Abstract
The extremely thermophilic anaerobic archaeon strain, HJ21, was isolated from a deep-sea hydrothermal vent, could produce hyperthermophilic α-amylase, and later was identified as Thermococcus from morphological, biochemical, and physiological characteristics and the 16S ribosomal RNA gene sequence. The extracellular thermostable α-amylase produced by strain HJ21 exhibited maximal activity at pH 5.0. The enzyme was stable in a broad pH range from pH 5.0 to 9.0. The optimal temperature of α-amylase was observed at 95°C. The half-life of the enzyme was 5 h at 90°C. Over 40% and 30% of the enzyme activity remained after incubation at 100°C for 2 and 3 h, respectively. The enzyme did not require Ca2+ for thermostability. This α-amylase gene was cloned, and its nucleotide sequence displayed an open reading frame of 1,374 bp, which encodes a protein of 457 amino acids. Analysis of the deduced amino acid sequence revealed that four homologous regions common in amylases were conserved in the HJ21 α-amylase. The molecular weight of the mature enzyme was calculated to be 51.4 kDa, which correlated well with the size of the purified enzyme as shown by the sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
α-Amylase(EC 3.2.1.1), endo-acting enzyme that hydrolyzes starch by cleaving α-1,4-glucosidic linkages at random sites, is one of the most important commercial enzymes widely used in starch-processing, brewing, alcohol production, textile, and other industries. The most thermostable α-amylase in industry is produced from Bacillus licheniformis (Violet and Meunier 1989). This enzyme operates optimally at 90°C and pH 6, and it requires addition of calcium ion (Ca2+) for its thermostability (Violet and Meunier 1989).
Starch bioprocessing usually involves two steps, liquefaction and saccharification, both of which run at high temperatures. B. licheniformis α-amylase is typically used during the liquefaction step where the semi-purified starch is digested to glucose oligomers. The ideal condition for this step is pH 4.5 and 105°C (Richardson et al. 2002). However, since the enzyme is unstable under these conditions, pH of starch slurry needs to be raised from 4.5–5.0 to 5.7–6.0, and calcium salt is added. In the saccharification step, the liquefied product is converted to glucose using a glucoamylase isolated from an Aspergillus sp. Because the glucoamylase has optimum activity at pH 4.2–4.5, pH of the slurry must be returned to about 4.5 for this step to proceed efficiently. These two pH adjustments not only increase chemical costs but also create additional steps for refining since the added salt in the final product has to be removed via ion exchange. An α-amylase that is able to work at pH 4.5 and 105°C without the addition of calcium would greatly reduce costs, simplify the process, and minimize the formation of high-pH by-product such as isomaltitol (Richardson et al. 2002; Vieille and Zeikus 2001).
Hyperthermophiles that optimally grow at temperatures above 80°C have attracted many researchers’ attention, as they are a source of enzymes with outstanding thermostability (Atomi 2005). The most thermoactive α-amylases have been characterized from the hyperthermophilic archaea, Pyrococcus woesei, Pyrococcus furiosus, Thermococcus profundus and Thermococcus hydrothermalis, and Thermotoga maritima (Vieille and Zeikus 2001). Their optimal activities range from 80 to 100°C at pH 4.0 to 7.5. An ideal catalyst for starch liquefaction should be optimally active at 100°C and pH 4.0 to 5.0 and should not require added Ca2+ for stability. None of the enzymes extracted from the above archaea meet these combined criteria (Vieille and Zeikus 2001).
In this paper, we report that the identification of the strain HJ21, which was isolated from a deep-sea hydrothermal vent and characterization of the extremely thermostable α-amylase by this strain.
Materials and methods
Strain and growth medium
Strain HJ21, isolated from a deep-sea hydrothermal vent was deposited in China Center for Type Culture Collection (accession no. CCTCCM207010). A modified YPS medium (Jolivet et al. 2004), in which 35 g sea salt per liter medium was replaced with 10 ml trace mineral solution, was used for culturing the archaea. The mineral solution contained 0.01 mg/ml CuSO4·5H2O, 0.1 mg/ml ZnSO4·7H2O, 0.005 mg/ml CoCl2·6H2O, 0.2 mg/ml MnCl2·4H2O, 0.1 mg/ml Na2MoO4·2H2O, 0.05 mg/ml KBr, 0.05 mg/ml KI, 0.1 mg/ml H3BO3, 0.05 mg/ml NaF, 0,05 mg/ml LiCl, 0.05 g Al2(SO4)3, 0.01 mg/ml NiCl2·6H2O, 0.005 mg/ml VOSO4·2H2O, 0.002 mg/ml H2WO4·2H2O, 0.005 mg/ml Na2SeO4, 0.005 mg/ml SrCl·6H2O, and 0.005 mg/ml BaCl2. pH was adjusted to 6.5 with 1 N NaOH before autoclaving. After addition of the medium, headspaces of sealed bottles were flushed with N2. Final anaerobiosis was achieved by adding sterile Na2S·9H2O 5% (w/v) to a final concentration of 0.025%.
Determination of growth conditions
To determine the optimal growth temperature, cells were cultivated in different temperatures from 40 to 100°C. The optimal pH and NaCl concentration for growth were determined at 88°C. To determine growth rates at different pHs, growth media was modified by addition of the following buffers (purchased from agency company of Sigma in China, Shanghai), each at a concentration of 10 mM: pH 3.5–4.5, citric acid buffer; pH 5.0–6.0, 2-(N-morpholino)ethanesulfonic acid buffer; pH 6.5–7.0, piperazine-1,4-bis(2-ethanesulfonic acid) buffer; pH 7.5–8.5, 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid buffer; and pH 9.0–10.0, Tris–Glycine buffer. To determine the optimal NaCl concentration, cells were cultured in growth media containing 0–10% NaCl. Samples were collected every 2 to 3 h. All experiments were performed in duplicate.
Various energy and carbon sources were added to a modified growth medium, in which the yeast extract and peptone were replaced by 0.43 mg/ml ammonium chloride and a filter-sterilized solution of vitamins (10 ml/l). These carbon and energy sources included the following: starch, maltose, glucose, sucrose, cellobiose, lactose, glycogen, xylose, gelatin, and cellulose [final concentration of 0.5% (w/v)] and yeast extract, peptone, tryptone, casein, casamino acid, succinate, pyruvate, acetate, citrate, lactate, ethanol, methanol, and glycerol [final concentration of 0.2% (w/v)].
The requirement for elemental sulfur was studied by comparing growth yields in standard medium vs. medium without elemental sulfur. The possibility for HJ21 to utilize alternative electron acceptors was studied by replacing the sulfate-containing compounds with sodium thiosulfate (10 mM), sodium sulfate (20 mM), or sodium sulfite (3 mM). Growth was determined by direct cell counts in a hemocytometer (Thoma, Hawksley, UK) with a phase-contrast microscope (Olympus, China) after incubation at 88°C.
Genomic DNA extraction
Cells were cultured in 1 l growth medium at 88°C and harvested at the end of the exponential phase of growth. Genomic DNA was isolated with Takara DNA extraction Kit (Takara Mirus Bio, Dalian, China).
16S rRNA gene amplification, sequencing, and phylogenetic analysis
The 16S ribosomal RNA (rRNA) gene was amplified by polymerase chain reaction from purified genomic DNA. The archaea-specific primers (forward primer, 5′TCCGGTTGATCCTGCCGG3′; reverse primer 5′CGGCTACCTTGTTACGAC3′) were used. Gene sequence was determined using the dideoxy chain termination method. Sequences were compared with other 16S rRNAs obtained from GenBank using the BLAST program (Altschul et al. 1990). Alignments and similarity comparison were initially conducted by the ClustalW method and final alignment was manually performed with the multiple sequence alignment editor MegAlign Program (DNAStar, Madison, WI, USA). A phylogenetic tree was constructed using MEGA with the neighbor-joining.
Amylase assay
A 10 μl aliquot of enzyme sample was mixed with 190 μl of 1% soluble starch in 50 mM sodium acetate buffer (pH 5.0) and incubated at 95°C for 30 min. One unit of amylase activity was defined as the amount of enzyme, which released 1 μmol of reducing sugar equivalent to glucose per minute (Chung et al. 1995).
Protein purification and gel electrophoresis analysis
The α-amylase enzymes [Thermococcus HJ21 amylase (THJA)] were purified from the fermentation broth produced by the strain. Amylase purification was performed at room temperature under aerobic conditions following protocols described previously (Kwak et al. 1998). Chromatography columns were controlled with a fast protein liquid chromatography system (Bio-Rad, Hercules, CA, USA). The molecular mass, homogeneity, and activity staining of the purified THJA from strain HJ21 were estimated by 0.1–10% sodium dodecyl sulfate (SDS) polyacrylamide gels containing 0.3% (w/v) starch (Brown et al. 1990). After electrophoresis, protein bands were detected by staining with 0.1% Coomassie brilliant blue R-250 (Bio-Rad). For amylase activity staining, the starch gels were washed for 1 h at room temperature in 50 mM Na2HPO4 (pH 7.5) buffer containing 2.5% (v/v) Triton X-100 before being incubated in acetate buffer (pH 5.0) for 1 h at 95°C. Gels were then stained in a solution containing 0.15% (w/v) I2 and 1.5% (w/v) KI for 2 min. Amylase activity was visualized as clear band.
Effects of pH and temperature on the activity and stability of THJA
To determine the optimal pH of THJA, the relative activities in various pHs ranging from 3 to 9 were examined. The buffers used were as follows: 50 mM sodium citrate buffer (pH 3.0 to 5.0), 50 mM sodium acetate buffer (pH 5.0 to 6.0), 50 mM sodium phosphate buffer (pH 6.0 to 7.5), and 50 mM Tris–HCl buffer (pH 7.5 to 9.0). To examine the pH stability, the enzyme was incubated in various pH solutions of 50 mM Britton–Robinson buffer (Rauen 1964). After incubation at different temperatures (80°C, 90°C, and 100°C) for 1 h, the remaining activity was measured at the optimal temperature under standard conditions as described above. The optimal temperature of the α-amylase activity was determined in 50 mM sodium acetate buffer (pH 5.0) in a range from 40°C to 120°C. To examine the thermostability, the enzyme was incubated either in the absence or the presence of 5 mM Ca2+ solutions at different temperatures (80°C, 90°C, and 100°C) for 1–5 h and the residual activity was measured as described above.
Determination of kinetics parameters
The kinetic properties were determined by measuring the initial rates at various soluble starch concentrations [0.2–4.0 (w/v)] for amylase under standard reaction conditions. K m and V max values were obtained from Lineweaver–Burk plot and expressed as the mean of the three different experiments (Li et al. 2007).
TLC of enzymatic hydrolysis products
The products released through hydrolysis of soluble starch by the amylase were identified by thin layer chromatography (TLC) using a silica gel 60 plate (Merck, Darmstadt, Germany) with the solvent system of n-butanol–pyridine–water (6:4:3) and a detection reagent comprising 2.0% (w/v) diphenylamine in acetone–2.0% (w/v) aniline in acetone–85% (w/v) phosphoric acid (5:5:1, v/v/v) (Li et al. 2007).
Isolation and nucleotide sequencing analysis of α-amylase gene
According to the conservative region of thermostable alpha-amylase gene from hyperthermophilic archaea, we synthesized degenerate primers to clone the gene between two conservative regions of THJA. The 3′- terminal and 5′-terminal sequence of THJA gene were acquired by site-finding method described by Tan (Tan et al. 2005). The complete THJA gene was obtained through the juncture of them. Based on the sequence result, the complete THJA gene was amplified from purified genomic DNA with primer: forward primer, 5′GCGCGAATTCATGAACAGGGGTATAT3′; reverse primer, 5′-TAGTCGACTCAGCCCACCCCACAG3′. Gene sequence was obtained using the same method as for the 16S rRNA. α-Amylase protein homologues were collected from the protein database using BLAST program (Altschul et al. 1990). Alignment of the translated HJ21 α-amylase amino acid sequence and the collected α-amylase homologues was performed using the MegAlign program (DNAStar).
Nucleotide sequence accession number
The nucleotide sequence of the 16S rRNA gene and the α-amylase encoding gene have been submitted to GenBank under accession number were EF198107 and EF634454, respectively.
Results
Characterization of strain HJ21
Strain HJ21 cells are irregular cocci of 1 to 1.2 μm in diameter (Fig. 1a). Data in Fig. 1b suggested that HJ21 was an organotrophic, thermophilic anaerobe growing at temperatures ranging from 60°C to 94°C with 88°C being the optimum. HJ21 could grow between pH 5.0 and 9.0, with an optimum around pH 6.5. No growth was detected at pH 4.0 and 10.0 (Fig. 1c). When different concentrations of NaCl were added to the culture medium, growth occurred in media containing 1–5% NaCl. Maximum growth rate was observed at 2% NaCl, and no growth occurred in medium without NaCl or concentrations higher than 6% (Fig. 1d). When different carbon and energy sources were tested for growth, it was found that HJ21 grew much better in medium containing yeast extract, peptone, or tryptone. HJ21 cells were capable of growing on casein, amino acid, starch, maltose, glucose, sucrose, cellobiose, lactose, glycogen, gelatin, succinate, pyruate, and acetate. No growth was observed when xylose, citrate, lactate, ethanol, methanol, or glycerol was used as the sole carbon source. When alternative electron acceptors were tested, neither 10 mM thiosulfate, 20 mM sulfate, nor 10 mM sulfite supported growth. The result suggested, however, that elemental sulfur considerably stimulated HJ21 growth. Lack of elemental sulfur in the culture medium caused a longer lag phase of growth and a much lower cell yield (Fig. 1e).
The 16S rRNA encoding gene of strain HJ21 was sequenced and aligned with other 16S rRNA sequences of Thermococcales strains. The phylogenetic tree, which was constructed for comparison of the 16S rRNA sequences, indicated that strain HJ21 belonged to the order of Thermococcales. The levels of similarity between the 16S rRNA of HJ21 and 16S rRNAs of other Thermococcus species are summarized in Fig. 2, and HJ21 has 99.6% similarity with Thermococcus siculi.
Phylogenetic tree based on 16S rRNA gene sequences. The tree was constructed by the neighbor-joining method. Numbers indicate bootstrap values of 1,000 trials. The scale bar represents ten nucleotide substitutions per 100 nucleotides. The accession numbers of 16S rRNA sequences used for the unrooted tree are as follows: Thermococcus celer strain DSM 2476 (AY099174); Thermococcus radiophilus (AF479013); Thermococcus barossii strain DSM 9535 (AY099173); Thermococcus coalescens (AB107767); Thermococcus hydrothermalis strain AL662 (AY099179); Thermococcus sp. OGL-20P (AF394925); Thermococcus siculi strain DSM 12349 (AJ291808); Thermococcus sp. strain HJ21(EF198107); Thermococcus profundus strain DSM 9503 (AY099184); Thermococcus marinus (AF479012); Thermococcus peptonophilus strain DSM 10343 (AY099183); Thermococcus kodakarensis KOD1 (D38650); Thermococcus gorgonarius strain DSM 10395 (AB055123); Thermococcus sp. NA1 (DQ167232); Thermococcus gammaltolerans strain FJ3 (AY206705); Thermococcus guaymasensis strain DSM 11113 (AY099178); Thermococcus fumicolans strain ST557 (AB055128); Thermococcus pacificus strain DSM 10394 (AY099182); Thermococcus sp. Rt3 (AF017455); Thermococcus waiotapuensis strain DSM 12768 (AY099187); Thermococcus zilligii strain AN1 (U76534); Pyrococcus glycovorans (AY099168); Thermococcus chitonophagus (X99570); Pyrococcus abyssi (AY099167); Pyrococcus horikoshii (D45214); Pyrococcus furiosus (AY099169); Pyrococcus woesei (AY519654)
Enzymatic properties of THJA
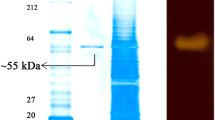
Extracellular thermostable α-amylase was found to be produced by strain HJ21.The amylase was purified from the concentrated supernatant prepared from the cell culture. The results in Table 1 shows that amylase was purified threefold with a yield of 9%. The purified enzyme exhibited a specific activity of 8.3 U/mg per protein and was electrophoretically homogeneous (Fig. 3a). The mobility of the active amylase band, as determined by activity staining, coincided with that of the single protein band stained with Coomassie brilliant blue. The molecular mass of the α-amylase was estimated to be 51.4 kDa by SDS polyacrylamide gel electrophoresis (PAGE; Fig. 3a).
Molecular weight and effects of temperature and pH on the activity and stability of THJA. a SDS-PAGE of THJA. The purified amylase was visualized by Coomassie brilliant blue staining (lane 1) and by activity staining (lane 2). Lane M Protein molecular weight marker. b Effect of temperature on the amylase activity (diamond). c Effect of temperature on stability of the amylase: 80°C (filled triangle, empty triangle), 90°C (filled square, open square), and 100°C (filled circle, open circle), the filled symbols and the empty symbols indicate the enzyme was incubated in the absence and the presence of 5 mM Ca2+ solutions, respectively. d Effect of pH on the amylase activity. The following buffers were used: pH 3.0 to 5.0, 50 mM sodium citrate (diamond); pH 4.5 to 6.0, 50 mM sodium acetate (square); pH 6.0 to 7.5, 50 mM sodium phosphate (triangle); and pH 7.5 to 9.0, 50 mM Tris–HCl (circle). e Effect of pH on stability of the amylase. For effect of temperature and pH on the amylase activity, the values are shown as percentages of the maximum activities, which were taken as 100%. For effect of temperature and pH on the stability of the amylase, the residual activity was assayed under standard conditions. Values shown are percentages of the original activities, which were taken as 100%
The optimum temperature for the THJA enzyme was observed at 95°C, and more than 60% of the maximum enzyme activity was detected at 100°C (Fig. 3b). The half-life of THJA was determined to be 5 h at 90°C. Over 40% and 30% of the enzyme activity remained after incubation at 100°C for 2 and 3 h, respectively (Fig. 3c). THJA did not require Ca2+ for thermostability. Maximum activity of THJA was observed at pH 5.0 and 95°C. More than 80% of the maximum activity remained at pH 4.5 and 95°C (Fig. 3d). In addition, the results indicated that the amylase was stable in a pH range from pH 5.0 to 7.0 at 80°C and 90°C for 1 h respectively, 5.0 to 6.0 at 100°C for 1 h (Fig. 3e).
Lineweaver–Burk plots showed that apparent K m and V max values of the purified amylase for soluble starch were 45 ± 0.23 and 9.0 ± 0.03 mg/ml/min, respectively. To clarify the action modes of THJA enzyme, soluble starch was hydrolyzed at various time durations, and the products were analyzed by TLC (Fig. 4). The sugar produced after the reaction from 1 h to 24 h were maltose (G2), maltotriose (G3), maltotetraose (G4), maltopentaose(G5), maltohexaose (G6), and maltoheptaose (G7). The main products by THJA enzyme from soluble starch were altooligosaccharides, indicating that THJA was a liquefying enzyme
Time course of maltooligosaccharide production by amylase activity with soluble starch as the substrate. Lanes 1 to 4 represent the end products at 1, 6, 16, and 24 h of incubation at 95°C. Lane M represents a standard mixture of maltooligosaccharides ranging from G1 to G7 (G1 glucose, G2 maltose, G3 maltotriose, G4 maltotetraose, G5 maltopentaose, G6 maltohexaose, G7 maltoheptaose)
The gene encoding α-amylase from strain HJ21
The gene encoding the highly thermostable THJA was cloned from the HJ21 genomic DNA, and sequencing results revealed an open reading frame (ORF) of 1,374 bp. The ORF encodes a protein of 457 amino acids (Fig. 5), with a calculated molecular weight of 51.4 kDa. Four conserved regions were present in THJA (Fig. 6) and showed great similarities with those of other thermophilic-secreted Thermococcus α-amylase.
Discussion
In the last decade, numerous hyperthermophilic Thermococcus strains have been isolated from various deep-sea hydrothermal vents, shallow marine hydrothermal areas, and high-temperature oil reservoirs (Jolivet et al. 2004). Presently, 16S rRNA gene sequences from 176 strains of the Thermococcus genus have been deposited in GenBank. However, only about 30 strains have been identified and described. These species include Thermococcus celer, Thermococcus stetteri, Thermococcus litoralis, Thermococcus profundus, Thermococcus peptonophilus, Thermococcus alcaliphilus, Thermococcus chitonophagus, Thermococcus fumicolans, Thermococcus hydrothermalis, Thermococcus zilligii, Thermococcus guaymasensis, Thermococcus aggregans, Thermococcus gorgonarius, Thermococcus pacificus, Thermococcus acidaminovorans, Thermococcus barossii, Thermococcus waiotapuensis, Thermococcus siculi, Thermococcus barophilus, Thermococcus aegaeicus, Thermococcus sibiricus, Thermococcus gammatolerans, Thermococcus marinus, Thermococcus radiotolerans (Jolivet et al. 2004), Thermococcus atlanticus (Cambon-Bonavita et al. 2003), Thermococcus kodakaraensis (Atomi et al. 2004), Thermococcus coalescens (Kuwabara et al. 2005), Thermococcus onnurineus (Bae et al. 2006), Thermococcus celericrescens (Kuwabara et al. 2007), and Thermococcus thioreducens (Pikuta et al. 2007). We have isolated strain HJ21 from deep-sea hydrothermal vents. Based on the biochemical test and 16s ribosomal DNA sequence comparison, strain HJ21 was identified as a hyperthermophilic archaeon, belonging to the Thermococcus sp.
Through more than 30 different species of Thermococcus have been isolated and identified, but only a few of these species have been reported to produce α-amylase and the α-amylase from these species were purified and characterized. The strain HJ21 has been found to produce the extracellular thermostable α-amylase.
BLA (α-amylase from B. licheniformis) has a wide application in industry today. We have purified the α-amylase produced by strain HJ21 and compared the isolated amylase with other amylases from Thermococcus strains and B. licheniformis, in the information from pH, temperature, Ca2+ requirement, the half-life, K m, V max, mode of action, end products, and so on (Table 2). THJA displayed a higher optimal temperature and thermostability than BLA and other amylases from other Thermococcus strains. The optimum temperature for the THJA was 95°C, much higher than those obtained from T. profundus (80°C), T. hydrothermalis (75–85°C), T. thioreducens (90°C), and B. licheniformis (90°C). The half-life of THJA was determined to be 5 h at 90°C. It was obvious that the thermostability of THJA was superior to those amylases from T. profundus (about 3 h at 80°C or 75 min at 90°C), B. licheniformis (<1 h at 98°C). Since Ca2+ is a strong inhibitor of glucose isomerase, performing starch liquefaction and saccharification in the absence of Ca2+ would be a significant improvement for high-fructose syrup production (Dong et al. 1997). The addition of Ca2+ had a significant effect on thermostability of the amylases from T. profundus, T. thioreducens, and B. licheniformis, but Ca2+ was not needed for THJA thermostability.
The optimum pH for the THJA was 5.5, lower than those amylases obtained from T. profundus (5.5 or 5.5–6.0), T. hydrothermalis (5.0–5.5), T. thioreducens (5.5), and B. licheniformis (6.0–8.0). The THJA was stable in a pH range from pH 5.0 to 7.0 at 80°C and 90°C for 1 h, respectively, and was superior to the amylase from T. profundus (5.5–9.8 at 60°C for 0.5 h) (Chung et al. 1995). Therefore, with THJA, starch liquefaction can be performed under pH conditions that reduce by-product formation.
The modes of action of the enzyme were studied and found that the main products of THJA hydrolyzing the soluble starch were maltose and maltotriose. Maltose was not hydrolyzed by the enzyme. These results indicated that THJA was a liquefying enzyme and cleaved α-1,4-linkages only and not α-1,6-linkages. High similarity results were found in α-amylase from T. profundus DT5432 (Chung et al. 1995).
The results showed that THJA under the conditions of pH 4.5–5.0 and 95°C without Ca2+, which is better than those α-amylases produced by other Thermococcus strains and B. licheniformis. Especially, THJA was thermostable without Ca2+, thus making it potentially profitable for industries as a liquefying enzyme.
The sequence of THJA gene was aligned with other available α-amylase gene sequences of Thermococcales strains and Bacillus. High similarity was found between the THJA gene sequence and α-amylase encoding genes from T. hydrothermalis (96%), T. onnurineus, and Thermococcus sp. Rt3 (94%) (Jone et al. 1999). Four conserved regions, probably forming the active site, have been identified in α-amylases (Nakajima et al. 1986). These regions were present in THJA and showed great similarities with those of other thermophilic secreted Thermococcus α-amylase (Lee et al. 1996; Léveque et al. 2000a), confirming that THJA was an α-amylase. The sequence of THJA gene was aligned with other available α-amylase gene sequences of other Thermococcus strains. There were about 4% of the amino acid sequence were different between the THJA gene sequence and that of other α-amylase genes from Thermococcus.
Compared with α-amylases secreted by bacteria, His residue in region II is replaced by a Gly residue in THJA and those of other Thermococcales. This His residue was thought to be involved in substrate binding (Nielsen and Borchert 2000). A conserved Trp residue was found in Domain II of all secreted Thermococcales α-amylases, which was suggested to possibly play the same role as the missing His residue (Lee et al. 1996; Léveque et al. 2000a). These replacements were also observed in high pI isozyme of barley α-amylase. Similarity in Domain II of the plant and archaeal enzymes could be due to the phylogenetical relatedness (Léveque et al. 2000b).
THJA showed properties promising advantages over the commercially available B. licheniformis α-amylase and might find an application in starch conversion biotechnologies. Currently, the α-amylase gene is being expressed in E. coli, and we are currently working on the purification and characterization of recombinant enzyme.
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Atomi H (2005) Recent progress towards the application of hyperthermophiles and their enzymes. Curr Opin Chem Biol 9:166–173
Atomi H, Fukui T, Tanaka T, Kanai T, Morikawa M, Imanaka T (2004) Description of Thermococcus kodakaraensis sp. nov., a well studied hyperthermophilic archaeon previously reported as Pyrococcus sp. KOD1. Archaea 1:263–267
Bae SS, Kim YJ, Yang SH, Lim JK, Jeon JH, Lee HS, Kang SG, Kim SJ, Lee JH (2006) Thermococcus onnurineus sp. nov., a hyperthermophilic archaeon Isolated from a deep-Sea hydrothermal vent area at the Pacmanus Field. J Microbiol Biotechnol 11:1826–1831
Bernhardsdotter ECMJ, Pusey ML, Ng JD, Garriott OK (2004) Cloning and characterization of an α-amylase gene from the hyperthermophilic archaeon Thermococcus thioreducens. Marshall Space Flight Center Database
Brown SH, Costantino HR, Kelly RM (1990) Characterization of amylolytic enzyme activities associated with the hyperthermophilic archaebacterium Pyrococcus furiosus. Appl Environ Microbiol 56:1985–1991
Cambon-Bonavita MA, Lesongeur F, Pignet P, Wery N, Lambert C, Godfroy A, Querellou J, Barbier G (2003) Extremophiles, thermophily section, species description Thermococcus atlanticus sp. nov., a hyperthermophilic archaeon isolated from a deep-sea hydrothermal vent in the Mid-Atlantic Ridge. Extremophiles 7:101–109
Chung YC, Kobayashi T, Kanai H, Akiba T, Kudo T (1995) Purification and properties of extracellular amylase from the hyperthermophilic archaeon Thermococcus profundus DT5432. Appl Environ Microbiol 61:1502–1506
Dong G, Vieille C, Savchenko A, Zeikus JG (1997) Cloning, sequencing, and expression of the gene encoding extracellular a-amylase from Pyrococcus furiosus and biochemical characterization of the recombinant enzyme. Environ Microbiol 63:3569–3576
Jolivet E, Corre E, L, Haridon S, Forterre P, Prieur D (2004) Thermococcus marinus sp. nov., and Thermococcus radiotolerans sp. nov., two hyperthermophilic archaea from deep-sea hydrothermal vents that resist ionizing radiation. Extremophiles 8:219–227
Jone RA, Jermiin LS, Easteal S, Patel BKC, Beacham IR (1999) Amylase and 16S rRNA genes from a hyperthermophilic archaebacterium. J Appl Microbiol 86:93–107
Kuwabara T, Minaba M, Iwayama Y, Inouye I, Nakashima M, Marumo K, Maruyama A, Sugai A, Itoh T, Ishibashi J, Urabe T, Kamekura M (2005) Thermococcus coalescens sp. nov., a cell-fusing hyperthermophilic archaeon from Suiyo Seamount. Int J Syst Evol Microbiol 55:2507–2514
Kuwabara T, Minaba M, Ogi N, Kamekura M (2007) Thermococcus celericrescens sp. nov., a fast-growing and cell-fusing hyperthermophilic archaeon from a deep-sea hydrothermal vent. Int J Syst Evol Microbiol 57:437–443
Kwak YS, Akiba T, Kudo T (1998) Purification and characterization of α-amylase from hyperthermophilic archaeon Thermococcus profundus, which hydrolyzes both α-1, 4 and α-1,6 glucosidic linkages. J Ferment Bioeng 86:363–367
Lee JT, Kanai H, Kobayashi T, Akiba T, Kudo T (1996) Cloning, nucleotide sequence, and hyperexpression of α-amylase Gene from an archaeon, Thermococcus profundus. J Ferment Bioeng 82:432–438
Léveque E, Haye B, Belarbi A (2000a) Cloning and expression of an α-amylase encoding gene from the hyperthermophilic archaebacterium Thermococcus hydrothermalis and biochemical characterization of the recombinant enzyme. FEMS Microbio Lett 186:67–71
Léveque E, Janeček S, Haye B, Belarbi A (2000b) Thermophilic archaeal amylolytic enzymes. Enz Microbiol Technol 26:3–14
Li HF, Chi ZM, Wang XH (2007) Purification and characterization of extracellular amylase from the marine yeast Aureobasidium pullulans N13d and its raw potato starch digestion. Enz Microbiol Technol 40:1006–1012
Nakajima R, Imanaka T, Aiba S (1986) Comparison of amino acid sequences of eleven different alpha -amylases. Appl Microbiol Biotechnol 23:355–360
Nielsen JE, Borchert TV (2000) Protein engineering of bacterial α–amylase. Biochimia et Biophysica Acta 1542:253–274
Pikuta EV, Marsic D, Itoh T, Bej AK, Tang J, Whitman WB, Ng JD, Garriott OK, Hoover RB (2007) Thermococcus thioreducens sp. nov., a novel hyperthermophilic, obligately sulfur-reducing archaeon from a deep-sea hydrothermal vent. Int J Syst Evol Microbiol 57:1612–1618
Rauen HM (1964) Biochemisches Taschenbuch, vol. 2. Springer, Berlin, Germany, pp 90–104
Richardson TH, Tan X, Frey G, Callen W, Cabell M, Lam D, Macomber J, Short JM, Robertson DE, Miller C (2002) A novel, high performance enzyme for starch liquefaction.discovery and optimization of a low pH, thermostable α-amylase. J Biol Chem 277:26501–26507
Tan GH, Gao Y, Shi M, Zhang XY, He SP, Chen ZL, An CC (2005) SiteFinding-PCR: a simple and efficient PCR method for chromosome walking. Nucleic Acids Res 33:2–7
Vieille C, Zeikus GJ (2001) Hyperthermophilic enzymes: sources, uses, and molecular mechanisms for thermostability. Microbiol Mol Biol Rev 65:1–43
Violet M, Meunier JC (1989) Kinetic study of the irreversible thermal denaturation of Bacillus Licheniformis α-amylase. Biochem J 263:665–670
Acknowledgements
The authors would like to thank Mr. Fuchao Li, Ms. Li Chen, and Mr. Jinyu Yang for their technical assistance. This work was financially supported by National Natural Science Foundation of China (40746030), Natural Science Foundation of the Education Department of Jiangsu Province (06KJB550004, 06-A-017), Jiangsu Key Laboratory of Marine Biotechnology (2006HS008), and Science and Technology Department of Lianyungang City (KK06076).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wang, S., Lu, Z., Lu, M. et al. Identification of archaeon-producing hyperthermophilic α-amylase and characterization of the α-amylase. Appl Microbiol Biotechnol 80, 605–614 (2008). https://doi.org/10.1007/s00253-008-1561-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-008-1561-8