Abstract
Cry4Aa produced by Bacillus thuringiensis is a dipteran-specific toxin and is, therefore, of great interest for developing a bioinsecticide to control mosquitoes. However, the expression of Cry4Aa in Escherichia coli is relatively low, which is a major disadvantage in its development as a bioinsecticide. In this study, to establish an effective production system, a 1,914-bp modified gene (cry4Aa-S1) encoding Cry4Aa was designed and synthesized in accordance with the G + C content and codon preference of E. coli genes without altering the encoded amino acid sequence. The cry4Aa-S1 gene allowed a significant improvement in expression level, over five-fold, compared to that of the original cry4Aa gene. The product of the cry4Aa-S1 gene showed the same level of insecticidal activity against Culex pipiens larvae as that from cry4Aa. This suggested that unfavorable codon usage was one of the reasons for poor expression of cry4Aa in E. coli, and, therefore, changing the cry4Aa codons to accord with the codon usage in E. coli led to efficient production of Cry4Aa. Efficient production of Cry4Aa in E. coli can be a powerful measure to prepare a sufficient amount of Cry4Aa protein for both basic analytical and applied researches.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The gram-positive soil bacterium Bacillus thuringiensis subsp. israelensis produces four major insecticidal crystalline proteins, Cry4Aa, Cry4Ba, Cry11Aa, and Cyt1Aa during sporulation. Cry4Aa, Cry4Ba, and Cry11Aa are dipteran-specific toxins and are, therefore, of great interest for developing a bioinsecticide to control mosquitoes, which are vectors of important tropical diseases like malaria and viral hemorrhagic fevers (Schnepf et al. 1998). Cry4Aa is especially toxic to Aedes and Culex larvae and slightly less toxic to Anopheles larvae (Poncet et al. 1995), and is an excellent candidate for an alternative or accessory to mosquitocidal chemical insecticide. However, in the most frequently used prokaryotic expression system for production of heterologous proteins (Hannig and Makrides 1998; Sorensen and Mortensen 2005), Escherichia coli, the expression level of Cry4Aa is relatively low, which is a major disadvantage in its development as a mosquitocidal insecticide.
It is frequently observed that some properties of heterologous genes preclude their efficient expression in E. coli. Switching to a different expression vector or a strain of E. coli may help to improve the production level of the recombinant proteins. However, if these fail to improve the situation, one of the most powerful approaches is to alter the codon usage of the heterologous gene to meet the codon preference of E. coli genes.
Poor expression of cry4Aa gene may be caused by the comparatively low G + C content of cry4Aa relative to E. coli genes, leading to a complete difference in codon preference between the cry4Aa gene and E. coli genes. To improve the production level of Cry4Aa in E. coli, we synthesized the cry4Aa-S1 gene, the G + C content and the codon usage of which is mostly in accordance with that in E. coli genes. We demonstrated that the expression level of cry4Aa-S1 was dramatically enhanced compared to that of cry4Aa.
In general, E. coli is most frequently used for genetic engineering of heterologous protein genes as well as an expression system. Therefore, our approach to establish efficient expression system in E. coli will facilitate both functional analyses of Cry4Aa in research and development of a new bioinsecticide.
Materials and methods
Synthesis of cry4Aa-S1 gene
For efficient expression, the synthetic cry4Aa gene (cry4Aa-S1) was designed in accordance with the G + C content and the codon preference of E. coli genes. The codon preference of E. coli B genes was taken from the Codon Usage database at http://www.kazusa.or.jp/codon/. The entire nucleotide sequence of cry4Aa-S1 was deposited in the DDBJ, GenBank, and EMBL databases under accession no. AB250380. A major part of cry4Aa-S1, which encodes the Cry4Aa active segment (G58–Q695), was synthesized by recursive PCR (Prodromou and Pearl 1992) using overlapping oligonucleotides containing portions of the nucleotide sequences of the sense and antisense strands of cry4Aa-S1. Oligonucleotides were 50 to 55 nucleotides long with similar melting temperatures, and the cry4Aa-S1 gene was tailed with Bam HI-Eco RI and Pst I-Eco RI cleavage sequences at the upstream and downstream termini, respectively. PCR was done with KOD plus DNA polymerase (Toyobo Life Science, Tokyo, Japan). The completed cry4Aa-S1 gene was cloned between the Bam HI and Pst I sites of pHSG398 (Takara Bio Inc., Ohtsu, Japan) to generate pHSG398-Cry4Aa-S1. The DNA sequence was confirmed using an ABI PRISM™ 310 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA).
Construction of expression plasmids
Cry4Aa was expressed as a fusion protein with glutathione-S-transferase (GST). The DNA segment of the original cry4Aa encoding the insecticidal Cry4Aa active fragment (G58–Q695) was amplified by PCR from plasmid vector pIS422 (Nishimoto et al. 1994) using forward primer 4A-N-f (GGGATCCGGTGGAGATTTTGAAAC) and reverse primer 4A-N-r (GGATCCTTATTGTTGTACTGTTTCTAATTTTTG). The amplified DNA segment, which was tailed with Bam HI cleavage sequences at both termini, was inserted in frame into the Bam HI site of pGEX 4T-2 (GE Healthcare UK Ltd, Little Chalfont, Buckinghamshire, UK) to generate pGST-Cry4Aa (Fig. 1).
pHSG398-Cry4Aa-S1 was cleaved with Eco RI and the cry4Aa-S1 segment encoding the Cry4Aa active fragment (G58–Q695) was inserted in frame into the Eco RI site of pGEX 4T-2 to generate pGST-Cry4Aa-S1 (Fig. 1).
Expression and protein analysis
The GST-Cry4Aa fusions were expressed in E. coli BL21, which was cultured in TB (24 g yeast extract, 12 g tryptone, 0.4% glycerol, 12.5 g K2HPO4, 2.3 g KH2PO4 l−1) supplemented with 100 μg ml−1 ampicillin. The expression was induced with 0.06 mM isopropyl β-d-thiogalactopyranoside (IPTG) at 20°C for 3 h. The GST-Cry4Aa fusions were purified using glutathione-Sepharose 4B (GE Healthcare UK Ltd, Little Chalfont, Buckinghamshire, UK) as described previously (Yamagiwa et al. 1999) and analyzed by SDS-10% PAGE followed by visualization with Coomassie Brilliant Blue staining. Protein concentration was estimated using a Bio-Rad Protein Assay (Bio-Rad Laboratories, Inc., Hercules, CA, USA) with bovine serum albumin as a standard. Purified GST-Cry4Aa was treated with trypsin to analyze the pattern of digestion. The processed proteins were analyzed by SDS-15% PAGE followed by visualization with Coomassie staining.
Bioassays
Bioassays were performed using 5-day-old, 3rd instar of C. pipiens larvae, reared from eggs that were supplied by the Research and Development Laboratory, Dainihon Jochugiku Co., Ltd., Osaka, Japan. About 2,000 larvae were reared in a container (35 × 25 × 3 cm) with 2 l of distilled water supplemented with a goldfish diet.
Bioassays were done as described previously with some modifications (Yamagiwa et al. 1999). Briefly, GST-Cry4Aa was adsorbed to latex beads (0.8 μm diameter, Sigma-Aldrich corp., St. Louis, MO, USA) for 1 h at room temperature to be administered as a diet to mosquito larvae; the adsorption was monitored by densitometric analysis using SDS-10% PAGE followed by Coomassie staining. Bioassays were done in a 96-well microtiter plate with one larva per well and 48 larvae per assay. Mortality was recorded 24 h after inoculation period with serially diluted latex beads bound with Cry4Aa toxin and the LC50 was determined using PROBIT analysis.
Results
Synthesis of codon-modified cry4Aa gene (cry4Aa-S1)
The G + C content and the codon usage of the four Bacillus thuringiensis insecticidal crystalline protein genes were compared with those of E. coli. The G + C contents of the selected cry genes were lower (less than 40%) than that of E. coli (51%), and cry4Aa was only 33% (Table 1). Furthermore, major differences were observed in the codon preference between cry4Aa and E. coli genes (Table 2). For example, among six synonymous codons for leucine, CTG was preferentially used (45%) in E. coli, whereas in the cry4Aa gene, TTA (43%) and CTT (32%) were preferentially used. These observations suggested that the differences in G + C content and codon preference were responsible for the poor expression of cry4Aa in E. coli.
The 1,914-bp synthetic gene (cry4Aa-S1) encoding the insecticidal Cry4Aa active segment was, therefore, designed and synthesized in accordance with the G + C content and the codon preference of E. coli genes without altering the amino acid sequence (Table 2). The cry4Aa-S1 was successfully constructed and its G + C content increased to 52%, which was similar to that of E. coli genes.
Expression of cry4Aa and cry4Aa-S1 genes
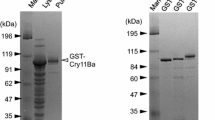
Production of Cry4Aa from pGST-Cry4Aa and pGST-Cry4Aa-S1 was analyzed. SDS-PAGE analysis of the E. coli cell lysate showed a GST-Cry4Aa fusion protein of 90 kDa expressed from both constructs (Fig. 2A). The production level from pGST-Cry4Aa-S1 was over five-fold higher than that of pGST-Cry4Aa based on estimates using densitometric scanning of the protein bands. This indicated that the low G + C content and the unfavorable codon of cry4Aa was at least one of the reasons for poor expression in E. coli. Changing the cry4Aa codons to accord with the codon usage in E. coli may contribute to translation efficiency and possibly to the stability of mRNA.
GST-Cry4Aa expressed by the recombinant vectors pGST-Cry4Aa and pGST-Cry4Aa-S1. a Sonicated E. coli cells were separated by SDS-10% PAGE and stained with Coomassie Brilliant Blue. Expression of GST-Cry4Aa was compared using a densitometric scanning method. b SDS-10% PAGE of purified GST-Cry4Aa. c GST-Cry4Aa was treated with trypsin and the processed polypeptides were separated by SDS-15% PAGE. The migration of molecular weight markers is indicated by arrowheads
GST-Cry4Aa was successfully purified using glutathione-Sepharose 4B (Fig. 2B). Few protein contaminants of lower molecular mass were detected, but these may have been caused by degradation of the purified proteins since these contaminants were minimized by adding a protease inhibitor (phenylmethylsulfonyl fluoride) to the solution. Recovery of purified GST-Cry4Aa from E. coli harboring pGST-Cry4Aa-S1 ranged from 0.4 to 0.6 mg from a 200-ml culture and was estimated as 2- to 3-fold that from E. coli harboring pGST-Cry4Aa (about 0.2 mg from 200 ml). Although there is a contradiction between the expression level and the recovery of GST-Cry4Aa, it may be caused by the slower proliferation of E. coli harboring pGST-Cry4Aa-S1 compared to that of E. coli harboring pGST-Cry4Aa. The hyper-expression of GST-Cry4Aa may affect the proliferation of E. coli.
Cry4Aa is processed in a susceptible insect midgut into protease-resistant segments of 20 and 45 kDa, which associate to form a complex for the toxicity (Yamagiwa et al. 1999). Thus, treatment with protease such as trypsin can be a presumptive test of folding fidelity of Cry4Aa. Upon treatment with trypsin, GST-Cry4Aa of 90 kDa was processed into 20 and 45 kDa polypeptides (Fig. 2C). No apparent difference was observed in the processing pattern between GST-Cry4Aa fusions expressed from pGST-Cry4Aa and pGST-Cry4Aa-S1, suggesting that both versions of GST-Cry4Aa were structurally the same; although the amount of 20 kDa polypeptide seemed to be lower than that expected from the amount of 45 kDa polypeptide, this may be caused by a difference in the stability of the 20 and 45 kDa polypeptides or preferable oligomerization of the 20 kDa polypeptide. A similar observation has been reported previously (Yamagiwa et al. 1999).
Insecticidal activity of GST-Cry4Aa
GST-Cry4Aa was purified and tested for biological activity against C. pipiens larvae. GST-Cry4Aa translated from pGST-Cry4Aa-S1 showed the same level of toxicity as that from pGST-Cry4Aa, and LC50 of the both proteins were estimated to be 0.24 μg ml−1 and 0.29 μg ml−1, respectively. Purified GST used as a negative control showed no toxicity.
Discussion
We constructed the cry4Aa-S1 gene encoding the Cry4Aa active polypeptide to meet the G + C content and the codon preference of E. coli genes without altering the original amino acid sequence. The cry4Aa-S1 gene allowed significant improvement of expression level, over five-fold, and the recovery of the purified product also increased two- to three-fold. Organisms exhibit different preferences for synonymous codons, resulting in a three- to six-fold difference in translation rates of heterologous genes (Robinson et al. 1984). This suggests that the improvement of expression level we achieved was caused by the change of cry4Aa codon usage to accord with that of E. coli genes. Improvement of Cry toxin production by adaptation of codon usage has also been reported for the expression of the cry4B gene in cyanobacteria (Soltes-Rak et al. 1995). The authors observed specific degradation of cry4B mRNA, possibly caused by difference in codon usage. Such codon usage problems are frequently encountered in higher plants and seem to be associated with mRNA degradation, which results in poor expression of cry toxin genes (Adang et al. 1993; Murray et al. 1991; Perlak et al. 1991).
No difference was observed in the processing pattern between the GST-Cry4Aa proteins derived from constructs containing either the original or synthetic coding sequence. Furthermore, the product of cry4Aa-S1 showed the same level of insecticidal activity against Culex pipiens larvae as that from cry4Aa. Therefore, we concluded that the synthetic cry4Aa-S1 gene was efficiently expressed in E. coli and the Cry4Aa protein produced was structurally and biologically similar to that from the original cry4Aa gene. The cry4Aa-S1 gene we constructed will be a powerful means of preparing sufficient Cry4Aa protein for both functional research analyses and application as a bioinsecticide.
Recently, Boonserm et al. (2004) have been achieved high-level expression of Cry4Aa in E. coli using dual promoter system of Ptac and Pcry4Ba. In this case, enhancement of expression in transcription and possibly translation level was likely to confer the high-level expression of Cry4Aa. Thus, their concept was completely different from ours; combination of these two techniques may help for further improvement of the production system of Cry4Aa.
References
Adang MJ, Brody MS, Cardineau G, Eagan N, Roush RT, Shewmaker CK, Jones A, Oakes JV, McBride KE (1993) The reconstruction and expression of Bacillus thuringiensis cryIIIA gene in protoplasts and potato plants. Plant Mol Biol 21:1131–1145
Boonserm P, Pornwiroon W, Katzenmeier G, Panyim S, Angsuthanasombat C (2004) Optimised expression in Escherichia coli and purification of the functional form of the Bacillus thuringiensis Cry4Aa δ-endotoxin. Protein Expr Purif 35:397–403
Hannig G, Makrides SC (1998) Strategies for optimizing heterologous protein expression in Escherichia coli. Trends Biotechnol 16:54–60
Murray EE, Rocheleau T, Eberle M, Stock C, Sekar V, Adang M (1991) Analysis of unstable RNA transcripts of insecticidal crystal protein genes of Bacillus thuringiensis in transgenic plants and electroporated protoplasts. Plant Mol Biol 16:1035–1050
Nishimoto T, Yoshisue H, Ihara K, Sakai H, Komano T (1994) Functional analysis of block 5, one of the highly conserved amino acid sequences in the 130-kDa CryIVA protein produced by Bacillus thuringiensis subsp. israelensis. FEBS Letters 348:249–254
Perlak FJ, Fuchs RL, Dean DA, McPherson SL, Fischhoff DA (1991) Modification of the coding sequence enhances plant expression of insect control protein genes. Proc Natl Acad Sci USA 88:3324–3328
Poncet S, Delécluse A, Klier A, Rapoport G (1995) Evaluation of synergistic interactions among the Cry IVA, Cry IVB, and Cry IVD toxic components of B. thuringiensis subsp. israelensis crystals. J. Invert Pathol 66:131–145
Prodromou C, Pearl LH (1992) Recursive PCR: A novel technique for total gene synthesis. Protein Eng 5:827–829
Robinson M, Lilley R, Little S, Emtage JS, Yarranton G, Stephens P, Millican A, Eaton M, Humphreys G (1984) Codon usage can affect efficiency of translation of genes in Escherichia coli. Nucleic Acids Res 12:6663–6671
Schnepf E, Crickmore N, Van Rie J, Lereclus D, Baum J, Feitelson J, Zeigler DR, Dean DH (1998) Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol Mol Biol Rev 62:775–806
Soltes-Rak E, Kushner DJ, Williams DD, Coleman JR (1995) Factors regulating cryIVB expression in the cyanobacterium Synechococcus PCC7942. Mol Gen Genet 246:301–308
Sorensen HP, Mortensen KK (2005) Soluble expression of recombinant proteins in the cytoplasm of Escherichia coli. Microb Cell Fact 4:1
Yamagiwa M, Esaki M, Otake K, Inagaki M, Komano T, Amachi T, Sakai H (1999) Activation process of dipteran-specific insecticidal protein produced by Bacillus thuiringiensis subsp. israelensis. Appl Environ Microbiol 65:3464–3469
Acknowledgement
We are grateful to the Dainihon Jochugiku Co., Ltd. for providing us with eggs of C. pipiens. This work was supported by a grant from the Program for Promotion of Basic Research Activities for Innovative Biosciences (PROBRAIN), Japan and by research grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Hayakawa, T., Howlader, M.T.H., Yamagiwa, M. et al. Design and construction of a synthetic Bacillus thuringiensis Cry4Aa gene: Hyperexpression in Escherichia coli . Appl Microbiol Biotechnol 80, 1033–1037 (2008). https://doi.org/10.1007/s00253-008-1560-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-008-1560-9