Abstract
The carbazole-catabolic plasmid pCAR1 isolated from Pseudomonas resinovorans strain CA10 was sequenced in its entirety; and it was found that pCAR1 carries the class II transposon Tn4676 containing carbazole-degradative genes. In this study, a new plasmid designated pCAR2 was isolated from P. putida strain HS01 that was a transconjugant from mating between the carbazole-degrader Pseudomonas sp. strain K23 and P. putida strain DS1. Southern hybridization and nucleotide sequence analysis of pCAR1 and pCAR2 revealed that the whole backbone structure was very similar in each. Plasmid pCAR2 was self-transmissible, because it was transferred from strain HS01 to P. fluorescens strain IAM12022 at the frequency of 2×10−7 per recipient cell. After the serial transfer of strain HS01 on rich medium, we detected the transposition of Tn4676 from pCAR2 to the HS01 chromosome. The chromosome-located copy of Tn4676 was flanked by a 6-bp target duplication, 5′-AACATC-3′. These results experimentally demonstrated the transferability of pCAR2 and the functionality of Tn4676 on pCAR2. It was clearly shown that plasmid pCAR2 and transposon Tn4676 are active mobile genetic elements that can mediate the horizontal transfer of genes for the catabolism of carbazole.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Carbazole (CAR) is a nitrogen-containing compound present in fuels derived from petroleum, oil shale, and tar sand sources (Mushrush et al. 1999). It is a recalcitrant azarene compound that is composed of a dibenzopyrrole ring (Fig. 1) and possesses mutagenic and toxic activity (Arcos and Argus 1968). Previously, we analyzed CAR degradation by Pseudomonas resinovorans strain CA10, which is able to grow on CAR as its sole source of carbon, nitrogen, and energy (Ouchiyama et al. 1993). The CAR-degradative car genes and their flanking regions have been cloned from strain CA10 and sequenced (Sato et al. 1997a, b; Nojiri et al. 2001). Also, the Car enzymes have been characterized and the degradation pathway for this compound has been completely characterized (Fig. 1; Nam et al. 2002; Nojiri et al. 2003; Habe et al. 2003; Iwata et al. 2003). CAR has a chemical structure similar to dioxin; and the degradation pathways of these two chemicals are highly homologous (Nojiri and Omori 2002). In fact, the Car enzymes can catalyze the respective degradation steps for dioxin (Sato et al. 1997a; Habe et al. 2001; Nojiri et al. 2003; Iwata et al. 2003).
Degradative pathway of CAR by Car enzymes encoded by car gene clusters distributed among Gram-negative bacteria, including P. resinovorans strain CA10. Compounds: I CAR, II 2′-aminobiphenyl-2,3-diol, III 2-hydroxy-6-oxo-6-(2′-aminobiphenyl)-hexa-2,4-dienoic acid, IV 2-hydroxypenta-2,4-dienoic acid, V anthranilic acid
Previously, we isolated other CAR/dioxin-degrading bacteria, which have car gene clusters that are nearly identical to that of strain CA10 and are carried either chromosomally or on plasmids. These bacteria were isolated from various sites in Japan, suggesting that the car genes have become broadly disseminated among different bacterial species, such as P. resinovorans strain CA06, Novosphingobium sp. strain J30, and Pseudomonas sp. strains K15, K22, and K23 (Ouchiyama et al. 1993, 1998; Habe et al. 2002; Inoue et al. 2004). Many degradative genes such as those for toluene, biphenyl, chlorobenzene, and so on have been detected from different environmental bacteria and most of these genes were shown to be contained within mobile genetic elements that include both plasmids and transposons (Tsuda and Iino 1987, 1988, 1990; Furukawa et al. 1989; van der Meer et al. 1991a, b; Springael et al. 1993; Davison 1999; Tsuda et al. 1999; Sentchilo et al. 2000; Tsuda and Genka 2001; Tsuda and Iino 1987). Based on the above facts, it is likely that the car genes are transferred horizontally by such genetic elements. In our previous study, we showed that the car operon of strain CA10 is localized on a large circular plasmid, designated pCAR1 (Nojiri et al. 2001). We determined the entire nucleotide sequence of pCAR1 by shotgun-sequencing and found genes that are homologous to the tra and trh genes whose products are involved in the conjugal transfer of plasmids R27 and Rts1 and/or the conjugative genomic island R391(Maeda et al. 2003). This indicates the possibility that pCAR1 is a self-transmissible plasmid. However, its transferability has not been experimentally proven. In contrast, as shown in Fig. 2, several genes were found on pCAR1 which were related to class II (Tn3 family) transposons (Grindley et al. 2002) carrying toluene/xylene-degradative xyl genes on the TOL plasmid pWW0 (Williams and Murray 1974; Burlage et al. 1989; Assinder and Williams 1990; Greated et al. 2002). The tnpRaAa and tnpRbAb cistrons on pCAR1 show homology with the tnpRA genes encoding the transposase (TnpA) and resolvase (TnpR) of Tn4656 (Tsuda and Genka 2001) and Tn4653 (Tsuda and Iino 1988), although the products of both tnpAa and tnpAb genes on pCAR1 seem to be nonfunctional because of the insertion of IS1162 and a point mutation, respectively. In addition, the tnpAc, tnpC, and tnpST genes on pCAR1 have more than 70% identity with genes encoding the transposase (tnpA), the repressor of tnpA gene expression (tnpC), and the cointegrate-resolution proteins (tnpST) of Tn4651 (Tsuda and Iino 1987; Hõrak and Kivisaar 1998), respectively. The relative order and orientation of the tnpAcCST genes (Fig. 2) are similar to those associated with Tn4651. Furthermore, in the vicinity of the tnpAcCST genes of pCAR1, there were six copies of 46-bp sequences that showed >67% identity to the inverted repeat (IR) of Tn4651, which were designated IR-a to IR-f (shown by arrowheads in Fig. 2). Generally, transposition of a transposon results in duplication of the target sequence and the insert is flanked by a direct repeat (DR). In pCAR1, the 5-bp DR sequence (5′-CAAAA-3′) was only observed at the immediate vicinity of the set of IR-a and IR-f (Maeda et al. 2003). Therefore, the region from IR-a to IR-f seemed to be a functional transposon and was tentatively designated as Tn4676 (Fig. 2). Consequently, the entire sequencing of pCAR1 revealed that two putative mobile genetic elements contained the car operon: (1) the putative conjugative plasmid pCAR1, and (2) the putative transposon Tn4676.
Genetic structure of the 73-kb putative class II transposon Tn4676 and its flanking region on pCAR1. Black pentagons indicate genetic elements on pCAR1 (tnpAa, tnpAb, tnpAc, tnpC, tnpRa, tnpRb, tnpS, tnpT) that showed >70% identity with those of the class II transposons Tn4651, Tn4653, and/or Tn4656 on TOL plasmids. The arrow represents the Tn4676 region found on pCAR1. Black arrowheads indicate the positions of the putative IRs (IR-a to IR-f) that showed >67% identity with those of Tn4651. The lengthwise bars attached beneath the physical map show the EcoRI sites. The solid bars and numbers represent the location of the probes used in Southern analyses
In this study, we performed mating assays with strain CA10 to test the transferability of pCAR1, but failed to detect the conjugative transfer of pCAR1. Then, we analyzed the transferability of CAR catabolic capacity in other CAR-degraders which carried car genes homologous to that of strain CA10. Using P. putida strain HS01, which carries a pCAR1-like plasmid designated as pCAR2, we demonstrated the conjugative transfer of pCAR2 and the Tn4676 transposition from pCAR2 into the host chromosome. We also compared the genetic organization of the flanking regions of the car gene clusters on the plasmid or chromosome of CAR-degraders with those of pCAR1 to determine the involvement of putative mobile genetic elements in the distribution of the CAR-degradative capacity among different bacterial strains and species.
Materials and methods
Bacterial strains, plasmids, media, and culture conditions
Bacterial strains and plasmids used in this study are listed in Table 1. For the extraction of total DNA or the large plasmid from CAR-degrading strains, bacteria were grown on Luria–Bertani (LB) medium (Sambrook and Russell 2001) at 30°C with reciprocal shaking (300 strokes/min) for 15 h. The minimal medium used was carbon-free mineral medium (CFMM) supplemented with CAR as a sole source of carbon and energy. The composition of CFMM and how to provide the CAR were described by Nojiri et al. (2001).
Escherichia coli strains JM109 (Sambrook and Russell 2001) and DH5α (Toyobo Co., Tokyo, Japan) were used as host strains of the plasmids pUC18/19 (Sambrook and Russell 2001) and pBluescript II SK(−) (Stratagene, La Jolla, Calif.) and their derivatives. E. coli strains were grown on 2×YT medium (Sambrook and Russell 2001) at 37°C with reciprocal shaking (300 strokes/min). Ampicillin (50 μg/ml), chloramphenicol (30 μg/ml), kanamycin (50 μg/ml), gentamicin (15 μg/ml), rifampicin (250 μg/ml), or tetracycline (12.5 μg/ml) was added to selective media.
For plate cultures, the above media solidified with 1.6% agar were used.
Matings and isolation of transconjugants
Before the mating experiment, natural resistance to antibiotics was checked (Table 1). When strain DS1 was used as a recipient strain, chloramphenicol and tetracycline were used for counter-selection against the donor strain; and the ability to grow on CAR was the selection marker of the transconjugants. When using P. fluorescens strain IAM12022 as a recipient strain in mating experiments, a gentamicin-resistant gene on a mini-transposon of pBSL202 was introduced into the spontaneously rifampicin-resistant strain IAM12022 by electroporation using a Gene Pulsar System II (Bio-Rad Laboratories, Glattbrugg, Switzerland) at 1.8 kV, 200 Ω, and 25 μF.
The donor and recipient strains were pregrown in 5 ml of LB liquid medium with and without an appropriate antibiotic, respectively, for 15–24 h. After washing these two cell lines with CFMM, cells were resuspended in 500 μl of CFMM and then mixed well by pipeting. These cells were transferred to 0.45-μm pore-size cellulose nitrate filters (Toyo Roshi Kaisha, Tokyo, Japan), and the resultant filter was incubated on a LB agar plate at 30°C for 24 h. Afterwards, the bacteria were resuspended in 1 ml of CFMM and appropriate dilutions of the cell suspension were spread on selective media. The number of transconjugants was determined by counting colonies on selective medium 5 days after mating. The frequency of plasmid transfer was expressed as the number of transconjugants per number of recipients.
DNA isolation and routine DNA manipulations
Plasmid DNA was prepared from the E. coli host strain by the alkaline lysis method (Sambrook and Russell 2001). The total DNA from bacterial strains was isolated as described by Sato et al. (1997b). Restriction endonuclease and DNA the Ligation kit ver. 2 (Takara Shuzo Co., Kyoto, Japan) were used according to the manufacturer’s instructions. DNA fragments were extracted from the agarose gel using the Concert rapid gel extraction system (Invitrogen Life Technologies Oriental, Tokyo, Japan) according to the manufacturer’s instructions. The large plasmids of CAR-degrading bacteria were extracted by a previously reported method (Ka and Tiedje 1994) or by using the Large-Construct kit (Qiagen, Tokyo, Japan) according to the manufacturer’s instructions in Appendix A (Special protocol for high yields of large-construct DNA without removal of genomic DNA). Other DNA manipulations were performed according to standard methods (Sambrook and Russell 2001).
Nucleotide sequence determination
The nucleotide sequence was determined by the chain-termination method, using a model 4200L-2 auto-DNA sequencer (Li-Cor, Lincoln, Neb.) and the ABI Prism 310 genetic analyzer (Applied Biosystems Japan, Tokyo, Japan) according to the manufacturers’ instructions; and the nucleotide sequences obtained were analyzed with DNASIS ver. 3.7 for Mac software (Hitachi Software Engineering Co., Yokohama, Japan).
Hybridization
Blotting for Southern hybridization was performed using the VacuGene XL vacuum blotting system (Amersham Biosciences, Amersham, UK) and a Biodyne B Nylon membrane (Pall Corp., New York, N.Y.) according to the recommendations of the manufacturer. The hybridization was done as described by Nojiri et al. (2001).
Pulsed-field gel electrophoresis
Large DNA was separated in agarose gels by pulsed-field gel electrophoresis (PFGE), using a GenePath system and the Clamped homogeneous electric fields bacterial genomic DNA plug kit (Bio-Rad Laboratories). Routinely, a 1% agarose gel and 0.5× Tris-borate/EDTA buffer (Sambrook and Russell 2001) were used for PFGE. The angle of pulse was 120° and the temperature was 14°C. In order to separate DNA within lengths of 0.1–23.0 kb, 1–50 kb, and 5–75 kb, the pulse time and the total running time were, respectively, 0.1 s and 5 h, 0.1–2.5 s and 8 h, and 1–6 s and 11 h.
PCR for the determination of the integrated region of Tn4676
For amplification of the DNA fragments containing the ends of Tn4676 in the chromosomes of strains K15, K22, and K23, the following primer pairs were used, based on the results of sequencing K23L1 and K23R1: primers K23L1-F (5′-TGAAGTTGGATTCTGCTGCG-3′) and K23L1-R (5′-CGAAAAATCACGTTGCAGCT-3′) for the left termini and primers K23R1-F (5′-CCTCATAACGCTCACCATCAT-3′) and K23R1-R (5′-AAAATCATGCCGGGGATGTT-3′) for the right termini. PCR was performed with denaturation at 94°C for 1 min, annealing at 60°C for 1.5 min and elongation at 72°C for 2 min, for 30 cycles.
Deposition of nucleotide sequences
The nucleotide sequence data of both terminals of the conserved regions in strains HS01 and K23 reported in this study were registered in the DDBJ, EMBL, and GenBank nucleotide sequence databases with accession numbers AB164629 (HS02L), AB164630 (HS02R), AB088759 (K23L1), AB088758 (K23R1), AB088751 (K23L2), and AB088752 (K23R2).
Results
Mating experiments between CAR-degraders and P. putida strain DS1
Prior analysis of the nucleotide sequence of pCAR1 showed that it contained a putative origin of transfer and a set of genes encoding putative proteins for conjugative transfer (Maeda et al. 2003). To confirm the transferability of pCAR1, we first performed cell-mating experiments with strain CA10 as donor. P. putida strain DS1 was selected as the recipient strain, which was originally isolated as a dimethylsulfide-utilizing bacterium (Endoh et al. 2003). Southern hybridization analysis confirmed that this strain did not possess any car genes. In addition, no plasmids were detected in this strain. As a result, no transconjugants were obtained from the selective media in experiments having a detection limit of 1.9×10−10 per number of recipient cells. Thus, it was concluded that pCAR1 is not transmissible from strain CA10 to strain DS1 at detectable frequencies.
Subsequent mating experiments were performed using other CAR-degraders as donors with strain DS1 again serving as the recipient. Previously, we isolated several CAR-degraders from different sites in Japan which have car genes homologous to those of strain CA10, such as P. resinovorans strain CA06 (Ouchiyama et al. 1993), Novosphingobium sp. strain J30, and Pseudomonas sp. strains K15, K22, and K23 (Inoue et al. 2004). Prior studies revealed that strain K23 possesses two types of car gene cluster, one of which contains duplicated carAa genes on a 6.9-kb EcoRI fragment (including carAaAaBaBbCAcAd) and the other contains only one carAa gene on a 5.6-kb EcoRI fragment (including carAaBaBbCAcAd; Inoue et al. 2004). Prior to the mating experiments, we compared EcoRI fragments containing the car gene cluster in the above strains by Southern hybridization analysis, using a car probe prepared from the 6.9-kb EcoRI fragment of strain CA10, which contains the carAaAaBaBbCAcAd genes. As shown in Fig. 3, strain J30 contained a DNA band that hybridized with this probe, which was the same in size as that in strain CA10 used as a reference. In contrast, strains CA06 and K15 contained a band that appeared in different sizes: 7.5 kb and 5.6 kb, respectively. In the case of strain K22 DNA, two hybridization bands were detected (5.6 kb, 9.8 kb; Fig. 3). The same results were obtained in similar experiments with small probes prepared from the carAa or carAc DNA fragment (data not shown). Mating experiments with various CAR-degraders as donors and strain DS1 as the recipient yielded putative transconjugants only in the mating with strain K23. The frequency was 5.0×10−7 per recipient cell and no colonies were detected using other strains as donors (the detection limits were 1.0–2.0×10−10 per number of recipient cells).
Southern hybridization analysis of EcoRI-digested total DNA of CAR-degrading bacteria, using a car probe prepared from the 6.9-kb DNA fragment containing the carAaAaBaBbCAcAd genes. The 6.9-kb and 5.6-kb hybridization bands in strain K23 correspond to the DNA fragments containing carAaBaBbCAcAd and carAaAaBaBbCAcAd, respectively (Inoue et al. 2004)
Genetic analysis of the transconjugants
Among the transconjugants from the mating between strains K23 and DS1, five colonies were randomly picked and genetically analyzed. Total DNA from each strain was digested with EcoRI and subjected to Southern analysis, using a car probe and a ssu probe prepared from the ssuD gene of strain DS1, which encodes a FMNH2-dependent monooxygenase involved in the desulfonation of methansulfonate, a metabolic intermediate of dimethylsulfide (Endoh et al. 2003). As shown in Fig. 4a, the car probe hybridized with the DNA of strain K23 and the five putative transconjugants, but not with that of strain DS1. Although the total DNA of strain K23 showed two hybridization bands, at 6.9 kb and 5.6 kb, the five putative transconjugants showed only the 6.9-kb hybridization band (Fig. 4a). In contrast, the ssu probe hybridized with the 10-kb EcoRI-digested total DNA of strain DS1 and five putative transconjugants, but not with that of strain K23 (Fig. 4b). These results clearly indicated that these five strains possessed car and ssu genes and thus were definitely transconjugants of strain DS1 that had recruited the car genes from strain K23.
Localization of the car gene following conjugative transfer
The cell lysates containing DNAs of strains K23, DS1 and CA10 and the five transconjugants prepared by the method by Ka and Tiedje (1994) were subjected to electrophoresis. The same results were obtained for all five transconjugants; and only the result from one transconjugant, which was designated as strain HS01, is shown in Fig. 5. There were two bands in strains HS01 and CA10 (reference strain carrying pCAR1, 199 kb), while only one band was detected in strains K23 and DS1. In our previous study, the upper band of strain CA10 was shown to correspond to pCAR1 and the lower band to the chromosome (Nojiri et al. 2001). Thus, the upper band of strain HS01 whose mobility on the gel was similar to that of pCAR1 was regarded as the plasmid of strain HS01 (Fig. 5) and was designated as pCAR2. After electrophoresis, these DNAs were subjected to Southern hybridization analysis with a car probe. As shown in Fig. 5 (lower panel), clear hybridization occurred only with the upper band in strain HS01, suggesting that the car gene of strain HS01 was on pCAR2. In contrast, in strain K23, the hybridization band was detected only on the lower band (Fig. 5 lower panel), implying that strain K23 had its car gene on the chromosome. Thus, there were two possibilities: first, that a part of pCAR2 (including the car gene cluster) had been transposed into the chromosome of strain K23, or second, that the whole pCAR2 region had been integrated into the K23 chromosome. In addition, when we cannot detect the plasmid in the cell lysates prepared according to this method, we cannot always conclude that this strain has no plasmid, because the lower band sometimes contains damaged and sheared plasmid DNA fragments, depending on the experimental conditions. Therefore, we tried to detect plasmid DNA from strain K23 by other available methods, but no plasmid DNA was detected (data not shown). At this point, we cannot conclude the localization of the car gene in strain K23.
Location of the car gene cluster in the genomes of strains K23, HS01, HS02, and CA10. Each panel shows the results of electrophoresis (upper) and Southern hybridization analysis using a car probe (lower) for cell lysates prepared by the methods of Ka and Tiedje (1994)
Genetic analysis of pCAR2 and the car gene cluster-flanking region in the strain K23 genome
To obtain information on the mechanism involved in the generation of strain HS01 in the filter-mating experiment, we compared the genetic structures of the whole region of pCAR2 with the car gene cluster-flanking region in the strain K23 genome, using the pCAR1 structure as a reference.
Plasmids from strains CA10 (pCAR1) and HS01 (pCAR2) were extracted using the Qiagen Large-Construct kit and digested with BamHI, EcoRI, HindIII, HpaI, NotI, SacI, SpeI, or XhoI. The resultant DNA fragments were subjected to PFGE and Southern hybridization analysis with a pCAR1 probe prepared from restriction endonuclease-digested pCAR1. Visual analysis indicated that there was no difference in the restriction fragment patterns for each plasmid (data not shown). Moreover, sequencing analysis revealed that the nucleotide sequence of the 2,276-bp putative ori region, including the repA gene and oriV (Maeda et al. 2003), were identical in both plasmids (data not shown). The terminal regions of putative Tn4676 on pCAR2 were also cloned and sequenced; and it was revealed that the nucleotide sequence of the transposition sites and target duplication on pCAR2 were also identical with those of pCAR1 (left end at 184,777–185,775 bp, right end at 59,208–59,834 bp in AB088420). From the above results, there does not appear to be any difference in the nucleotide sequence and genetic organization of these two plasmids; and thus it can be predicted that the whole genetic structure of pCAR2 is quite similar to that of pCAR1.
To determine whether the strain K23 had the whole pCAR2 DNA in its genome, Southern hybridization analysis was performed against EcoRI- and SacI-digested total DNA of strain K23. The probes were prepared from pCAR1, as shown in Fig. 2. The DNAs of strains HS01 and CA10 were also analyzed as references. As expected, the same-sized hybridization signals were detected both in strains HS01 and CA10 with all of the probes (Table 2). In contrast, although probe02–probe08 all hybridized to the DNAs of both strains K23 and HS01, surprisingly probe01 and probe09–probe11 did not hybridized to the DNA of strain K23. In addition, plural and different-sized hybridization bands from that of strain HS01 were detected in strain K23 using probe02 and probe08, while the other hybridization bands with probe03–probe07 were the same size in both strains (Table 2). The 6.8-kb EcoRI fragment from DNA of strain HS01 was detected with probe02, whereas the sizes of the target EcoRI fragments of strain K23 were 4.2 kb and 7.8 kb (Fig. 6a, Table 2). In contrast, probe08 hybridized to 10.5-kb and 9.5-kb SacI fragments in strain K23, but hybridized to an 8.1-kb fragment from CA10 DNA (Fig. 6b) and HS01 DNA (data not shown). These results definitely indicate that strain K23 possessed only a part of the DNA of pCAR2 and implied that there were two copies of a similar part of the DNA region of pCAR2, which was detected by probe02 and probe08. Since, in the pCAR1 sequence, these DNA regions were contained by Tn4676 and the loci of probe02 and probe08 corresponded to the termini of Tn4676, it was inferred that these two additional hybridized fragments contained the termini of the putative Tn4676 region located on the K23 genome. To confirm this hypothesis, we cloned 4.2-kb and 7.8-kb EcoRI fragments from the left termini in the strain K23 genome (Fig. 6a) into pUC18, to give pUK23301 and pUK23501, respectively. Similarly, the 10.5-kb and 9.5-kb SacI fragments, including the right termini (Fig. 6b), were cloned into pUC18, to give pUK23401 and pUK23601, respectively. The inserts of these plasmids were sequenced using the internal primers prepared from the pCAR1 sequence; and the borders of the conserved regions were observed in pUK23301, pUK23501, pUK23401, and pUK23601, which were designated as K23L1, K23L2, K23R1, and K23R2, respectively.
As shown in Fig. 7a, two sets of IR-a and IR-f were found. Sequence comparisons revealed that the 4-bp common sequence (5′-GATA-3′) was located in the immediate vicinity of the IR-a and IR-f sequences in the K23L1 and K23R1 regions, respectively. Similarly, 5-bp common sequences (5′-TACCA-3′) were found in K23L2 and K23R2 (Fig. 7a). Because common DR sequences are thought to be the result of target-site duplications, it was suggested that K23L1 and K23R1 contain the respective ends of a copy of Tn4676 and that K23L2 and K23R2 contain the respective ends of another copy of Tn4676. After connecting the DNA regions of the K23L2 and K23R2 and removing both the Tn4676 region and one of the DRs, there was a part of the open reading frame (ORF) whose deduced amino acid sequence showed 76% identity with that of the putative flavin-dependent oxidoreductase of P. fluorescens strain Pf0-1 (accession ZP_00086468), although no homologous proteins were detected in the corresponding region of K23L1 and K23R1. This fact also supported that K23L2 and K23R2 included the ends of Tn4676, which was one of the Tn4676s of strain K23.
Comparison of both ends of Tn4676 in pCAR1 with the corresponding ends of those in the strain K23 chromosome (a) and the strain HS02 chromosome (b). Asterisks indicate nucleotides identical with those in pCAR1. The underlined stretches indicate the target duplications (DRs) observed in each set of termini. The arrows indicate the 46-bp imperfect IR-a and IR-f. The sequences of K23L1 and K23L2 are derived from the left termini of two Tn4676s in the strain K23 genome. Similarly, those of K23R1 and K23R2 are from the right termini of two Tn4676s
In conclusion, it was revealed that the whole genetic structure of pCAR2 in strain HS01 is highly similar to that of plasmid pCAR1, in addition to the DNA region of the car gene cluster. In the case of strain K23, it was clearly proven that this strain possesses two copies of Tn4676 on its genome and that it does not have the other DNA region of pCAR2 (pCAR1) in its genome.
Transferability of pCAR2
A mating experiment to confirm the transferability of pCAR2 from strain HS01 was performed with P. fluorescens strain IAM12022 and P. putida strain Dfi175 as recipients. The latter strain is a DS1 mutant having a Tn5 insertion in the ssuA gene encoding a putative component of an ABC-type transporter for sulfonate uptake (Endoh et al., 2003). Putative IAM12022-based and Dfi175-based transconjugants were observed at frequencies of 2×10−7 and 7×10−2 per recipient, respectively. These putative transconjugants were genetically analyzed as follows. Southern analyses were performed with total DNAs of the putative transconjugants of strain Dfi175, using a probe prepared from a 3.4-kb HindIII fragment of pSUP2021 including Tn5. As a result, the probe hybridized with the DNAs of both strain Dfi175 and the putative transconjugants but not with that of strain HS01 (data not shown). As to the putative transconjugants of strain IAM12022, the 16S rDNA sequences of selected transconjugants were sequenced and shown to be identical that of the recipient strain (data not shown), confirming that these colonies were definitely derived from strain Dfi175 and IAM12022. In addition, pCAR2 was again detected from these transconjugants by the methods described above (data not shown). These results clearly indicated that pCAR2 was a self-transmissible plasmid.
Analysis of the chromosomal car gene in the HS01 derivative strain
After several transfers of strain HS01 in rich media (LB media), we fortuitously obtained a derivative strain of HS01, named HS02. Because the car probe clearly hybridized with both the upper and lower bands of DNA from HS02 (Fig. 5), it was suggested that strain HS02 carried the car genes both on the plasmid (pCAR2) and within its chromosome. As described above, the genetic structure of pCAR2 was supposed to be highly similar to that of pCAR1 and thus it was predicted that pCAR2 also carried Tn4676. Generating strain HS02 from strain HS01 suggested the possibility that Tn4676 on pCAR2 was a functional transposon and that the putative chromosomal car gene of strain HS02 was carried by Tn4676 that had transposed from pCAR2 into the strain HS01 chromosome during cultivation. To confirm this, Southern analyses were performed using probe01–probe11 with the EcoRI- or SacI-digested total DNA of strain HS02. Although the same-sized hybridization signals were detected with all probes in both strain HS01 and strain HS02, additional different-sized hybridization bands were detected in strain HS02 for probe02 (2.5-kb EcoRI fragment) and probe08 (6.0-kb EcoRI fragment; Fig. 6a, Table 2). Since it was implied that these two additional fragments contained the termini of the putative Tn4676 that might transpose from pCAR2 into the HS02 chromosome, these were cloned to pUC18 (pUHS02101, pUHS02201, respectively). Then, the inserts of the resultant plasmids were sequenced with the same primers used in the analysis of strain K23. In this analysis, IR-a and IR-f were found as the termini of the transposed DNA region from pUHS02101 (HS02L) and pUHS02201 (HS02R), respectively; and the 6-bp target duplication (5′-AACATC-3′) was observed (Fig. 7b). After connecting the nucleotide sequence of HS02L and HS02R and removing both the Tn4676-homologous region and one of the DRs, one ORF was found, whose deduced amino acid sequence showed 85% identity with part of the probable acyl-CoA synthetase of P. putida strain S5 (accession BAD07369) and 46% identity with part of the medium-chain fatty acid CoA ligase of P. putida strain KT2440 (accession NP_742924). Since these homologous proteins were thought to be encoded on the chromosome of each host strain, the data suggested that the ORF found on the outer region of the transposed region was also located on the chromosome of strain HS02. Furthermore, Southern analyses were performed with EcoRI-digested total DNA of strains DS1, HS01, and HS02, using a probe prepared from the above ORF. In this case, the probe hybridized with DNAs of all three strains, although the size of the signal detected in strain HS02 DNA was different from those in the other strains (data not shown). This indicated that the EcoRI fragment that hybridized with the probe in strain HS02 was located on the chromosome and was disrupted by Tn4676. Therefore, it was concluded that Tn4676 had transposed from pCAR2 into the chromosome of strain HS02.
Other CAR-degraders related to strain K23
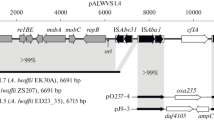
Strains K15 and K22 were isolated from the same activated sludge with strain K23 (Inoue et al. 2004) and were thought to have car genes in their genome (data not shown). The 16S rDNA sequences of strains K15, K22, and K23 showed >99% identity and were related to one another with close phylogenetic affiliation (data not shown). Southern analyses with probe01–probe11 revealed that these two strains also had Tn4676 in their genome (Table 2). In detail, probe02 hybridized with a 4.2-kb fragment in strain K15 and 4.2- and 7.8-kb EcoRI fragments in strain K22 (Fig. 6a), while probe08 hybridized to SacI-digested DNA of strains K15 and K22, which were detected at 10.5 kb and 9.5 kb, and 10.5 kb and 12.0 kb, respectively (Fig. 6b). A set of 4.2-kb EcoRI and 10.5-kb SacI fragments containing the respective ends, K23L1 and K23R1, of a single copy of Tn4676 in strain K23 chromosome, were also detected in strains K15 and K22 (Fig. 6), although we cannot explain the 9.5-kb hybridization band with probe08 to SacI-digested K15 DNA (Fig. 6b). Thus, it was suggested that these three strains might have Tn4676 at the same locus in their genomes. To examine this hypothesis, we performed PCR amplification using the primer sets spanning the respective ends of Tn4676 (K23L1-F, K23L1-R, K23R1-F, K23R1-R in Fig. 8a), designed from the nucleotide sequence of K23L1 and K23R1. As a result, same-sized PCR products were amplified from the total DNAs of strains K15, K22, and K23, while no amplification or nonspecific amplification was observed when total DNA of strain CA10 was used as thetemplate DNA (Fig. 8b,c). Together with the facts that: (1) only the 5.6-kb EcoRI fragment of strain K15 genome hybridized with the car probe (Fig. 3), and (2) the 5.6-kb EcoRI fragment of strain K23 genome contains a single copy of the carAa gene (Inoue et al. 2004), it was concluded that these three strains possessed an insertion of Tn4676 containing a single copy of the carAa gene at the same site in their genome.
PCR analysis using primer sets to amplify the spanning region from inside to immediately outside the termini of Tn4676s. a The genetic map of Tn4676 (bold bar) is shown on the chromosome of strain K23; and the amplified regions at both terminals are indicated by solid blocks on the bar. The black arrows represent IR-a and IR-f on Tn4676. The arrowheads represent primers (K23L1-F, K23L1-R, K23R1-F, K23R1-R). The PCR primer sets for the amplification of K23L1 (K23L1-F, K23L1-R) and K23R1 (K23R1-F, K23R1-R) observed in strain K23 are used, respectively, in b and c
Discussion
In our results, the conjugative transfer of pCAR1 from strain CA10 to strain DS1 was not detected. However, we obtained P. putida strain HS01 as a transconjugant of P. putida strain DS1, which recruited a car gene cluster on plasmid pCAR2 (Fig. 4). The whole genetic structure of pCAR2 was proposed to be quite similar to that of pCAR1; and it was proven that pCAR2 was a self-transmissible plasmid. In contrast, we also detected the transposition of Tn4676, whose termini were IR-a and IR-f, from pCAR2 to the chromosome of strain HS02 (Fig. 7); and strains K15, K22, and K23 had Tn4676 at the same loci in their genomes. These facts clearly indicated the transferability of pCAR2 and the transposability of Tn4676 and suggested that these functional mobile genetic elements have an important role in the distribution of the car gene in different bacteria. Many other bacteria carrying a car gene homologue have been isolated, such as Pseudomonas sp. strain LD2 (Riddle et al. 2003), Janthinobacterium sp. strain J3 (Inoue et al. 2004), P. stutzeri strain OM1 (Ouchiyama et al. 1998; Shintani et al. 2003), and so on; and it was implied that their car gene homologues were also distributed by this machinery. This hypothesis is also supported by our observation that the CAR-degrading strains CA06 and J30 possessed CAR-degradative plasmids whose genetic structures were similar to pCAR2 (and pCAR1; data not shown).
The dual mechanism (conjugation, transposition) to transfer the car gene into the genome of the host strain(s) is similar to those used for other catabolic genes. The molecular diversity of the TOL plasmid in Pseudomonas strains isolated from different sites has been reported (Sentchilo et al. 2000). Some TOL plasmids, such as pWW0 and pWW53, carry large transposable elements that contain the whole xyl genes (Keil et al. 1987; Tsuda and Iino 1987, 1988; Burlage et al. 1989; Greated et al. 2002). The plasmid pWW0 was unstable in E. coli when cultured at 42°C; and thus it was possible to detect the transposition of Tn4651 and Tn4653 from pWW0 into the chromosome of E. coli (Tsuda and Iino 1988). It is likely that the car gene cluster could exist on pCAR2 or its derivative plasmids in some hosts, while the plasmid can be maintained in the host cell environment (strains CA06, CA10, J30, HS01). In other strains, such as strains K15, K22, and K23, pCAR2 might be unstable and the car gene cluster could be maintained by transposition of the Tn4676 into the chromosome (or other stable plasmid). Although the plasmid instability and frequency of transposition should be estimated, the above-mentioned dual transfer (maintenance) system of car gene cluster by transposon and plasmid is thought to be largely advantageous for distributing CAR-catabolic capacity in nature.
As to the conjugation between strains K23 and DS1, we first hypothesized that strain DS1 recruited plasmid pCAR2 by conjugal transfer from strain K23. However, we failed to detect any plasmids in strain K23 after a mating experiment, and instead, we could detect only two copies of Tn4676 and no other regions of pCAR2 in the strain K23 genome. Secondly, there was a possibility of retrotransfer (a plasmid from strain DS1 going into strain K23, “picking up” Tn4676, and returning into DS1) for generating strain HS01. However, this possibility could be excluded because of the following reasons. First, we could not detect any plasmids from strain DS1 by any available methods. Second, we could not detect pCAR2 DNA, including both the inside and outside regions of Tn4676 from strain DS1 by Southern hybridization using probe01–probe11 (Table 2). Third, the integration sites of Tn4676 in both pCAR1 and pCAR2 were identical; and thus it was difficult to suppose the possibility that a plasmid had picked up Tn4676 in strain K23 (data not shown). Since Tn4676 did not carry the genes related to conjugative transposition (transfer, integration), this excluded the possibility of conjugative transposition of Tn4676 into the strain DS1 genome. The only possible hypothesis to explain this phenomenon was that at least one copy of Tn4676 transposed from pCAR2 to the K23 chromosome after the conjugal transfer of pCAR2 from strain K23 to strain DS1 and pCAR2 was then lost from strain K23 during culture in our laboratory, resulting in the present genomic status of strain K23.
When strain HS01 was obtained in the mating experiment, the whole nucleotide sequence of pCAR1 had not been determined and we had only the information about the 44-kb nucleotide sequence of the car gene clusters and flanking regions of strains CA10 and K23 (Nojiri et al. 2001; Inoue et al. 2004). Nor was there any information on the occurrence of the 73-kb transposon that contains the whole above-mentioned 44-kb DNA region. At that time, we had cultured these strains on minimum media with CAR; and CAR catabolic capacity was used as a selective marker. Thus, we could not detect whether Tn4676 could have transposed into the chromosome of strain K23, with the subsequent loss of pCAR2 during culture in the laboratory. The proposed genetic versatility of the “original” strain K23 is in accordance with the fact that the CAR-metabolizing capacity of strain K23 was liable to be lost at that time. (Now, the CAR-metabolism of strain K23 is stable because the car genes have been transposed into its chromosome by Tn4676.) The above speculation is supported by the fact that strains K15 and K22, which were isolated from the same activated sludge with strain K23 and were related to one another with close phylogenetic affiliation, had Tn4676s containing single carAa genes at the same location in their genome as did strain K23.
One strain having two copies of a class II catabolic transposon seems rare. To the best of our knowledge, the only other report detecting the duplication of Tn4651 was for the cointegrate intermediate of Tn4651 on the chromosome of P. aeruginosa PAO1, which was stably maintained and was not resolved by resolvase (Sinclair and Holloway, 1991). Strain K23 was found to have two copies of Tn4676, which are not the cointegrate intermediate, because no hybridizations were detected with the probes from a location outside of Tn4676 on pCAR1 (Table 2). Since it is known that the Tn3 family transposon is immune to further insertions of the same transposon (Grindley et al. 2002), the fact that the transpositions of Tn4676 had occurred twice in strain K23 is noteworthy. Because the increment in the copy number of the degradative gene may result in the enhancement of degrading ability, this phenomenon is of interest.
Previously, we reported that the putative genes for replication of pCAR1 were similar to those of Pseudomonas group plasmids, while the putative genes for transfer showed a high degree of homology with those of Enterobacteriaceae group plasmids (Maeda et al. 2003). The frequency of conjugative transfer of pCAR2 from strain HS01 to P. fluorescens strain IAM12022 was about 10−7 per recipient cell, which is not very high when compared with transfer rates reported for plasmids from Pseudomonas group bacteria such as RP4 (about 10−2 to 10−1 per recipient cell; Maher and Taylor 1993), pWW0 (about 10−1 to 10−5 per donor cell; Bradley and Williams 1982), and Enterobacteriaceae group plasmids such as R27 (6.0×10−4 per recipient; Maher and Taylor 1993). However, when strain Dfi175 (which is a P. putida strain DS1 derivative strain like strain HS01) was used as a recipient, the frequency of conjugation was about 105-fold higher than with strain IAM12022. The frequency of conjugation is regulated by various factors from the plasmid itself and the host cell (Zechner et al. 2000). In order to reveal the reason why the conjugative frequency to strain IAM12022 of pCAR2 was lower than these plasmids and why the frequency was quite different in changing the recipients, it is necessary to analyze the functionality of the genes related to conjugative transfer.
Recently, we performed transfer assays of pCAR1 from strain CA10 into its plasmid-cured strain by filter-mating and succeeded in detecting its transconjugants (data not shown). We concluded that pCAR1 is also a self-transmissible plasmid, although it did not transfer from strain CA10 to strain DS1 at a detectable frequency. Thus, the transferability of pCAR1 from strain CA10 and that of pCAR2 from strain HS01 were markedly different in conjugation with strain DS1 as the recipient. However, as described above, we could not detect any difference in the genetic structure between pCAR1 and pCAR2 by Southern hybridization and sequencing analysis. To reveal what made the difference in transferability, an investigation into the effect(s) of host factor(s), or gene product(s) of plasmids to conjugative transfer is now underway and the data will be published elsewhere. However, the functionalities of the tnpAa and tnpAb genes on pCAR2 are still not clear, whereas tnpAc gene product was revealed to be functional because a mini-transposon containing tnpAc, tnpC, tnpS, and tnpT genes could transpose into other replicons (data not shown). A more detailed characterization and a comparison of pCAR1 and pCAR2 are necessary to understand the mechanism of car gene transfer in different hosts.
By artificially controlling the distribution of the car gene by mobile genetic element(s), we might be able to direct car gene-containing bacteria to be dominant in bacterial consortia. This strategy is a potential tool for developing new techniques for the bioremediation of CAR or dioxin contamination. In order to improve the efficiency of horizontal gene transfer as a tool in bioremediation, the questions still remain as to when, where, at what rates, and between what range of “species” these horizontal gene transfer events actually occur (Top et al. 2002; Top and Springael 2003; Nojiri et al. 2004). Ilves et al. (2001) reported that the frequency of transposition of Tn4652, which is a 17-kb derivative of toluene/xylene degradative Tn4651, was elevated at the stationary growth phase of P. putida. It will be necessary to investigate the relations between the environmental conditions or the types of host/recipient cells and the frequencies of the conjugal transfer of pCAR2 and the transposition of Tn4676 in more detail. We also know very little about the natural host range to maintain such plasmids and transposons and to express car genes in both the absence and presence of selective pressure. In P. resinovorans strain CA10, we have not detected the pCAR1-cured strain under normal conditions, suggesting that the plasmid pCAR1 is highly stable in a CA10 cell background. We are now determining the stability of pCAR1 (or pCAR2) in various cell backgrounds, including strain K23. To understand this natural distribution/maintenance mechanism of the car gene cluster, the above basic information on pCAR1 behavior in nature is quite important.
References
Alexeyev MF, Shokolenko IN, Croughan TP (1995) New mini-Tn5derivatives for insertion mutagenesis and genetic engineering inGram-negative bacteria. Can J Microbiol 41: 1053–1055
Arcos JC, Argus MF (1968) Molecular geometry and carcinogenic activity of aromatic compounds. Adv Cancer Res 11:305–471
Assinder SJ, Williams PA (1990) The TOL plasmids: determinants of the catabolism of toluene and the xylenes. Adv Microb Physiol 31:1–69
Bradley DE, Williams PA (1982) The TOL plasmid is naturally derepressed for transfer. J Gen Microbiol 128:3019–3024
Burlage RS, Hooper SW, Sayler GS (1989) The TOL (pWW0) catabolic plasmid. Appl Environ Microbiol 55:1323–1328
Davison J (1999) Genetic exchange between bacteria in the environment. Plasmid 42:73–91
Endoh T, Kasuga K, Horinouchi M, Yoshida T, Habe H, Nojiri H, Omori T (2003) Characterization and identification of genes essential for dimethyl sulfide utilization in Pseudomonas putida strain DS1. Appl Microbiol Biotechnol 62:83–91
Furukawa K, Hayase N, Taira K, Tomizuka N (1989) Molecular relationship of chromosomal genes encoding biphenyl/polychlorinated biphenyl catabolism: some soil bacteria possess a highly conserved bph operon. J Bacteriol 171:5467–5472
Greated A, Lambertsen L, Williams PA, Thomas CM (2002) Complete sequence of the IncP-9 TOL plasmid pWW0 from Pseudomonas putida. Environ Microbiol 4:856–871
Grindley NDF (2002) The movement of Tn3-like elements: Transposition and cointegrate resolution. In: Craig LNR, Craigie R, Gellert M, Lambowitz AM (eds) Mobile DNA II. ASM, Washington, D.C., pp 272–302
Habe H, Chung J-S, Lee J-H, Kasuga K, Yoshida T, Nojiri H, Omori T (2001) Degradation of chlorinated dibenzofurans and dibenzo-p-dioxins by two types of bacteria having angular dioxygenases with different features. Appl Environ Microbiol 67:3610–3617
Habe H, Ashikawa Y, Saiki Y, Yoshida T, Nojiri H, Omori T (2002) Sphingomonas sp. strain KA1, carrying a carbazole dioxygenase gene homologue, degrades chlorinated dibenzo-p-dioxins in soil. FEMS Microbiol Lett 211:43–49
Habe H, Morii K, Fushinobu S, Nam J-W, Ayabe Y, Yoshida T, Yamane H, Nojiri H, Omori T (2003) Crystal structure of a histidine-tagged serine hydrolase involved in the carbazole degradation (CarC enzymes). Biochem Biophys Res Commun 303:631–639
Hõrak R, Kivisaar M (1999) Regulation of the transposase of Tn4652 by the transposon-encoded protein TnpC. J Bacteriol 181:6312–6318
Ilves H, Hõrak R, Kivisaar M (2001) Involvement of σs in starvation-induced transposition of Pseudomonas putida transposon Tn4652. J Bacteriol 183:5445–5448
Inoue K, Widada J, Nakai S, Endoh T, Urata M, Ashikawa Y, Shintani M, Saiki Y, Yoshida T, Habe H, Omori T, Nojiri H (2004) Divergent structures of carbazole degradative car operons isolated from gram-negative bacteria. Biosci Biotechnol Biochem 68:1467–1480
Iwata K, Nojiri H, Shimizu K, Yoshida T, Habe H, Omori T (2003) Expression, purification, and characterization of 2′-aminobiphenyl-2,3-diol 1,2-dioxygenase from carbazole-degrader Pseudomonas resinovorans strain CA10. Biosci Biotechnol Biochem 67:300–307
Ka JO, Tiedje JM (1994) Integration and excision of a 2,4-dichlorophenoxyacetic acid-degradative plasmid in Alcaligenes paradoxus and evidence of its natural intergeneric transfer. J Bacteriol 176:5284–5289
Keil H, Keil S, Williams PA (1987) Molecular analysis of regulatory and structural xyl genes of the TOL plasmid pWW53-4. J Gen Microbiol 133:1149–1158
Maeda K, Nojiri H, Shintani M, Yoshida T, Habe H, Omori T (2003) Complete nucleotide sequence of carbazole/dioxin-degrading plasmid pCAR1 in Pseudomonas resinovorans strain CA10 indicates its mosaicity and the presence of large catabolic transposon Tn4676. J Mol Biol 326:21–33
Maher D, Taylor DE (1993) Host range and transfer efficiency of incompatibility group HI plasmids. Can J Microbiol 39:581–587
Meer JR van der, Neerven ARW van, Vries EJ de, Vos WM de, Zehnder AJB (1991a) Cloning and characterization of plasmid-encoded genes for the degradation of 1,2-dechloro-, 1,4-dichloro-, and 1,2,4-trichlorobenzene of Pseudomonas sp. P51. J Bacteriol 173:6–15
Meer JR van der, Zehnder AJB, Vos WM de (1991b) Identification of a novel composite transposable element, Tn5280, carrying chlorobenzene dioxygenase genes of Pseudomonas sp. strain P51. J Bacteriol 173:7077–7083
Mushrush GW, Beal EJ, Hardy DR, Hughes JM (1999) Nitrogen compound distribution in middle distillate fuels derived from petroleum, oil shale, and tar sand sources. Fuel Process Technol 61:197–210
Nam J-W, Nojiri H, Noguchi H, Uchimura H, Yoshida T, Habe H, Yamane H, Omori T (2002) Purification and characterization of carbazole 1,9a-dioxygenase, a three-component dioxygenase system of Pseudomonas resinovorans strain CA10. Appl Environ Microbiol 68:5882–5890
Nojiri H, Omori T (2002) Molecular bases of aerobic bacterial degradation of dioxins: involvement of angular dioxygenation. Biosci Biotechnol Biochem 66:2001–2016
Nojiri H, Sekiguchi H, Maeda K, Urata M, Nakai S, Yoshida T, Habe H, Omori T (2001) Genetic characterization and evolutionary implications of a car gene cluster in the carbazole degrader Pseudomonas sp. strain CA10. J Bacteriol 183:3663–3679
Nojiri H, Taira H, Iwata K, Morii K, Nam J-W, Yoshida T, Habe H, Nakamura S, Shimizu K, Yamane H, Omori T (2003) Purification and characterization of meta-cleavage compound hydrolase from a carbazole degrader Pseudomonas resinovorans strain CA10. Biosci Biotechnol Biochem 67:36–45
Nojiri H, Shintani M, Omori T (2004) Divergence of mobile genetic elements involved in the distribution of xenobiotic–catabolic capacity. Appl Microbiol Biotechnol 64:154–174
Ouchiyama N, Zhang Y, Omori T, Kodama T (1993) Biodegradation of carbazole by Pseudomonas spp CA06 and CA10. Biosci Biotechnol Biochem 57:455–460
Ouchiyama N, Miyachi S, Omori T (1998) Cloning and nucleotide sequence of carbazole catabolic genes from Pseudomonas stutzeri strain OM1, isolated from activated sludge. J Gen Appl Microbiol 44:57–63
Riddle RR, Gibbs PR, Willson RC, Benedik MJ (2003) Recombinant carbazole-degrading strains for enhanced petroleum processing. J Ind Microbiol Biotechnol 30:6–12
Sambrook J, Russell DW (2001) Molecular cloning. A laboratory manual, 3rd edn. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
Sato S, Nam J-W, Kasuga K, Nojiri H, Yamane H, Omori T (1997a) Identification and characterization of genes encoding carbazole 1,9a-dioxygenase in Pseudomonas sp. strain CA10. J Bacteriol 179:4850–4858
Sato S, Ouchiyama N, Kimura K, Nojiri H, Yamane H, Omori T (1997b) Cloning of genes involved in carbazole degradation of Pseudomonas sp. strain CA10: nucleotide sequences of genes and characterization of meta-cleavage enzymes and hydrolase. J Bacteriol 179:4841–4849
Sentchilo VS, Perebituk AN, Zehnder AJB, Meer JR van der (2000) Molecular diversity of plasmids bearing genes that encode toluene and xylene metabolism in Pseudomonas strains isolated from different contaminated sites in Belarus. Appl Environ Microbiol 66:2842–2852
Shintani M, Nojiri H, Yoshida T, Habe H, Omori T (2003) Carbazole/dioxin-degrading car gene cluster is located on the chromosome of Pseudomonas stutzeri strain OM1 in a form different from the simple transposition of Tn4676. Biotechnol Lett 25:1255–1261
Simon R, Priefer U, Pühler A (1983) A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Biotechnology 1:784–791
Sinclair MI, Holloway BW (1991) Chromosomal insertion of TOL transposons in Pseudomonas aeruginosa PAO. J Gen Microbiol 137:1111–1120
Springael D, Kreps S, Mergeay M (1993) Identification of catabolic transposon, Tn4371, carrying biphenyl and 4-chlorobiphenyl degradation genes in Alcaligenes eutrophus A5. J Bacteriol 175:1674–1681
Top EM, Springael D (2003) The role of mobile genetic elements in bacterial adaptation to xenobiotic organic compounds. Curr Opin Biotechnol 14:262–269
Top EM, Springael D, Boon N (2002) Catabolic mobile genetic elements and their potential use in bioaugumentation of polluted soils and waters. FEMS Microbiol Ecol 42:199–208
Tsuda M, Genka H (2001) Identification and characterization of Tn4656, a novel class II transposon carrying a set of toluene-degrading genes from TOL plasmid pWW53. J Bacteriol 183:6215–6224
Tsuda M, Iino T (1987) Genetic analysis of a transposon carrying toluene degrading genes on a TOL plasmid pWW0. Mol Gen Genet 210:270–276
Tsuda M, Iino T (1988) Identification and characterization of Tn4653, a transposon covering the toluene transposon Tn4651 on TOL plasmid pWW0. Mol Gen Genet 213:72–77
Tsuda M, Iino T (1990) Naphthalene degrading genes on plasmid NAH7 are on a defective transposon. Mol Gen Genet 223:33–39
Tsuda M, Tan HM, Nishi A, Furukawa K (1999) Mobile catabolic genes in bacteria. J Biosci Bioeng 87:401–410
Williams PA, Murray K (1974) Metabolism of benzoate and the methylbenzoates by Pseudomonas arvilla mt-2: evidence for the existence of a TOL plasmid. J Bacteriol 120:416–423
Zechner EL, Cruz F de la, Eisenbrandt R, Grahn AM, Koraimann G, Lanka E, Muth G, Pansegrau W, Thomas CM, Wilkins BM, Zatyka M (2000) Conjugative-DNA transfer processes. In: Thomas CM (ed) The horizontal gene pool. Harwood, Amsterdam, pp 87–174
Acknowledgements
We thank Dr. Masataka Tsuda, Tohoku University, for helpful discussion. We are most grateful to Dr. David E. Crowley for careful reading of the manuscript. This work was supported by the Program for the Promotion of Basic Research Activity for Innovative Bioscience (PROBRAIN) and by a Grant-in-aid (Hazardous Chemicals) from the Ministry of Agriculture, Forestry and Fisheries of Japan (HC-04-2325-1)
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Shintani, M., Yoshida, T., Habe, H. et al. Large plasmid pCAR2 and class II transposon Tn4676 are functional mobile genetic elements to distribute the carbazole/dioxin-degradative car gene cluster in different bacteria. Appl Microbiol Biotechnol 67, 370–382 (2005). https://doi.org/10.1007/s00253-004-1778-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-004-1778-0