Abstract
The flexible peptides (GGGGS)n (n ≤ 3), the α-helical peptides (EAAAK)n (n ≤ 3) and two other peptides were used as linkers to construct bifunctional fusions of β-glucanase (Glu) and xylanase (Xyl) for improved catalytic efficiencies of both moieties. Eight Glu-Xyl fusion enzymes constructed with different linkers were all expressed as the proteins of ca. 46 kDa in Escherichia coli BL21 and displayed the activities of both β-glucanase and xylanase. Compared to all the characterized fusions with the parental enzymes, the catalytic efficiencies of the Glu and Xyl moieties were equivalent to 304–426% and 82–143% of the parental ones, respectively. The peptide linker (GGGGS)2 resulted in the best fusion, whose catalytic efficiency had a net increase of 326% for the Glu and of 43% for the Xyl. The two moieties of a fusion with the linker (EAAAK)3 also showed net increases of 262 and 31% in catalytic efficiency. Our results highlight, for the first time, the enhanced bifunctional activities of the Glu-Xyl fusion enzyme by optimizing the peptide linkers to separate the two moieties at a reasonable distance for beneficial interaction.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Polysaccharides, such as arabinoxylans, mixed-linked-β-glucans, celluloses, mannans, and galactans are rich in the endosperm cell walls of cereal grains (Longland et al. 1995) and can be converted into animal nutrition and fuel ethanol by microbial enzymes (Dien et al. 2003; Levasseur et al. 2005). Hydrolysis of the endosperm cell walls requires actions of different enzymes to degrade the polysaccharides into utilizable oligo- or monosaccharides. Since fibrolytic enzymes tend to evolve toward bi- or multifunctional directions for more efficient biodegradation of plant fibre (Xue et al. 1992; Flint et al. 1993), an interest in construction of bi- or multifunctional fusions of the enzymes for the degradation of cereal polysaccharides has been increasing.
Plenty of fusion enzymes have been developed by means of end-to-end fusion technique (Trujillo et al. 1997; Hong et al. 2006). With this technique, individual genes encoding intact functional proteins are fused into a chimeric gene that may encode a larger protein to perform two or more functions (Doi and Yanagawa 1999). Previous studies have shown that proper linker peptides can be inserted between individual enzymes to reduce folding interference from each other so that the two moieties of a fusion enzyme may function as independently as possible (Crasto and Feng 2000). Importantly, selected linkers are required to have a certain degree of flexibility and hydrophilicity (Gustavsson et al. 2001; Volkel et al. 2001). A web server (Xue et al. 2004) has been established to generate an array of peptide sequences as linker candidates, whose extended formations are usually known in public protein data bank. To date, a large number of peptides have been used as linkers for constructing fusion enzymes. Of those, the flexible linkers (GGGGS)n (usually n ≤ 6) are often used to separate functional domains of bi- or multifunctional fusion proteins or engineered antibodies (Lois et al. 1998; Remy et al. 1999; Shan et al. 1999; Wang et al. 2007). However, the moieties of some fusions constructed with selected linkers are not always expressed as expected and often lose their functions (Maeda et al. 1997). Apart from the flexible linkers, the rigid peptides (EAAAK)n (n ≤ 6) are also used to take the mission. For instance, an α-helical conformation is utilized to link two domains of a fusion protein by controlling a distance between them (Arai et al. 2004; Chang et al. 2005).
Arabinoxylans and β-glucans constitute major parts of the endosperm cell walls of cereal grains as animal feed and can be degraded by endo-1,4-β-xylanase (E.C. 3.2.1.8) and 1,3-1,4-β-glucanase (E.C. 3.2.1.73), respectively. Both enzymes have been separately applied in feed industry for breakdown of internal β-1,4-linkages of 1,4-β-d-xylan backbone (Salobir 1998) and specific cleavage of 1,4-β-d-glucosidic bonds adjacent to β-1,3-linkages in mixed-linked-β-glucans (Mathlouthi et al. 2002). It would be ideal to integrate both enzymes for enhanced biodegradation of β-glucans and xylans and thus for higher nutrition availability to animals (Salobir 1998; Mathlouthi et al. 2002). Previously, we constructed a bifunctional fusion of Bacillus amyloliquefaciens β-glucanase (Glu) and B. subtilis xylanase (Xyl) using the method of end-to-end fusion with no linker (Lu et al. 2006). However, this Glu-Xyl fusion was featured with decreased Xyl activity despite improved Glu activity. This study sought to enhance the activities of both moieties by optimizing peptide linkers in the Glu-Xyl fusion. Fusion enzymes with different linkers were characterized in comparison to the linker-free fusion and their parental enzymes.
Materials and methods
Strains, vectors, reagents, and enzymes
All plasmids constructed in this study were propagated in E. coli TOP10F (Invitrogen, Carlsbad, CA, USA). Two recombinant plasmids harboring β-glucanase gene Glu and xylanase gene Xyl were constructed previously (Lu et al. 2006). The inducible T7 expression vector, plasmid pET-30a(+), was purchased from Invitrogen. For protein expression, the plasmids were transformed into E. coli BL21(DE3)pLysS (Invitrogen), called BL21 herein, following the manufacturer’s manual. Cells were grown in LB medium supplemented with 100 μg ml−1 ampicillin or kanamycin when necessary.
Restriction endonucleases, T4 DNA ligase and Taq DNA polymerase from Promega (Madison, WI, USA) were used for gene cloning. Birchwood xylan and barley β-glucan from Sigma (St. Louis, MO, USA) were used as substrates in all activity assays of xylanase and β-glucanase throughout the study.
Construction of chimeric genes and recombinant plasmids
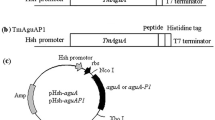
The chimeric genes were constructed using the method of splicing-by-overlap extension (Lu et al. 2006). Synthetic oligonucleotides encoding difference sequences of peptide linkers (Table 1) were introduced between the Glu excluding the stop codon and the Xyl. To express the fusion enzymes, the cleavage site of the restriction enzyme EcoRI and XholI were introduce into the Glu 5′ end and Xyl 3′ end respectively, followed by digestion with EcoRI and XholI and subsequent ligation with EcoRI I/XholI-linearized pET-30a(+). The constructed plasmids included pET-Glu-S1-Xyl, pET-Glu-S2-Xyl, pET-Glu-S3-Xyl, pET-Glu-S-Xyl, pET-Glu-α1-Xyl, pET-Glu-α2-Xyl, pET-Glu-α3-Xyl, and pET-Glu-H-Xyl, and harbored the chimeric genes Glu-S1-Xyl, Glu-S2-Xyl, Glu-S3-Xyl, Glu-S-Xyl, Glu-α1-Xyl, Glu-α2-Xyl, Glu-α3-Xyl, and Glu-H-Xyl, respectively.
Expression of the chimeric genes
The above plasmids were transformed into BL21. Ten positive transformants taken from each of the transformations were shaken at 30°C in 6 ml kanamycin-inclusive LB medium. When the OD600 of the cultures reached 0.6 (usually 6–8 h after initiation), expression of each of the chimeric genes was induced for 6 h by adding 0.5 mM isopropyl-β-d-thiogalactopyranoside to the BL21 cultures. The transformant over-expressing each of the chimeric genes (i.e., showing maximal enzymatic activity) was used in the following assays.
Purification and zymogram of the fusion enzymes
Each of the fusion enzymes for purification was produced in 500 ml baffled shake flask. Every 80-ml cell broth was sonicated for 3 s at 220–240 W in ice bath at 3-s interval during a 20-min period. The sonicated suspension was then centrifuged for 20 min at 4°C and 10,000×g. Subsequently, the fusion protein was fractioned on ice by slowly adding solid (NH4)2SO4 to the supernatant being stirred until 30% saturation (164 mg ml−1) was achieved (within 30 min). The resultant solution was kept overnight at 4°C. The precipitate was discarded and the supernatant was further saturated with solid (NH4)2SO4 to 60% saturation (345 mg ml−1). The solution was centrifuged again as above after overnight maintenance at 4°C. The precipitate was used for further purification by chromatography.
The precipitates of the fusion enzymes Glu-S1-Xyl, Glu-S2-Xyl, Glu-S3-Xyl, Glu-S-Xyl, Glu-α1-Xyl, Glu-α2-Xyl, Glu-α3-Xyl, and Glu-H-Xyl were separately dissolved in 50 mM sodium acetate buffer (pH 5.0), followed by desalting with a Sephadex™ G-25 column. Each active fraction was loaded onto a cation exchange column of HiTrap CM FF (GE Healthcare Bioscience, Shanghai, China), which was pre-equilibrated with the sodium acetate buffer (loading buffer). The column was washed with ten-bed volumes of the elution buffer (the loading buffer plus 1 M NaCl) in the gradient solutions of 0–1.0 M NaCl. The active fractions were then desalted, pooled, and determined for purity via the analysis of sodium dodecyl sulfate-polyarcylamine gel electrophoresis (SDS-PAGE). Each of the purified solutions was dried in a vacuum-freeze drier. The concentration of each active protein was determined using bovine serum albumin as standard (Bradford 1976).
Zymograms analysis was performed using the method of Morag et al. (1990). Samples of the purified fusion enzymes were run through SDS-PAGE (12.5% polyacrylamide co-polymerized with 0.5% (w/v) β-glucan or xylan). Subsequently, the gels were washed four times for 30 min at 4°C in the buffer of 0.1 mM glycin–NaOH (pH 9.0) with gentle shaking. The first two washes were conducted with 25% (v/v) isopropanol. The gels with the renatured protein were further incubated for 30 min in the same buffer at 37°C, soaked in 0.1% (w/v) Congo red for 15 min at room temperature and subsequently washed with 1 M NaCl until excessive dye was removed from the active bands. The resultant bands became clear zones after 0.5% (v/v) acetic acid was added to the gel.
Enzymatic activity assays
For all the fusion enzymes, the Xyl activity was assayed at 50°C for 10 min using 1.8 ml 1.0% (w/v) birchwood xylan in glycin–NaOH buffer (pH 9.0) and 0.2 ml enzyme solution (diluted as necessary). The amount of xylose released from the reaction solution was determined at OD540 after 3,5-dinitrosalicylic acid was added to terminate the reaction for 5-min boiling (Miller 1959). One unit of xylanase activity was defined as 1 mM xylose released per min. The Glu activity was assayed using the same method but the birchwood xylan was replaced by β-glucan as substrate for the reaction at 60°C. One unit of β-glucanase activity was expressed as 1 mM glucose released per minute.
To determine optimal temperatures for the Glu and Xyl activities, all the fusion enzymes were assayed at the gradient temperatures of 40–90°C in the above reaction system. The heat stabilities of both moieties were also assayed at 40–90°C by incubating the enzyme solution for 10 min at a given temperature. The residual activity was determined after the enzyme solution was immediately quenched on ice for 5 min. Similarly, pH effects on the enzymatic activities and stabilities were determined at pH 3.0–10.0 under the optimal temperature of 60°C for the Glu and 50°C for the Xyl, respectively. Each enzyme solution for reaction was adjusted to pH 3.0–7.0 with 0.2 M Na2HPO4–citric, to pH 8.0 with 0.2 M Na2HPO4–NaH2PO4, and to pH 9.0 and 10.0 with 0.2 M glycin–NaOH, respectively. The relative activity of each moiety was assessed as the ratio of its activity at a concerned temperature or pH over that of the same enzyme at the optimal temperature or pH. The pH stability was assayed at optimal pH 9.0 after a 4-h incubation of the pH-specific enzyme solutions at 25°C.
The kinetic parameters of all the fusion enzymes, including the Michaelis constant K m (mg ml−1), the catalytic constant K cat (min−1), and the catalytic efficiency K cat/K m (ml mg−1 min−1), were measured. The Xyl-catalyzed reaction was performed at 50°C using 1.0–10.0 mg ml−1 birchwood xylan as substrate in glycin–NaOH buffer (pH 9.0) while the Glu-catalyzed reaction was carried out at optimal 50 or 60°C using 1.0–10.0 mg ml−1 β-glucan as substrate. The K m values were determined using Lineweaver–Burk plots. All the above assays were repeated three times.
DNA sequencing and accession numbers
All DNA samples were sent to Invitrogen (Shanghai, China) for sequencing. The sequences of Glu-S1-Xyl, Glu-S2-Xyl, Glu-S3-Xyl, Glu-S-Xyl, Glu-α1-Xyl, Glu-α2-Xyl, Glu-α3-Xyl, and Glu-H-Xyl were determined based on bi-directional sequencing data from three different positive clones and were registered in GenBank, respectively (accession numbers: EU181172, EU181173, EF648220, DQ100312, EU183362, EU183363, EU365623, and DQ100311).
Results
Expression of the constructed chimeric genes
The chimeric genes constructed by linking the genes Glu and Xyl with eight of the peptide linkers were expressed well in the selected BL21 transformants. In the SDS-PAGE profiles of the crude extracts from their cell cultures, all products of the chimeric genes expressed in different transformants were the fusion enzymes of ca. 47 kDa (Fig. 1). However, the chimeric genes including the linkers S3–1, S3–2, S3–3, and S3–4 were not successfully expressed in BL21 and thus excluded for further assays.
The SDS-PAGE profiles of the Glu-Xyl fusion enzymes expressed in E. coli BL21. Lanes 3–10 were loaded with the crude extracts of the cell cultures expressing the Glu-Xyl fusion enzyme (white arrow) in the selected transformants Glu-S3-Xyl, Glu-S2-Xyl, Glu-S1-Xyl, Glu-S-Xyl, Glu-H-Xyl, Glu-α3-Xyl, Glu-α2-Xyl, and Glu-α1-Xyl, respectively. Lane 1 parental β-glucanase (Glu). Lane 2 parental xylanase (Xyl). M mol marker
Purification and zymogram of the fusion enzymes
The purified fusion enzymes Glu-S1-Xyl, Glu-S2-Xyl, Glu-S3-Xyl, Glu-S-Xyl, Glu-α1-Xyl, Glu-α2-Xyl, Glu-α3-Xyl, and Glu-H-Xyl were ca. 46 kDa in mol wt. (Fig. 2a). To confirm their bifunctional activities, the three fusions Glu-S3-Xyl, Glu-S-Xyl, and Glu-H-Xyl (Lanes 1, 4, and 5 in Fig. 2a) were selected for zymogram analysis. Their activities were confirmed for both Glu (Fig. 2b) and Xyl (Fig 2c).
The SDS-PAGE profiles (A) of the Glu-Xyl fusion enzymes purified from the cell cultures of the E. coli BL21 transformants and their bifunctional activities of β-glucanase (B) and xylanase (C) detected by zymogram analysis. Lane 1 Glu-S3-Xyl. Lane 2 Glu-S2-Xyl. Lane 3 Glu-S1-Xyl. Lane 4 Glu-S-Xyl. Lane 5 Glu-H-Xyl. Lane 6 Glu-α3-Xyl. Lane 7 Glu-α2-Xyl. Lane 8 Glu-α1-Xyl. Lane Glu parental β-glucanase. Lane Xyl parental xylanase. M mol marker. A 0.08 μM purified protein loaded in Lanes 1–3 and 0.05 μM in Lanes 4–8. B and C 0.04 μM purified protein loaded in each of the lanes
Characterization of the fusion enzymes
Among the bifunctional fusion enzymes, the Glu was most active at 60°C (Glu-S3-Xyl, Glu-S1-Xyl, Glu-S-Xyl, and Glu-H-Xyl) or 50°C (Glu-S2-Xyl, Glu-α1-Xyl, Glu-α2-Xyl, and Glu-α3-Xyl). The relative activity of this moiety decreased rapidly at ≥70°C but the declining rates differed among the fusions (Fig. 3a). In contrast, the optimal temperature for the Xyl activity was 50°C for all the fusions (Fig. 3b). However, the Xyl was much less active at 60°C for the fusions with the α-helix motif linkers. Both moieties shared an optimum of pH 9.0 for peak activity (Fig. 3c,d), being well in accordance with their parents.
The effects of temperature (A Glu; B Xyl) and pH (C Glu; D Xyl) on the activities of the moieties of the fusion enzymes (empty circle Glu-S3-Xyl; filled circle Glu-S2-Xyl; empty upright triangle Glu-S1-Xyl; filled upright triangle Glu-α3-Xyl; empty downward triangle Glu-α2-Xyl; filled downward triangle Glu-α1-Xyl; empty square Glu-S-Xyl; filled square Glu-H-Xyl; empty diamond no linker; filled diamond, parental Glu or Xyl). Error bars SD
The thermal and pH stabilities of the active moieties are illustrated for all the fusion enzymes in Fig. 4. Based on the residual activities assayed at 40–90°C for 10 min, the Glu of the fusions constructed with most of the peptide linkers showed greater stability at 40–60°C than the parent and the same moiety of the linker-free fusion (Fig. 4a). The fusion Glu-α3-Xyl had even more stable Glu at ≥60°C than the parent. All the Xyl moieties displayed thermal stabilities similar or close to the parental enzyme at ≥60°C but some of them were less stable at 40–50°C (Fig. 4b). For both Glu and Xyl, the pH stabilities of the linker-inclusive fusions were less variable at the tested pH range compared to those of their parents and the moieties of the linker-free fusion (Fig. 4c,d).
The thermal (A Glu; B Xyl) and pH (C Glu; D Xyl) stabilities of the moieties of the fusion enzymes (empty circle Glu-S3-Xyl; filled circle Glu-S2-Xyl; empty upright triangle Glu-S1-Xyl; filled upright triangle Glu-α3-Xyl; empty downward triangle Glu-α2-Xyl; filled downward triangle Glu-α1-Xyl; empty square Glu-S-Xyl; filled square Glu-H-Xyl; empty diamond no linker; filled diamond, parental Glu or Xyl). Error bars SD
Kinetic parameters for the fusion enzymes
Listed in Table 2 are the kinetic parameters of the Glu and Xyl moieties of all the fusion enzymes in comparison to those of the linker-free fusion and their parents. All the Glu moieties were significantly superior to the parental Glu in catalytic efficiency (F 9,18 = 8,146, P < 0.01). The linker S2 resulted in the best Glu efficiency (K cat/K m = 6,658), which equaled to 4.26 fold of the parental efficiency (K cat/K m = 1,563), followed by S1 (K cat/K m = 6,012) and S (K cat/K m = 5,864), respectively. The Glu moieties associated with the three linkers S3, α3, and H also displayed high efficiencies ca. 3.5 fold of the parent.
For the Xyl moiety, the catalytic efficiencies differed significantly among the fusion enzymes (F 9,18 = 1,214, P < 0.01). Their Xyl all exhibited better efficiencies (K cat/K m: 1,073–1,875) than that of the linker-free fusion (K cat/K m = 903) but only those associated with the linkers S2, α3, and α2 performed better than the parent (K cat/K m = 1,311). The efficiencies of the three Xyl moieties were equivalent to 1.43, 1.31, and 1.05 fold of the parental efficiency, respectively.
Discussion
Based on the results presented above, the eight Glu-Xyl fusion enzymes constructed with peptide linkers were all bifunctional despite conspicuous variations in optimal temperature or thermal stability and in catalytic efficiency. Among the fusion enzymes, the best Glu-S2-Xyl was characterized with the net increase of catalytic efficiency by 326% for the Glu moiety and by 43% for the Xyl compared to the parental enzymes. The fusions associated with the peptide linkers α3 and α2 also yielded the net increases of catalytic efficiencies for the Glu by 262 and 247% and for the Xyl by 31 and 5%, respectively. Both moieties of the fusion enzymes with other linkers did not show better efficiencies than their parents although a given moiety could perform better than its parent and the linker-free fusion. This is the first report on the enhanced performance of both moieties in the Glu-Xyl fusion with optimized peptide linkers. The effects of the used linkers on the performances of the fusion enzymes are discussed below.
The linkers (GGGGS)n (1 ≤ n ≤ 6) have proved to be flexible and often used in antibody engineering, which included the construction of single-chain antibody (scFv) molecules by joining the variable regions of light- and heavy-chain (V L and V H) domains (Hoedemaeker et al. 1997) and of diabodies by linking both chains (V L–V H or V H–V L) with peptides (Volkel et al. 2001) and other targeting molecules, such as the fusion of scFv and antitumor by linkers (Rosenblum et al. 2003). They are also used in the construction of bifunctional proteins, including cellular localization probes achieved by fusing green fluorescent protein to the ends of some anchoring molecules (Petriz et al. 2004) and the fusions of two enzymes that may catalyze sequential reactions (Seo et al. 2000). For a specific linker, the repeated times of the basic amino acid sequence (GGGGS) may affect a distance between the two moieties to be fused and thus vary with the different fusions. This could be chosen according to the structure and size of a concerned fusion. Shan et al. (1999) engineered four scFv molecules by linking the heavy- and light-chain variable regions of murine anti-human CD20 mAb 1F5 with the spacer peptides (GGGGS)n (n ≤ 3) and found that the short linker (GGGGS) was best. However, only the chimeric with the linker (GGGGS)6 was desired when the extracellular and transmembrane domains of murine EpoR were fused to one of two complementary fragments of murine dihydrofolate reductase (Remy et al. 1999). In this study, (GGGGS)2 was best for linking the Glu and Xyl among the tested peptides (GGGGS)n (n ≤ 3).
Moreover, it should be cautious when a nucleotide sequence encoding (GGGGS)n was designed. Since the codon of glycin is GGN (N = A, T, C, or G), the most parts of the designed nucleotide sequences may consist of G and C. The GC-rich sequences back-translated on the basis of codon-usage biases may not be suitable for direct use because the linker section in a fusion could form stable mRNA secondary structure, which can depress the translation of an intact chimeric gene (Gray and Hentze 1994). Another concern is the effect of codon pair utilization bias that may decrease the translation efficiency (Gutman and Hatfield 1989). Due to the compatibility of adjacent aminoacyl-tRNA isoacceptors at sites A and P of a translating ribosome, the codon context could affect the decoding accuracy and speed of mRNA (Irwin et al. 1995; Moura et al. 2005). This is evident with significantly increased expression of a fusion antibody by adjusting the nucleotide sequences of the (GGGGS)3 with mammalian codon pairing rules (Trinh et al. 2004). Among the five oligonucleotide sequences (S3, S3–1, S3–2, S3–3, and S3–4) tested in this study, only the chimeric of the Glu and Xyl genes linked with S3 expressed a fusion protein as was expected to be bifunctional. Interestingly, the chimeric gene fused with the linker S3–1 optimized by Trinh et al. (2004) for a constructed fusion antibody was not successfully expressed in our study. The S3–1 was originally optimized based on the codon pair usage rule of mammalian cells and thus did not suit E. coli. In fact, the S3–1 codon pairs would be superior to the S3 counterparts if the pairs were evaluated based on the same rule of E. coli (Gutman and Hatfield 1989). This suggests that not only the codon context but other factors, such as the codon-usage bias and the mRNA secondary structure, influence the translation.
The linker S (MGSSSN) designed with the web server LINKER (Xue et al. 2004) resulted in a fusion with greatly enhanced Glu, but slightly reduced Xyl, efficiency in this study. Blazyk and Lippard (2004) used four linkers to generate the fusions of the reductase soluble methane monooxygenase (sMMO) from Methylococcus capsulatus and found that a fusion with the LINKER-designed peptide consisting of 14 amino acids exhibited the best function of electronic transfer. A desired probe for dual localization of mitochondrial proteins was also achieved by fusing the α-fragment of β-galactoside to the end of a fumarase with a short linker designed with the web server (Karnielya et al. 2006). Since the linker length may affect the fusion efficiency (Crasto and Feng 2000), the linker S consisting of only six amino acids in length could be too short to separate the two moieties at a reasonable distance, which allows to enhance the activities of both Glu and Xyl. Thus, it is convenient to design peptide linkers with the web server but it is necessary to optimize their lengths for practical use.
Amino acid sequences to form α-helix or β-strand structures were not considered as favorable linkers until an α-helical bundle domain of staphylococcal protein A was successfully used in the fusion of protein G and luciferase (Maeda et al. 1997). The linker H used in this study was a bundle of two α-helices from Humicola insolens endocellulase (PDB ID: 1a39) and generated a characterized fusion similar to those fused by the flexible peptides. It is possible that the partial sequence of this linker could interact with the C and N terminal of the Glu and Xyl domains, respectively, thus disturbing the formation of the α-helical structure (Arai et al. 2004). The peptides (EAAAK)n (n ≤ 6) are monomeric α-helical sequences, which can be stabilized by Glu−-Lys+ salt bridges (Arai et al. 2001), and are less frequently used as linkers than the (GGGGS)n. However, the efficiencies of their fusions have proven to be comparable to those of the flexible linkers (Arai et al. 2001, 2004; Werner et al. 2006). Interestingly, the Glu thermal stabilities of the three fusions linked with the peptides (EAAAK)n (1 ≤ n ≤ 3) increased with the repeated times of the basic sequence; all of them were more thermally stable than those fused with the flexible linkers. Noticeably, the Glu moiety of Glu-α3-Xyl even showed higher thermal stability than the parental enzyme. This is perhaps due to the rigidity of the α-helical linker that may cause less overall interference between the linked parts. Moreover, the α3-linked fusion was secondary to the best fusion Glu-S2-Xyl when the catalytic efficiencies of both moieties were concerned. This is well in accordance with a report that helical spacers could work better than flexible ones by keeping reasonable distances between functional domains in fusion proteins (Bai and Shen 2006).
Conclusively, the performance of the Glu-Xyl fusion can be greatly improved by inserting optimized peptide linkers between the moieties. Among the tested peptides, both of the flexible glycin-rich S2 and the rigid helical α3 are excellent to link the Glu and the Xyl at a reasonable distance for beneficial interaction and thus for enhanced bifunctional activities. Changes in linker length and structure would affect overall performance of both moieties. Thus, it is essential to optimize the peptide linkers for construction of expected fusion proteins.
References
Arai R, Ueda H, Kitayama A, Kamiya N, Nagamune T (2001) Design of the linkers which effectively separate domains of a bifunctional fusion protein. Protein Eng 14:529–532
Arai R, Wriggers W, Nishikawa Y, Nagamune T, Fujisawa T (2004) Conformations of variably linked chimeric proteins evaluated by synchrotron X-ray small-angle scattering. Proteins 57:829–838
Bai Y, Shen WC (2006) Improving the oral efficacy of recombinant granulocyte colony-stimulating factor and transferrin fusion protein by spacer optimization. Pharm Res 23:2116–2121
Blazyk JL, Lippard SJ (2004) Domain engineering of the reductase component of soluble methane monooxygenase from Methylococcus capsulatus (Bath). J Biol Chem 279:5630–5640
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Chang HC, Kaiser CM, Hartl FU, Barral JM (2005) De novo folding of GFP fusion proteins: high efficiency in eukaryotes but not in bacteria. J Mol Biol 353:397–409
Crasto CJ, Feng J (2000) LINKER: a program to generate linker sequences for fusion proteins. Protein Eng 13:309–312
Dien BS, Cotta MA, Jeffries TW (2003) Bacteria engineered for fuel ethanol production: current status. Appl Microbiol Biotechnol 63:258–266
Doi N, Yanagawa H (1999) Insertional gene fusion technology. FEBS Letters 457:1–4
Flint HJ, Martin J, McPherson CA, Daniel AS, Zhang JX (1993) A bifunctional enzyme, with separate xylanase and β(1, 3-1, 4)-glucanase domains, encoded by the xynD gene of Ruminococcus flavefaciens. J Bacteriol 175:2943–2951
Gray NK, Hentze MW (1994) Regulation of protein synthesis by mRNA structure. Mol Biol Rep 19:195–200
Gustavsson M, Lehtio J, Denman S, Teeri TT, Hult K, Martinelle M (2001) Stable linker peptides for a cellulose-binding domain-lipase fusion protein expression in Pichia pastoris. Protein Eng 14:711–715
Gutman GA, Hatfield GW (1989) Nonrandom utilization of codon pairs in Escherichia coli. Proc Natl Acad Sci USA 86:3699–3703
Hoedemaeker FJ, Signorelli T, Johns K, Kuntz DA, Rose DR (1997) A single chain Fv fragment of p-glycoprotein-specific monoclonal antibody C219 design, expression, and crystal structure at 2.4 Å resolution. J Biol Chem 272:29784–29789
Hong SY, Lee JS, Cho KM, Math RK, Kim YH, Hong SJ, Cho YU, Kim H, Yun HD (2006) Assembling a novel bifunctional cellulose-xylanase from Thermotoga maritima by end-to-end fusion. Biotechnol Lett 28:1857–1862
Irwin B, Heck JD, Hatfield GW (1995) Codon pair utilization biases influence translational. J Biol Chem, 270:22801–22806
Karnielya S, Rayznera A, Sass E, Pines O (2006) a-Complementation as a probe for dual localization of mitochondrial proteins. Exp Cell Res 312:3835–3846
Levasseur A, Navarro D, Punt PJ, Belaich JP, Asther M, Record E (2005) Construction of engineered bifunctional enzymes and their overproduction in Aspergillus niger for improved enzymatic tools to degrade agricultural by-products. Appl Environ Microbiol 71:8132–8140
Lois AC, Barbara AS, John RM, Janet EP, Sharon LA, Andrew B, Glenn FP, Houston LL (1998) Targeting tumor cells via EGF receptors: selective toxicity of an HBEGF-toxin fusion protein. Int J Cancer 78:106–111
Longland AC, Theodorou MK, Sanderson R, Lister SJ, Powell CJ, Morris P (1995) Non-starch polysaccharide composition and in vitro fermentability of tropical forage legumes varying in phenolic content. Anim Feed Sci Technol 55:161–177
Lu P, Feng MG, Li WF, Hu CX (2006) Construction and characterization of a bifunctional fusion enzyme of Bacillus-sourced β-glucanase and xylanase expressed in Escherichia coli. FEMS Microbiol Lett 261:224–230
Maeda Y, Ueda H, Kazami J, Kawano G, Suzuki E, Nagamune T (1997) Engineering of functional chimeric protein G-Vargula luciferase. Anal Biochem 249:147–152
Mathlouthi N, Saulnier L, Quemener B, Larbier M (2002) Xylanase, b-glucanase, and other side enzymatic activities have greater effects on the viscosity of several feedstuffs than xylanase and b-glucanase used alone or in combination. J Agric Food Chem 50:5121–5127
Miller GL (1959) Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem 31:426–428
Moura G, Pinheiro M, Silva R, Miranda I, Afreixo V, Dias G, Freitas A, Oliveira JL, Santos MAS (2005) Comparative context analysis of codon pairs on an ORFeome scale. Genome Biol 6:R28
Petriz J, Gottesman MM, Aran JM (2004) An MDR-EGFP gene fusion allows for direct cellular localization, function and stability assessment of p-glycoprotein. Curr Drug Deliv 1:43–56
Morag E, Bayer EA, Lamed R (1990) Relationship of cellulosomal and noncellulosomal xylanases of Clostridium thermocellum to cellulose-degrading enzymes. J Bacteriol 172:6098–6105
Remy I, Wilson IA, Stephen WM (1999) Erythropoietin receptor activation by a ligand-induced conformation change. Science 283:990–993
Rosenblum MG, Cheung LH, Liu Y, Marks JW (2003) Design, expression, purification, and characterization, in vitro and in vivo, of an antimelanoma single-chain Fv antibody fused to the toxin gelonin. Cancer Res 63:3995–4002
Salobir J (1998) Effect of xylanase alone and in combination with β-glucanase on energy utilization, nutrient utilization and intestinal viscosity of broilers fed diets based on two wheat samples. Arch Gefluegelkund 62:209–213
Seo HS, Koo YJ, Lim JY, Song JT, Kim CH, Kim JK, Lee JS, Choi YD (2000) Characterization of a bifunctional enzyme fusion of trehalose-6-phosphate synthetase and trehalose-6-phosphate phosphatase of Escherichia coli. Appl Environ Microbiol 66:2484–2490
Shan D, Press OW, Tsu TT, Hayden MS, Ledbetter JA (1999) Characterization of scFv-Ig constructs generated from theAnti-CD20 mAb 1F5 using linker peptides of varying lengths. J Immunol 162:6589–6595
Trinh R, Gurbaxani B, Morrison SL, Seyfzadeh M (2004) Optimization of codon pair use within the (GGGGS)3 linker sequence results in enhanced protein expression. Mol Immunol 40:717–722
Trujillo M, Duncan R, Santi DV (1997) Construction of a homodimeric dihydrofolate reductase-thymidylate synthase bifunctional enzyme. Protein Eng 10:567–573
Volkel T, Korn T, Bach M, Muller R, Kontermann RE (2001) Optimized linker sequences for the expression of monomeric and dimeric bispecific single-chain diabodies. Protein Eng 14:815–823
Wang WWS, Das D, McQuarrie SA, Suresh MR (2007) Design of a bifunctional fusion protein for ovarian cancer drug delivery: single-chain anti-CA125 core-streptavidin fusion protein. Eur J Pharm Biopharm 65:398–405
Werner S, Marillonnet S, Hause G, Klimyuk V, Gleba Y (2006) Immunoabsorbent nanoparticles based on a tobamovirus displaying protein A. Proc Natl Acad Sci USA 103:17678–17683
Xue GP, Goblus KS, Orpin CG (1992) A novel polysaccharide hydrolase cNAD (celD) from Neocallimastix pareiciarum encoding three multi-functional catalytic domains with high endoglucanase, cellobiohydrolase and xylanase activities. J Gen Microbiol 138:2397–2403
Xue F, Gu Z, Feng J (2004) LINKER: a web server to generate peptide sequences with extended conformation. Nucleic Acids Res 32:562–565
Acknowledgements
Chang HC (Department of Cellular Biochemistry, Max Planck Institute of Biochemistry, Martinsried, Germany) is thanked for providing the nucleotide sequences of α-helical peptide linkers. This study was supported jointly by the Ministry of Science and Technology of China (2007DFA3100), the Natural Science Foundation of China (30571250), the Ministry of Education of China (IRT0535), and Zhejiang R&D Program (2007C12035).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lu, P., Feng, MG. Bifunctional enhancement of a β-glucanase-xylanase fusion enzyme by optimization of peptide linkers. Appl Microbiol Biotechnol 79, 579–587 (2008). https://doi.org/10.1007/s00253-008-1468-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-008-1468-4