by resting cells of acetic acid bacteria (Ahmed et al. 1999; De Myunck et al. 2006, Kylmä et al. 2004). Woodyer et al. (2008) have reported a one-step conversion of ribitol to l-ribose with a production rate of 0.73 g l−1 h−1 and a conversion of over 70% using E. coli strain expressing a mannitol-1-dehydrogenase from Apium graveolens. A problem with using ribitol as the raw material is that it is presently very expensive and that it cannot be isolated from any natural sources. The production of ribitol from glucose by fermentation using the fungus Trichosporonoides megachillensis has been reported (Kawaguchi et al. 2001). However, the yield of ribitol from glucose is low in this process (less than 30%), since a large fraction of the glucose is lost to other polyols.
The results of the present study suggest that biotechnological l-ribose production from l-arabinose by resting cells or by protein precipitates containing l-arabinose and l-ribose isomerase activities holds promise of becoming an alternative for chemical l-ribose production from l-arabinose. Unlike in the chemical process, also lower purity grade l-arabinose or even crude or fractionated plant material can be utilized as a raw material in the current bioprocess. However, the production conditions should be studied further and a downstream process developed in order to fully evaluate the feasibility of the current approach.
References
Aarnikunnas J, von Weymarn N, Rönnholm K, Leisola M, Palva A (2003) Metabolic engineering of Lactobacillus fermentum for production of mannitol and pure L-lactic acid and pyruvate. Biotechnol Bioeng 82:653–663
Adachi O, Fujii Y, Ano Y, Moonmangmee D, Toyama H, Shinagawa E, Theeragool G, Lotong N, Matsushita K (2001) Membrane-bound sugar alcohol dehydrogenase in acetic acid bacteria catalyzes L-ribulose formation and NAD-dependent ribitol dehydrogenase is independent of the oxidative fermentation. Biosci Biotechnol Biochem 65:115–125
Ahmed Z, Shimonishi T, Bhuiyan SH, Utamura M, Takada G, Izumori K (1999) Biochemical preparation of L-ribose and L-arabinose from ribitol: a new approach. J Biosci Bioeng 88:444–448
Cho EA, Lee DW, Cha YH, Lee SH, Jung HC, Pan JG, Pyun YR (2007) Characterization of a novel D-lyxose isomerase from Cohnella laevoribosii RI-39 sp. nov. J Bacteriol 189:1655–1663
De Muynck C, Pereira C, Soetaert W, Vandamme E (2006) Dehydrogenation of ribitol with Gluconobacter oxydans: production and stability of L-ribulose. J Biotechnol 125:408–415
De Muynck C, Van der Borght J, De Mey M, De Maeseneire SL, Van Bogaert IN, Beauprez J, Soetaert W, Vandamme E (2007) Development of a selection system for the detection of L-ribose isomerase expressing mutants of Escherichia coli. Appl Microbiol Biotechnol 76:1051–1057
Doten RC, Mortlock RP (1985) Production of D- and L-xylulose by mutants of Klebsiella pneumoniae and Erwinia uredovora. Appl Environ Microbiol 49:158–162
Eijsink VG, Brurberg MB, Middelhoven PH, Nes IF (1996) Induction of bacteriocin production in Lactobacillus sake by a secreted peptide. J Bacteriol 178:2232–2237
Englesberg E (1961) Enzymatic characterization of 17 L-arabinose negative mutants of Escherichia coli. J Bacteriol 81:996–1006
Hanahan D, Jessee J, Bloom FR (1991) Plasmid transformation of Escherichia coli and other bacteria. Methods Enzymol 204:63–113
Hayn M, Steiner W, Klinger R, Steinmüller H, Sinner M, Esterbauer H (1993) Basic research and pilot studies on the enzymatic conversion of lignocellulosics. In: Saddler JN (ed) Bioconversion of forest and agricultural plant residues. CAB International, Wallingford, UK, pp 33–72
Heath EC, Horecker BL, Smyrniotis PZ, Takagi Y (1958) Pentose fermentation by Lactobacillus plantarum. II. L-arabinose isomerase. J Biol Chem 231:1031–1037
Helanto M, Kiviharju K, Leisola M, Nyyssölä A (2007) Metabolic engineering of Lactobacillus plantarum for production of L-ribulose. Appl Environ Microbiol 73:7083–7091
Holo H, Nes IF (1989) High-frequency transformation by electroporation of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl Environ Microbiol 55:3119–3123
Jumppanen J, Nurmi J, Pastinen O (2000) High purity L-ribose from L-arabinose. WO0029417
Jung ME, Xu Y (1997) Efficient synthesis of L-ribose and 2-deoxy L-ribose from D-ribose and L-arabinose. Tetrahedron Lett 38:4199–4202
Kawaguchi T, Hara M, Ueda M (2001) Process for producing L-ribose. EP1083234
Kuipers OP, de Ruyter PG, Kleerebezen M, de Vos WM (1998) Quorum sensing controlled gene expression in lactic acid bacteria. J Biotechnol 64:15–21
Kylmä AK, Granström T, Leisola M (2004) Growth characteristics and oxidative capacity of Acetobacter aceti IFO 3281: implications for L-ribulose production. Appl Microbiol Biotechnol 63:584–591
Mizanur RM, Takada G, Izumori K (2001) Cloning and characterization of a novel gene encoding L-ribose isomerase from Acinetobacter sp. strain DL-28 in Escherichia coli. Biochim Biophys Acta 1521:141–145
Nyyssölä A, Pihlajaniemi A, Palva A, von Weymarn N, Leisola M (2005) Production of xylitol from D-xylose by recominant Lactococcus lactis. J Biotechnol 118:55–66
Seo M, An J, Shim JH, Kim G (2003) One-pot inversion of D-mannono-1,4-lactone for the practical synthesis of L-ribose. Tetrahedron Lett 44:3051–3052
Shi Z-D, Yang B-H, Wu Y-L (2001) A stereospecific synthesis of L-ribose and L-ribosides from D-galactose. Tetrahedron Lett 42:7651–7653
Shimonishi T, Izumori K (1996) A new enzyme, L-ribose isomerase from Acinetobacter sp. strain DL-28. J Ferment Bioeng 81:493–497
Sorvig E, Mathiesen G, Naterstad K, Eijsink VGH, Axelsson L (2005) High-level, inducible gene expression in Lactobacillus sakei and Lactobacillus plantarum using versatile expression vectors. Microbiology 151:2439–2449
Spagnuolo M, Crecchio C, Pizzigallo MDR, Ruggiero P (1999) Fractionation of sugar beet pulp into pectin, cellulose, and arabinose by arabinases combined with ultrafiltration. Biotechnol Bioeng 64:685–691
Takahashi H, Iwai Y, Hitomi Y, Igegami S (2002) Novel synthesis of L-ribose from D-mannono-1,4-lactone. Organic Lett 4:2401–2403
Wang P, Hong JH, Cooperwood JS, Chu CK (1998) Recent advances in L-nucleosides: chemistry and biology. Antiviral Res 40:19–44
Woodyer RD, Wymer NJ, Racine M, Khan SN, Saha BC (2008) Efficient production of L-ribose with a recombinant Escherichia coli biocatalyst. Appl Environ Microbiol 74:2967–2975
Yun M, Moon HR, Kim HO, Choi WJ, Kim Y-C, Park C-S, Jeong LS (2005) A highly efficient synthesis of unnatural L-sugars from D-ribose. Tetrahedron Lett 46:5903–5905
Acknowledgments
The authors thank Dr. Ossi Pastinen for excellent advice. Kalle Salonen, Marjaana Rytelä, and Auli Murrola are acknowledged for technical support. The research was funded by the Academy of Finland (210778).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Helanto, M., Kiviharju, K., Granström, T. et al. Biotechnological production of l-ribose from l-arabinose. Appl Microbiol Biotechnol 83, 77–83 (2009). https://doi.org/10.1007/s00253-008-1855-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-008-1855-x