Abstract
To develop a system for combinatorial biosynthesis of glycosylated macrolides, Streptomyces venezuelae was genetically manipulated to be deficient in the production of its macrolide antibiotics by deletion of the entire biosynthetic gene cluster encoding the pikromycin polyketide synthases and desosamine biosynthetic enzymes. Two engineered deoxysugar biosynthetic pathways for the biosynthesis of thymidine diphosphate (TDP)-d-quinovose or TDP-d-olivose in conjunction with the glycosyltransferase–auxiliary protein pair DesVII/DesVIII derived from S. venezuelae were expressed in the mutant strain. Feeding the representative 12-, 14-, and 16-membered ring macrolactones including 10-deoxymethynolide, narbonolide, and tylactone, respectively, to each mutant strain capable of producing TDP-d-quinovose or TDP-d-olivose resulted in the successful production of the corresponding quinovose- and olivose-glycosylated macrolides. In mutant strains where the DesVII/DesVIII glycosyltransferase–auxiliary protein pair was replaced by TylMII/TylMIII derived from Streptomyces fradiae, quinovosyl and olivosyl tylactone were produced; however, neither glycosylated 10-deoxymethynolide nor narbonolide were generated, suggesting that the glycosyltransferase TylMII has more stringent substrate specificity toward its aglycones than DesVII. These results demonstrate successful generation of structurally diverse hybrid macrolides using a S. venezuelae in vivo system and provide further insight into the substrate flexibility of glycosyltransferases.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Macrolides include some of the most clinically important antibiotics and derive their activities from the presence of a polyketide macrolactone ring with one or more deoxysugars attached. The number of cloned and sequenced genes involved in macrolide antibiotic biosynthesis has increased rapidly during the past decades, and expanded knowledge regarding genetic information has enabled manipulation of the polyketide synthase (PKS) and deoxysugar biosynthetic pathways to create novel macrolide antibiotics (Rodriguez and McDaniel 2001). Therefore, the combinatorial biosynthetic approach of modifying macrolide structures by combining genes responsible for the biosynthesis of macrolactone rings, deoxysugar constituents, and their transfer has been pursued (Méndez and Salas 2001; Blanchard and Thorson 2006).
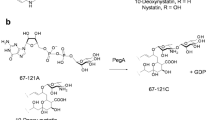
Streptomyces venezuelae has recently been developed as a host for combinatorial biosynthesis and heterologous expression of macrolide antibiotic pathways originated from other species of Streptomyces (Yoon et al. 2002; Hong et al. 2004; Jung et al. 2006). In addition to being amenable to genetic manipulation, S. venezuelae has a relatively fast growth rate (doubling time, approximately 1 h) and requires a short culture period (3–4 days) for metabolite production (Xue and Sherman 2001; Jung et al. 2006). This strain also has another favorable feature for combinatorial biosynthesis in that it can produce both 12- and 14-membered ring macrolides (YC-17 and its hydroxylated forms, ∼10 mg/l; narbomycin and its hydroxylated forms, ∼40 mg/l) by the action of a single PKS (Fig. 1a). Another advantage of using S. venezuelae as a host for combinatorial biosynthesis is the presence of its native glycosyltransferase, DesVII (Fig. 1a), together with its auxiliary protein, DesVIII (Borisova et al. 2004), which have unusual substrate flexibility toward both aglycones and deoxysugars. Recent reports demonstrating the flexibility of DesVII at utilizing non-native deoxysugars (Zhao et al. 1998a, b, 2001; Yamase et al. 2000; Hong et al. 2004) and non-native aglycones (Tang and McDaniel 2001; Borisova et al. 2006; Jung et al. 2006) as substrates have made this enzyme an important tool for combinatorial biosynthesis (Table 1). These characteristics make S. venezuelae an attractive host for generating novel macrolide antibiotics by combinatorial biosynthetic approaches. However, a combinatorial biosynthesis system based on S. venezuelae, which is able to attach non-native deoxysugars to non-native aglycones, has not yet been developed.
In this study, we engineered a strain of S. venezuelae that had the entire biosynthetic gene cluster encoding pikromycin (Pik) PKS and desosamine (des) biosynthetic enzymes deleted. Subsequently, engineered biosynthetic pathways for thymidine diphosphate (TDP)-d-quinovose or TDP-d-olivose together with glycosyltransferases were expressed in the S. venezuelae deletion mutant. Representative 12-, 14-, and 16-membered ring macrolactones such as 10-deoxymethynolide, narbonolide, and tylactone were fed to the mutant strains, which in turn successfully produced quinovosyl and olivosyl derivatives by the action of DesVII/DesVIII. Additionally, we replaced DesVII/DesVIII with another substrate-flexible glycosyltransferase TylMII (Fig. 1b) coupled with its partner protein TylMIII (Melançon et al. 2004), derived from tylosin-producing Streptomyces fradiae to investigate the substrate flexibility of TylMII. This work demonstrates successful production of hybrid macrolide antibiotics in engineered strains of S. venezuelae and provides a new tool for investigating substrate flexibility of glycosyltransferases.
Materials and methods
Bacterial strains, culture conditions, and genetic manipulation
S. venezuelae ATCC15439 was used for the construction of mutant strains. S. venezuelae mutant strains were cultured in SCM liquid medium (Yoon et al. 2002) containing appropriate antibiotics. S. fradiae ATCC19609 grown in tryptic soy broth (Kieser et al. 2000) was used for the preparation of genomic DNA. Genetic manipulation was performed in Escherichia coli DH5α according to standard procedures (Sambrook et al. 2001). Litmus 28 (New England Biolabs), T-easy vector (Promega), and pGEM-3Zf(+) (Promega) were used for subcloning, and Southern blot hybridization was conducted using a digoxigenin-labeling and detection kit following the manufacturer’s protocols (Roche).
Construction of a clean host bearing a deletion of Pik PKS and des cluster
To construct a Pik PKS and des gene cluster deletion mutant of S. venezuelae, a deletion plasmid, pYJ188, derived from pKC1139 (Bierman et al. 1992) that contained an apramycin resistant marker was constructed by cloning two 1-kb fragments homologous to the upstream and downstream regions of the des cluster. The HindIII–PstI fragment of the upstream region and the KpnI–EcoRI fragment of the downstream region of the des cluster were obtained from pYJ100 (Hong et al. 2004) and subsequently subcloned into pGEM-3Zf(+). The resulting HindIII–EcoRI fragment containing the segments upstream and downstream of the des cluster was transferred to pKC1139 to form a plasmid that was designated pYJ188. This plasmid was then introduced into protoplasts of the Pik PKS deletion mutant DHS2001 (Jung et al. 2006) for plasmid-mediated homologous recombination. Double-crossover mutants were identified on the basis of their phenotype of apramycin sensitivity and their genotype by Southern blot hybridization. The resulting strain was designated YJ028.
Construction of expression plasmids and mutant strains
To synthesize a key intermediate sugar, TDP-4-keto-6-deoxy-d-glucose (Fig. 2a) and confer self-resistance against the macrolide antibiotics, the integrative plasmid pYJ136 containing desIII, desIV, and desR was used as previously described (Hong et al. 2004). This plasmid is a derivative of pSET152 (Bierman et al. 1992), which carries the ΦC31 attachment region (attP) and integrates at the bacterial attachment site (attB). The plasmid pYJ136 was integrated into the chromosomal DNA of the mutant strain YJ028 to generate YJ069. The plasmids pYJ142, which contains desVII and desVIII for the transfer of TDP-d-quinovose (Fig. 2a), and pYJ146, which contains oleV, oleW, urdR, desVII, and desVIII for biosynthesis and transfer of TDP-d-olivose (Fig. 2a), were constructed as previously described (Hong et al. 2004). Four plasmids, pYJ320, pYJ243, pYJ321, and pYJ249, were constructed for the biosynthesis of TDP-d-quinovose and TDP-d-olivose and the transfer of them by TylMII/TylMIII, as described below. The tylMII gene was amplified from the genomic DNA of S. fradiae by polymerase chain reaction (PCR) using the following primers: 5′-GAGGATCCGTCTGAGGGCGCTCGCCGA-3′ and 5′-TGCGGGATCCCCGTGGCGGATGAATGGGCC-3′ (BamHI sites are italicized). The PCR product of tylMII was cut with BamHI and cloned into the same site of pSE34 (Yoon et al. 2002), generating pYJ320. The BamHI fragment containing tylMII was also transferred to the same site of pYJ196 (Hong et al. 2007), which is a pSE34 derivative inserted with a BamHI–XbaI fragment containing tylMIII, yielding pYJ243. The XbaI–HindIII fragment containing oleV, oleW, and urdR genes isolated from pYJ144 (Hong et al. 2004), in which desVII, oleV, oleW, and urdR genes had been cloned, was subcloned into pYJ320 to generate pYJ321. The tylMIII gene was amplified by PCR using the oligonucleotides 5′-ACCGTTCTAGACGGCGGGGAGAGAGGAGAGGCC-3′and5′-CGGCGTCTAGACAGTGCCCTTCTCACTCGGGGA-3′ (XbaI sites are italicized). The tylMIII gene was excised by XbaI and cloned into pYJ321 to construct pYJ249. The strains and plasmids used in this study are summarized in Table 2.
Production and purification of 10-deoxymethynolide, narbonolide, and tylactone
The des deletion mutant YJ003 (Hong et al. 2004) accumulating 10-deoxymethynolide and narbonolide was cultured with appropriate antibiotics on SPA solid medium (Yoon et al. 2002) at 30 °C for 3 days. The mutant strain YJ005 (Jung et al. 2006) accumulating tylactone was cultured on R2YE solid medium (Kieser et al. 2000) at 30 °C for 4 days under appropriate antibiotic selection. Agar-grown cultures of YJ003 and YJ005 were diced and extracted with 2 vol of methanol. The extracts of YJ003 and YJ005 were washed with 1 vol of water and then extracted with 1 vol of ethylacetate. The crude extracts of YJ003 and YJ005 were purified as described previously (Lee et al. 2006) to obtain purified 10-deoxymethynolide, narbonolide, and tylactone. The purity of these compounds was checked by liquid chromatography/mass spectrometry (LC/MS) and tandem mass spectrometry (MS/MS). The tylactone standard was obtained from Professor Eric Cundliffe (University of Leicester).
Bioconversion and analysis of macrolides in the mutant S. venezuelae
Mutant strains were cultured in 50 ml of SCM liquid medium at 30 °C for 24 h under appropriate antibiotic selection. Five hundred micrograms equivalent to 10 mg/l of purified 10-deoxymethynolide, narbonolide, or tylactone was added into each growing medium and then incubated for additional 48 h. The resulting culture broth was extracted using 1 vol of chloroform and concentrated with methanol. The products were analyzed using a combination of LC/MS and MS/MS. LC was performed with a Waters (Milford, MA) Model 2690 separation module, using an Xterra column (4.6 × 15 mm, 5 μm) with a binary mobile phase (solvent A: acetonitrile/methanol = 4:1 with 5 mM ammonium acetate; solvent B: 5 mM ammonium acetate in H2O; A: 20–75%, 30 min, 75%, 10 min; 75–20%, 10 min) at a flow rate of 0.15 ml/min. The effluent was applied to the Micromass (Baverly, MA) Quattro LC triple-tandem quadrupole mass spectrometer and mass spectrometric data were acquired in the positive-ion mode. The relative amount of each compound produced was compared using the peak intensity obtained from the LC/MS chromatogram. The production level of each glycosylated compound was calculated by averaging the yield from five separate cultivations and extractions.
Results
Construction of the mutant YJ028 bearing Pik cluster deletion and the mutant YJ069 synthesizing TDP-4-keto-6-deoxy-d-glucose
The des cluster was deleted in-frame from the chromosome of the Pik PKS deletion mutant DHS2001 (Jung et al. 2006) by homologous recombination to generate YJ028 (Fig. 3a). Southern hybridization analysis of the genomic DNA from several recombinant clones was performed to identify the mutant strain with the desired double-crossover recombination (Fig. 3b). The culture extract of a Pik PKS and des deletion mutant YJ028 was analyzed by LC/MS, and the production of macrolactone aglycone and glycosylated macrolides was not detected. Feeding 10-deoxymethynolide and narbonolide to the growing medium of YJ028 did not lead to the generation of the corresponding desosaminyl compounds, indicating that the des cluster was inactivated (data not shown). The integrative plasmid pYJ136 (Hong et al. 2004), which contains desIII, desIV for the biosynthesis of a key intermediate sugar TDP-4-keto-6-deoxy-d-glucose (Fig. 2a) and desR for conferring self-resistance against macrolide antibiotics, was integrated into the chromosomal DNA of YJ028 to obtain YJ069 (Fig. 3a). A previous study has shown that this intermediate sugar is converted to TDP-d-quinovose by a specific pathway-independent reduction capability of the wild-type S. venezuelae (Borisova et al. 1999); thus, the reintroduction of two genes (desIII, desIV) into the strain YJ028 is sufficient for biosynthesis of TDP-d-quinovose (Fig. 2a). Southern hybridization analysis of the genomic DNA from YJ069 revealed that the above three genes (desIII, desIV, desR) are integrated into the chromosome (Fig. 3c).
Construction of the mutant YJ028 bearing Pik cluster deletion and desR-, desIII-, and desIV- integrated mutant YJ069. a Genetic organization of pikA deletion mutant DHS2001, pikA and des deletion mutant YJ028, and desR-, desIII-, and desIV-integrated mutant YJ069. The endonuclease restriction sites are abbreviated as follows: Bg BglII; P PstI; Ba BamHI, X XbaI. b Southern blot analysis of PstI- and BglII-digested genomic DNA of DHS2001 and YJ028. Lanes 1 (DHS2001) and 2 (YJ028) were probed by a pikC gene. c Southern blot analysis of BamHI- and XbaI-digested genomic DNA of YJ028 and YJ069. Lanes 1 (YJ028) and 2 (YJ069) were probed by a desIII gene
Generation of quinovose- and olivose-glycosylated macrolides by DesVII/DesVIII
The plasmids pYJ142 containing two genes (desVII, desVIII) involved in the transfer of TDP-d-quinovose and pYJ146 containing five genes (oleV, oleW, urdR, desVII, desVIII) involved in the biosynthesis and transfer of TDP-d-olivose (Hong et al. 2004) were transformed into the mutant YJ069 to generate YJ069/pYJ142 and YJ069/YJ146, respectively. To validate the biosynthesis of TDP-d-quinovose and TDP-d-olivose in these engineered strains of S. venezuelae, 10-deoxymethynolide or narbonolide was fed to each growing medium of YJ069/pYJ142 and YJ069/pYJ146. Quinovosyl derivatives of 10-deoxymethynolide and narbonolide (Fig. 2b) were observed at retention times of 22.5 min with m/z = 465 (Fig. 4a) and 24.7 min with m/z = 521 (Fig. 4b), respectively. The compounds with masses corresponding to olivosyl derivatives of 10-deoxymethynolide and narbonolide (Fig. 2b) were detected with retention times of 25.5 min with m/z = 449 (Fig. 4d) and 27.4 min with m/z = 505 (Fig. 4e), respectively. The masses given here and below correspond to the sodium adduct of each compound. The MS/MS fragmentation patterns of these compounds were identical to those of quinovosyl and olivosyl derivatives previously reported (Hong et al. 2004), validating that the S. venezuelae system developed in this study was capable of bioconverting aglycone to quinovose- and olivose-glycosylated compounds. When the production of glycosylated macrolides in the mutant strains was assessed at each 24 h interval up to 4 days, the conversion yield of quinovosyl and olivosyl compounds measured after 72 h reached a maximum of approximately 4 and 3% for 10-deoxymethynolide, respectively, and 9 and 6% for narbonolide, respectively, based on the comparison of the peak intensity on the LC/MS chromatogram. To investigate whether quinovosyl and olivosyl tylactone (Fig. 2b) was synthesized by the action of DesVII, tylactone was supplemented to the growing medium of YJ069/pYJ142 and YJ069/pYJ146. The organic extract of YJ069/pYJ142 fed with tylactone was analyzed by LC/MS, and a peak with m/z = 563 was observed at a retention time of 28.5 min, corresponding to quinovosyl tylactone (Fig. 4c). A peak with m/z = 547 was detected in the organic extract of YJ069/pYJ146 fed with tylactone at a retention time of 30.9 min, corresponding to olivosyl tylactone (Fig. 4f). Quinovosyl tylactone with m/z = 563 fragmented into characteristic ions at m/z = 545 corresponding to the dehydrated form of quinovosyl tylactone, at m/z = 417 for the loss of quinovose, and at m/z = 399 corresponding to the dehydrated form of tylactone from the parent ion upon MS/MS spectrometry (Fig. 5a). Olivosyl tylactone with m/z = 547 also resulted in characteristic ions at m/z = 528, corresponding to the dehydrated form of olivosyl tylactone, at m/z = 417 for the loss of olivose, and at m/z = 399 corresponding to the dehydrated form of tylactone in MS/MS analysis (Fig. 5b). The MS/MS fragmentation pattern of standard tylactone was shown in Fig. 5c. Approximately 8 and 3% of tylactone was converted to quinovosyl and olivosyl tylactone, respectively.
LC/MS analyses of quinovosyl and olivosyl derivatives produced by DesVII/DesVIII. a LC/MS selected for m/z = 465, corresponding to quinovosyl 10-deoxymethynolide. b LC/MS selected for m/z = 521, corresponding to quinovosyl narbonolide. c LC/MS selected for m/z = 563, corresponding to quinovosyl tylactone. d LC/MS selected for m/z = 449, corresponding to olivosyl 10-deoxymethynolide. e LC/MS selected for m/z = 505, corresponding to olivosyl narbonolide. f LC/MS selected for m/z = 547, corresponding to olivosyl tylactone. Abbreviations were as follows: 10-DML 10-deoxymethynolide, NL narbonolide, TL tylactone
Generation of quinovose- and olivose-glycosylated macrolides by TylMII/TylMIII
Another glycosyltransferase, TylMII, and its partner protein, TylMIII, derived from S. fradiae have been used to transfer non-native deoxysugars (Gaisser et al. 2000; Melançon et al. 2005) to its native aglycone, tylactone, and its native deoxysugar, mycarminose to non-native hybrid aglycones (Reeves et al. 2004). In this study, we investigated the substrate flexibility of TylMII toward non-native deoxysugars including TDP-d-quinovose and TDP-d-olivose and aglycones including 10-deoxymethynolide, narbonolide, and tylactone. The growing medium of YJ069/pYJ243 and YJ069/pYJ249 in which TDP-d-quinovose or TDP-d-olivose deoxysugar pathway was expressed, respectively, together with genes encoding for TylMII/TylMIII, were fed with 10-deoxymethynolide or narbonolide. Extracts obtained from the strains YJ069/pYJ243 and YJ069/pYJ249 fed with 10-deoxymethynolide and narbonolide were analyzed by LC/MS; however, no peaks corresponding to 10-deoxymethynolide and narbonolide derivatives were detected, indicating the absence of the corresponding glycosylated compounds in the growing medium. Tylactone was also fed to the growing medium of YJ069/pYJ243 or YJ069/pYJ249. In LC/MS analyses, the peaks corresponding to quinovose- and olivose-glycosylated tylactone were detected at the same retention time of these compounds produced by YJ069/pYJ142 and YJ069/pYJ146, respectively, in which DesVII/DesVIII was expressed as a glycosyltransferase and auxiliary protein pair, demonstrating the presence of quinovosyl and olivosyl tylactone in the growing medium of YJ069/pYJ243 and YJ069/pYJ249, respectively. The conversion yields of quinovose- and olivose-glycosylated tylactone by TylMII were approximately 7 and 6%, respectively.
Discussion
The antibacterial activity of macrolide antibiotics often depends on the presence and type of deoxysugar moiety attached to the aglycone. Efforts to alter the glycosylation pattern of macrolide antibiotics could have practical implications because they lead to the creation of structurally diverse antibiotics with the potential of enhanced biological activity (Méndez and Salas 2001). In this study, we developed genetically engineered mutants of S. venezuelae expressing enzymes for the synthesis of nucleotide-activated sugars and glycosyltransferase–auxiliary protein pairs, which can generate novel glycosylated macrolides when fed with an exogenous source of macrolactone. Two mutant strains YJ069/pYJ142 (producing TDP-d-quinovose) and YJ069/pYJ146 (producing TDP-d-olivose) expressing the DesVII/DesVIII glycosyltransferase–auxiliary protein pair were cultured in the growing medium supplemented by 10-deoxymethynolide and narbonolide, leading to the successful production of quinovose- and olivose-glycosylated macrolides. Using these two mutant strains, we also successfully converted the 16-membered ring aglycone, tylactone to quinovosyl or olivosyl tylactone. Quinovosyl tylactone was previously synthesized in vitro using the purified enzymes of DesVII/DesVIII (Borisova et al. 2006), while biosynthesis of olivosyl tylactone has not been reported. The olivosyl tylactone is another example of a novel glycosylated 16-membered macrolide produced by the substrate-flexible glycosyltransferase DesVII, further expanding the usefulness of this glycosyltransferase as a tool for the attachment of non-native deoxysugars to non-native aglycones. In the mutant strain YJ069/pYJ142, the conversion yields of 10-deoxymethynolide, narbonolide, and tylactone to the corresponding quinovosyl derivatives were approximately 4, 9, and 8%, respectively. Olivosyl 10-deoxymethynolide, narbonlide, and tylactone were also synthesized in the YJ069/pYJ146 at the conversion yields of approximately 3, 6, and 3%, respectively. These results suggest that the 14-membered macrolide, narbonolide, is the best aglycone substrate of glycosyltransferase DesVII among the representative 12-, 14-, and 16-membered macrolides when non-native deoxysugar TDP-d-quinovose or TDP-d-olivose was used as a deoxysugar donor.
We also tested the flexibility of TylMII glycosyltransferase derived from S. fradiae toward deoxysugars and aglycones. The supplementation of 10-deoxymethynolide and narbonolide to two mutant strains YJ069/pYJ243 (producing TDP-d-quinovose) and YJ069/pYJ249 (producing TDP-d-olivose) expressing TylMII/TylMIII resulted in the failure to generate glycosylated macrolides. On the other hand, the 16-membered ring aglycone, tylactone, was glycosylated by TylMII to produce quinovosyl and olivosyl tylactone. These results indicated that TylMII has more stringent substrate specificity toward aglycone than DesVII because it can transfer non-native deoxysugars only to its native aglycone substrate, tylactone.
One of the major limitations on the combinatorial biosynthesis of hybrid macrolides is the significantly low yield of hybrid compounds (Tang and McDaniel 2001; Yoon et al. 2002). The yield of new macrolides, olivosyl tylactone produced from YJ069/pYJ146 and YJ069/pYJ249 fed with tylactone, required sensitive analytical methods such as LC/MS for detection. Although the glycosylation site of olivosyl tylactone was not determined because of this low conversion yield, it is likely that TDP-d-olivose was introduced to the oxygen at position C-5 of tylactone, according to the intrinsic catalytic specificity of TylMII (Fig. 1b) and DesVII (Borisova et al. 2006). Further analysis, such as by nuclear magnetic resonance spectroscopy (NMR), is required to confirm the glycosylation site of tylactone.
The recent studies have shown that a number of glycosyltransferase including DesVII and TylMII require their auxiliary proteins for the efficient transfer of deoxysugar to aglycone (Melançon et al. 2004; Borisova et al. 2004; Hong et al. 2005), and interspecies complementation was successfully carried out by expressing DesVIII homologs including EryCII from Saccharopolyspora erythraea, OleP1 from Streptomyces antibioticus, and DnrQ from Streptomyces peucetius in the desVIII deletion mutant of S. venezuelae (Hong et al. 2007). We carried out two additional studies to examine this requirement of auxiliary protein for efficient glycosylation and possibility of interspecies complementation between the two auxiliary proteins DesVIII and TylMIII. First, the expression of DesVII or TylMII lacking their partner protein DesVIII or TylMIII, respectively, did not lead to the production of glycosylated products (data not shown), confirming the requirement of the auxiliary protein for the catalytic activities of DesVII and TylMII. Second, changing the partner proteins by expressing DesVII/TylMIII or TylMII/DesVIII also resulted in the failure to produce any glycosylated compound (data not shown). These results strongly suggested that the native auxiliary proteins, TylMIII and DesVIII, were necessarily required for the in vivo activity of their native partner glycosyltransferase TylMII and DesVII, respectively.
The newly developed S. venezuelae mutants described in this study provide a simple system for bioconversion of aglycones to structurally diverse glycosylated macrolides. In addition, we can investigate the flexibility of glycosyltransferase toward aglycone and deoxysugar and the function of the auxiliary protein to activate its glycosyltransferase enzyme. Although the glycosylated macrolides were synthesized at a low conversion yield similar to other attempts for combinatorial biosynthesis (Tang and McDaniel 2001; Pérez et al. 2006), S. venezuelae has still an attractive feature in that it grows faster than other Streptomyces species and requires a relatively short culture period (3 days) for complete conversion. Further studies on enhancing the conversion yield of glycosylated macrolides will facilitate the development of S. venezuelae as an efficient host for combinatorial biosynthesis of glycosylated compounds.
References
Bierman M, Logan R, O’Brien K, Seno ET, Rao RN, Schoner BE (1992) Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43–49
Blanchard S, Thorson JS (2006) Enzymatic tools for engineering natural product glycosylation. Curr Opin Chem Biol 10:263–271
Borisova SA, Zhao L, Sherman DH, Liu HW (1999) Biosynthesis of desosamine: construction of a new macrolide carrying a genetically designed sugar moiety. Org Lett 1:133–136
Borisova SA, Zhao L, Melançon CE, Kao CL, Liu HW (2004) Characterization of the glycosyltransferase activity of DesVII: analysis and implication for the biosynthesis of macrolide antibiotics. J Am Chem Soc 126:6534–6535
Borisova SA, Zhang C, Takahashi H, Zhang H, Wong AW, Thorson JS, Liu HW (2006) Substrate specificity of the macrolide-glycosylating enzyme pair DesVII/DesVIII: opportunity, limitations, and mechanistic hypotheses. Angew Chem Int Ed 45:2748–2753
Gaisser S, Reather J, Wirtz G, Kellenberger L, Staunton J, Leadlay P (2000) A defined system for hybrid macrolide biosynthesis in Saccharopolyspora erythraea. Mol Microbiol 36:391–401
Hong JSJ, Park SH, Choi CY, Sohng JK, Yoon YJ (2004) New olivosyl derivatives of methymycin/pikromycin from an engineered strain of Streptomyces venezuelae. FEMS Microbiol Lett 238:291–399
Hong JSJ, Kim WS, Lee SK, Koh HS, Park HS, Park SJ, Kim YS, Yoon YJ (2005) The role of a second protein (DesVIII) in glycosylation for the biosynthesis of hybrid macrolide antibiotics in Streptomyces venezuelae. J Microbiol Biotechnol 15:640–645
Hong JSJ, Park SJ, Parajuli N, Park SR, Koh HS, Jung WS, Choi CY, Yoon YJ (2007) Functional analysis of DesVIII homologues involved in glycosylation of macrolide antibiotics by interspecies complementation. Gene 386:123–130
Jung WS, Lee SK, Hong JSJ, Rark SR, Jeong SJ, Han AR, Sohng JK, Kim BG, Choi CY, Sherman DH, Yoon YJ (2006) Heterologous expression of tylosin polyketide synthase and production of a hybrid macrolide in Streptomyces venezuelae. Appl Microbiol Biotechnol 72:763–769
Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hoopwood DA (2000) Practical Streptomyces genetics. John Innes Centre, Norwich
Lee SK, Park JW, Kim JW, Jung WS, Park SR, Choi CY, Kim ES, Kim BS, Ahn JS, Sherman DH, Yoon YJ (2006) Neopikromycin and novapikromycin from the pikromycin biosynthetic pathway of Streptomyces venezuelae. J Nat Prod 69:847–849
Melançon CE, Liu HW (2007) Engineered biosynthesis of macrolide derivatives bearing the non-natural deoxysugars 4-epi-D-mycaminose and 3-N-monomethylamino-3-deoxy-D-fucose. J Am Chem Soc 129:4896–4897
Melançon CE, Takahashi H, Liu HW (2004) Characterization of tylM3/tylM2 and mydC/mycB pairs required for efficient glycosyltransfer in mcrolide antibiotic biosynthesis. J Am Chem Soc 126:16725–16727
Melançon CE, Yu WL, Liu HW (2005) TDP-mycaminose biosynthetic pathway revised and conversion of desosamine pathway to mycaminose pathway with one gene. J Am Chem Soc 127:12240–12241
Méndez C, Salas JA (2001) Altering the glycosylation pattern of bioactive compounds. Trends Biotechnol 11:449–456
Pérez M, Lombó F, Baig I, Braña AF, Rohr J, Salas JA, Méndez C (2006) Combinatorial biosynthesis of antitumor deoxysugar pathways in Streptomyces griseus: reconstitution of “unnatural natural gene cluster” for the biosynthesis of four 2,6-D-dideoxyhexoses. Appl Environ Microbiol 72:6644–6652
Reeves CD, Ward SL, Revill WP, Suzuki H, Marcus M, petrakovsky OV, Marquez S, Fu H, Dong SD, Katz L (2004) Production of hybrid 16-membered macrolides by expressing combinations of polyketide synthease genes in engineered Streptomyces fradiae hosts. Chem Biol 11:1465–1472
Rodriguez E, McDaniel R (2001) Combinatorial biosynthesis of antimicrobials and other natural products. Cur Opin Microbiol 4:526–534
Sambrook J, Fritsch EF, Maniatis T (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY
Tang L, McDaniel R (2001) Construction of desosamine containing polyketide libraries using a glycosyltransferase with broad substrate specificity. Chem Biol 8:547–555
Xue Y, Sherman DH (2001) Biosynthesis and combinatorial biosynthesis of pikromycin-related macrolides in Streptomyces venezuelae. Metab Eng 3:15–26
Yamase H, Zhao L, Liu HW (2000) Engineering a hybrid sugar biosynthetic pathway: production of L-rhamnose and its implication on dihydrostreptose biosynthesis. J Am Chem Soc 122:12397–12398
Yoon YJ, Beck JB, Kim BS, Kang HY, Reynolds KA, Sherman DH (2002) Generation of multiple bioactive macrolides by hybrid modular polyketide synthases in Streptomyces venezuelae. Chem Biol 9:203–214
Zhao L, Sherman DH, Liu HW (1998a) Biosynthesis of desosamine: construction of a new methymycin/neomethymycin analogue by deletion of a desosamine biosynthetic gene. J Am Chem Soc 120:10256–10257
Zhao L, Que NLS, Xue Y, Sherman DH, Liu HW (1998b) Mechanistic studies of desosamine biosynthesis: C-4 deoxygenation precedes C-3 transamination. J Am Chem Soc 120:12159–12160
Zhao L, Borisova SA, Yeung SM, Liu HW (2001) Study of C-4 deoxygenation in the biosynthesis of desosamine: evidence implicating a novel mechanism. J Am Chem Soc 123:7909–7910
Acknowledgments
We thank Professor Eric Cundliffe (University of Leicester) for kindly providing a tylactone standard. This work was supported by a grant (20050401-034-682-006-02-00) from the BioGreen 21 Program and grants from the National R&D Program for Cancer Control (0620300-1) and the SRC program of KOSEF through the Center for Intelligent Nano-Bio Materials at Ewha Womans University (grant R11-2005-008-00000-0). The authors wish to thank the Ministry of Education for the Brain Korea 21 fellowship.
Author information
Authors and Affiliations
Corresponding author
Additional information
Won Seok Jung and Ah Reum Han contributed equally to this work.
Rights and permissions
About this article
Cite this article
Jung, W.S., Han, A.R., Hong, J.S.J. et al. Bioconversion of 12-, 14-, and 16-membered ring aglycones to glycosylated macrolides in an engineered strain of Streptomyces venezuelae . Appl Microbiol Biotechnol 76, 1373–1381 (2007). https://doi.org/10.1007/s00253-007-1101-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-007-1101-y