Abstract
In Enterobacteriaceae, the blaOXA-48-like genes have been identified on plasmids in different regions of the world. The OXA-370 is a plasmid-encoded OXA-48-like enzyme reported in two distinct regions of Brazil. Recently, we demonstrate that the blaOXA-370 gene is disseminated among several Enterobacteriaceae species and clones, indicating a high potential for dissemination. In this work, we described for the first time the complete nucleotide sequence of six plasmids harboring the blaOXA-370 gene. Complete DNA sequencing using the Illumina platform and annotation of the plasmids showed that they belonged to incompatibility groups IncX and had in average 70 kbp. The blaOXA-370 gene is located in a composite transposon containing four genes encoding transposases, named Tn6435. In this study, highly similar plasmids were detected in different Enterobacteriaceae genera.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
OXA-type β-lactamases are currently classified in three different subgroups based on their hydrolytic profile [1]. The group 2df includes enzymes able to hydrolyze carbapenems, including OXA-48. OXA-370, a plasmid-encoded OXA-48-like enzyme, differs from the former by a single amino acid substitution and was firstly reported from Porto Alegre, south of Brazil [2] and subsequently reported from Rio de Janeiro, on the southeast of Brazil [3]. Dissemination of blaOXA-370 has been demonstrated to occur by clonal dissemination [3], but recently the detection of this gene was demonstrated in different enterobacterial species [4]. However, there is no data regarding the complete sequence of plasmids harboring the blaOXA-370 gene so far. The aim of this work was to determine the complete nucleotide of plasmids involved in the dissemination of the blaOXA-370 gene among different species of Enterobacteriaceae.
Methods
Strains used in this study and species identification
The Enterobacteriaceae isolates in this study are listed in Table 1. All strains were isolated from rectal swab samples in the same hospital in Porto Alegre, RS, Brazil. Identification to the species level was achieved by partial sequencing of the rrs gene [5]. Enterobacter strains were also identified at the species level by partial sequencing of the hsp60 gene, as previously described [6, 7], with modifications proposed by Campos et al. [8].
Plasmid extraction, transformation, and conjugation assays
Extraction of plasmid DNA from wild-type strains was carried out as previously described by Birnboim and Doli [9], except that sodium acetate solution was refrigerated. Plasmid DNA solutions were used to transform electrocompetent E. coli TOP10 (Thermo) using E. coli Pulser (BioRad). Transformants were selected on LB agar containing ampicillin (100 mg/L). Plasmids were subsequently extracted from transformants grown in LB broth containing ampicillin (100 mg/L). Conjugation experiments were performed using the wild-type strains as donors and E. coli J53 as the recipient, as described previously [10]. Transconjugants were selected on LB agar containing ampicillin (100 mg/L) plus sodium azide (125 mg/L). The presence of the blaOXA-370 gene in transformants and transconjugants was confirmed by PCR as previously described by Poirel et al. [11] with modifications proposed by Campos et al. [8]. The presence of plasmids was confirmed by 0.7% agarose gel electrophoresis applying 2.5 V/cm during 7 h. E. coli 39R861 was used as a reference for estimating plasmid size [12]. Plasmid bands were visualized under UV transillumination after GelRed (Biotium) staining.
Antimicrobial susceptibility profile of transformants
Antimicrobial susceptibility profiles for carbapenems were determined by broth microdilution [13] using cation-adjusted Mueller-Hinton broth (Becton-Dickinson). E. coli ATCC 25922 was used as a control. Results were interpreted according to the M100-S25 document from CLSI [13].
Complete plasmid sequencing, assembly, annotation, and analysis
Plasmid DNA samples were tagmented using the Nextera DNA sample preparation kit before fragments of ∼ 2000 bp were captured, purified, and sequenced using a MiSeq Reagent kit, v2 (500 cycles), in MiSeq system (Illumina). Sequences were assembled de novo in contigs using the SeqMan NGen program and subsequently aligned using SeqMan Pro, both in version 14.1.0 (DNAStar). Open reading frames (ORFs) were predicted and annotated using RAST (http://rast.nmpdr.org/) [14]. Manual curation and sequence similarity searches directed against the GenBank database were carried out using the ARTEMIS [15] genome browser and annotation tool. Insertion sequences were manually reviewed, directing searches against the IS Finder database (https://www-is.biotoul.fr/) [16]. The full plasmid sequences were compared to those available at GenBank using BLAST. Plasmid incompatibility group was evaluated using the Plasmid Finder software (https://cge.cbs.dtu.dk/services/PlasmidFinder/) [17]. Plasmid nucleotide sequences were aligned using BioEdit [18].
Results
The plasmids carrying the blaOXA-370 gene were successfully transferred by conjugation from wild-type strains to E. coli J53. Carbapenem MICs for the transformants carrying the blaOXA-370 gene are described in Table 2. A total of six plasmids had the complete sequence determined. They were all circular and their GC content varied from 46.7 to 47.2%. All sequenced plasmids belonged to the IncX incompatibility group and had in average 70 kbp (Table 1). When the full nucleotide sequences of the 70-kbp plasmids were analyzed using the PlasmidFinder 1.3 Server, no plasmid replicons were found even using a 50.0% threshold. Contrasting to this finding, annotation revealed the presence of genes encoding replication-associated proteins (pir, bis, parA, hns, and topB), characteristically found in IncX plasmids. These genes are highlighted in blue color in Figure 1S.
The largest IncX plasmid was designated p1368 and all others had their nucleotide sequences compared to it. Similarity indexes among them varied from 98 to 99%.
The p1368 plasmid, as well as the others described in this study, has a virB operon-encoding proteins of a type IV secretion apparatus implicated in conjugal transfer, in which virB3 is fused with virB4. virB7 is absent, but there is a gene immediately upstream, virB8, encoding for a 42 amino acid hypothetical protein 89% similar to PilX7 protein from E. coli (GenBank EIQ73345.1). It has vapC-like and vapD-like genes, known to encode a toxin-antitoxin system implicated in plasmid stability [19]. The plasmid also has the frmA, frmB, and frmR genes implicated in formaldehyde metabolism and tolerance [20]. Concerning antimicrobial resistance genes, only genes encoding β-lactamases were found. The blaCTX-M-8 and the blaOXA-370 genes were found located 24 kbp apart from each other (Figure 1S).
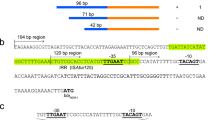
The blaOXA-370 gene is located in a 3,710-bp composite transposon containing four genes encoding transposases; it is bracketed by IS26 at the 5′ and tnpA gene at 3′ (Figure 2S). This transposon was registered as Tn6435 at the Transposon Registry Database (http://transposon.lstmed.ac.uk). This mobile element is present in all IncX plasmids sequenced in this study and also an IncF plasmid p87F from Enterobacter hormaechei, which partial sequence was previously described by our group (GenBank accession KJ488493.1). When the Tn6435 complete nucleotide sequence was compared to other sequences available at GenBank, the maximum query cover was 63%.
When the p1368 nucleotide complete nucleotide sequence was compared to that available at GenBank, the maximum query cover (52%) and similarity index (99%) were observed with GenBank deposit CP011588.1, an isolate of Enterobacter asburiae from human sample detected in VA, USA, in 2008.
The IncX plasmids harboring blaOXA-370 differ from one another by insertion sequences. In the p1368 plasmid, isolated from E. hormaechei in October 2013, the uvrB gene is interrupted by the ISKpn24, but this interruption is absent in the other five plasmids sequenced (Figure 1S). The p1233 plasmid, isolated from E. hormaechei in October 2013, has a ISKpn40 between two genes encoding hypothetical proteins but this insertion is absent in all other plasmids. In plasmids p1032 and p1182, the IS5075, observed downstream the Tn6435 in p1368, p1233, p844, and p2048, is absent (Figure 1S). Of note, highly similar plasmids (99%) were detected in different Enterobacteriaceae genera.
Discussion
In this paper, we described for the first time the complete nucleotide sequence of six plasmids harboring the blaOXA-370 gene. All of them belong to the IncX incompatibility group. IncX plasmids have been described to harbor different carbapenemase genes and to be responsible for the dissemination of these genes in different countries around the world, as is the case for blaNDM-1 [8, 21, 22], blaKPC-2 [23,24,25,26], blaKPC-3 [27], and blaKPC-4 and blaKPC-5 [28]. More recently, these groups of plasmids have been described to harbor the mcr-1 gene [29, 30]. There is a report of blaOXA-370 in plasmids of variable sizes in Klebsiella pneumoniae from Rio de Janeiro [3], but the complete nucleotide sequences and the incompatibility groups have not been reported.
All plasmid sequenced had a virB operon with virB3 fused with virB4, as previously reported in plasmids from Campylobacter [31] and apparently lacked the virB7 gene. The virB operon encodes a type IV secretion system (T4SS) that enables plasmid transfer through conjugation. If we consider that plasmids harboring blaOXA-370 were successfully transferred by conjugation from wild-type strains to E. coli J53, that the small VirB7 protein is essential for T4SS function and there are more than ten VirB7 homologues [32], it is possible that the hypothetical protein encoded by the gene located immediately upstream the virB8 gene is in fact a new Virb7 homologue.
When we compared the carbapenem MICs obtained for transformants containing different plasmids described in this study, they differed by one or two dilutions. Consequently, transposon position on the plasmid does not seem to interfere in carbapenem MICs.
The plasmids that we sequenced in this work were detected in isolates cultivated from rectal swabs during the period from August 2013 to February 2014. In a 6-month period, we observed various occurrences of insertion sequences in the same K. pneumoniae and E. hormaechei clonal groups Kp1 and Eh1. These findings suggest a high frequency of genetic events in a plasmid in a short period of time.
We found that the same Tn6435 was present in plasmids pertaining to IncX and IncF incompatibility groups. This finding evidences the capacity of this transposon to be transferred from one plasmid to another.
The complete sequence of IncX plasmids containing the blaOXA-370 gene showed that blaCTX-M-8 was also present. Considering that OXA-370 is a weak carbapenemase that is not active against third- and fourth-generation cephalosporins [2], the presence of the blaCTX-M-8 may facilitate the dissemination of clones harboring the IncX plasmids we sequenced.
In summary, we demonstrate that the blaOXA-370 gene is disseminated among Enterobacteriaceae both by plasmid transfer and mobilization by a newly describe transposon herein designated Tn6435.
References
Bush K, Jacoby GA (2010) Updated functional classification of β-lactamases. Antimicrob Agents Chemother 54:969–976
Sampaio JL, Ribeiro VB, Campos JC, Rozales FP, Magagnin CM, Falci DR, da Silva RC, Dalarosa MG, Luz DI, Vieira FJ, Antochevis LC, Barth AL, Zavascki AP (2014) Detection of OXA-370, an OXA-48-related class D β-lactamase, in Enterobacter hormaechei from Brazil. Antimicrob Agents Chemother 58:3566–3567
Pereira PS, Borghi M, de Araujo CF, Aires CA, Oliveira JC, Asensi MD, Carvalho-Assef AP (2015) Clonal dissemination of OXA-370-producing Klebsiella pneumoniae in Rio de Janeiro, Brazil. Antimicrob Agents Chemother 59:4453–4456
Magagnin CM, Rozales FP, Antochevis L, Nunes LS, Martins AS, Barth AL, Sampaio JM, Zavascki AP (2017) Dissemination of bla OXA-370 gene among several Enterobacteriaceae species in Brazil. Eur J Clin Microbiol Infect Dis 36:1907–1910
Maiwald M (2004) Broad-range PCR for detection and identification of bacteria. In: Persing DH (ed) Molecular microbiology diagnostic principles and practice, 2rd edn. American Society for Microbiology, Washington, DC, pp 379–390
Hoffmann H, Stindl S, Ludwig W, Stumpf A, Mehlen A, Monget D, Pierard D, Ziesing S, Heesemann J, Roggenkamp A, Schleifer KH (2005) Enterobacter hormaechei subsp. oharae subsp. nov., E. hormaechei subsp. hormaechei comb. nov., and E. hormaechei subsp. steigerwaltii subsp. nov., three new subspecies of clinical importance. J Clin Microbiol 43:3297–3303
Hoffmann H, Roggenkamp A (2003) Population genetics of the nomenspecies Enterobacter cloacae. Appl Environ Microbiol 69:5306–5318
Campos JC, da Silva MJ, dos Santos PR, Barros EM, Pereira Mde O, Seco BM, Magagnin CM, Leiroz LK, de Oliveira TG, de Faria-Junior C, Cerdeira LT, Barth AL, Sampaio SC, Zavascki AP, Poirel L, Sampaio JL (2015) Characterization of Tn3000, a transposon responsible for bla NDM-1 dissemination among Enterobacteriaceae in Brazil, Nepal, Morocco, and India. Antimicrob Agents Chemother 59:7387–7395
Birnboim HC (1983) A rapid alkaline extraction method for the isolation of plasmid DNA. Methods Enzymol 100:243–255
Casali N, Preston A(ed) (2003) E. coli plasmid vectors: methods and applications. Humana Press, Totowa
Poirel L, Walsh TR, Cuvillier V, Nordmann P (2011) Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis 70:119–123
Macrina FL, Kopecko DJ, Jones KR, Ayers DJ, McCowen SM (1978) A multiple plasmid-containing Escherichia coli strain: convenient source of size reference plasmid molecules. Plasmid 1:417–420
Clinical and Laboratory Standards Institute (2016) Performance standards for antimicrobial susceptibility testing. Document M100-S26. CLSI, Wayne, PA
Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O (2008) The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75
Carver T, Harris SR, Berriman M, Parkhill J, McQuillan JA (2012) Artemis: an integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics 28:464–469
Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M (2006) ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res 34:D32–D36
Carattoli A, Zankari E, Garcia-Fernandez A, Voldby Larsen M, Lund O, Villa L, Moller Aarestrup F, Hasman H (2014) In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser 41
Pullinger GD, Lax AJ (1992) A Salmonella dublin virulence plasmid locus that affects bacterial growth under nutrient-limited conditions. Mol Microbiol 6:1631–1643
Hibi M, Sonoki T, Mori H (2005) Functional coupling between vanillate-O-demethylase and formaldehyde detoxification pathway. FEMS Microbiol Lett 253:237–242
Ho PL, Li Z, Lo WU, Cheung YY, Lin CH, Sham PC, Cheng VC, Ng TK, Que TL, Chow KH (2012) Identification and characterization of a novel incompatibility group X3 plasmid carrying bla NDM-1 in Enterobacteriaceae isolates with epidemiological links to multiple geographical areas in China. Emerg Microbes Infect 1:e39
Sonnevend A, Al Baloushi A, Ghazawi A, Hashmey R, Girgis S, Hamadeh MB, Al Haj M, Pal T (2013) Emergence and spread of NDM-1 producer Enterobacteriaceae with contribution of IncX3 plasmids in the United Arab Emirates. J Med Microbiol 62:1044–1050
Cerdeira LT, Cunha MPV, Francisco GR, Bueno MFC, Araujo BF, Ribas RM, Gontijo-Filho PP, Knobl T, de Oliveira GD, Lincopan N (2017) IncX3 plasmid harboring a non-Tn4401 genetic element (NTEKPC) in a hospital-associated clone of KPC-2-producing Klebsiella pneumoniae ST340/CG258. Diagn Microbiol Infect Dis 89:164–167
Shen P, Zhang Y, Li G, Jiang X (2016) Characterization of the genetic environment of the bla KPC-2 gene among Klebsiella pneumoniae isolates from a Chinese hospital. Braz J Infect Dis 20:384–388
Ho PL, Cheung YY, Lo WU, Li Z, Chow KH, Lin CH, Chan JF, Cheng VC (2013) Molecular characterization of an atypical IncX3 plasmid pKPC-NY79 carrying bla KPC-2 in a Klebsiella pneumoniae. Curr Microbiol 67:493–498
Kassis-Chikhani N, Frangeul L, Drieux L, Sengelin C, Jarlier V, Brisse S, Arlet G, Decre D (2013) Complete nucleotide sequence of the first KPC-2- and SHV-12-encoding IncX plasmid, pKpS90, from Klebsiella pneumoniae. Antimicrob Agents Chemother 57:618–620
Garcia-Fernandez A, Villa L, Carta C, Venditti C, Giordano A, Venditti M, Mancini C, Carattoli A (2012) Klebsiella pneumoniae ST258 producing KPC-3 identified in Italy carries novel plasmids and OmpK36/OmpK35 porin variants. Antimicrob Agents Chemother 56:2143–2145
Chen L, Chavda KD, Fraimow HS, Mediavilla JR, Melano RG, Jacobs MR, Bonomo RA, Kreiswirth BN (2013) Complete nucleotide sequences of bla KPC-4 and bla KPC-5 harboring IncN and IncX plasmids from Klebsiella pneumoniae strains isolated in New Jersey. Antimicrob Agents Chemother 57:269–276
Fernandes MR, McCulloch JA, Vianello MA, Moura Q, Perez-Chaparro PJ, Esposito F, Sartori L, Dropa M, Matte MH, Lira DP, Mamizuka EM, Lincopan N (2016) First report of the globally disseminated IncX4 plasmid carrying the mcr-1 gene in a colistin-resistant Escherichia coli sequence type 101 isolate from a human infection in Brazil. Antimicrob Agents Chemother 60:6415–6417
Sellera FP, Fernandes MR, Sartori L, Carvalho MP, Esposito F, Nascimento CL, Dutra GH, Mamizuka EM, Perez-Chaparro PJ, McCulloch JA, Lincopan N (2017) Escherichia coli carrying IncX4 plasmid-mediated mcr-1 and bla CTX-M genes in infected migratory Magellanic penguins (Spheniscus magellanicus). J Antimicrob Chemother 72:1255–1256
Batchelor RA, Pearson BM, Friis LM, Guerry P, Wells JM (2004) Nucleotide sequences and comparison of two large conjugative plasmids from different Campylobacter species. Microbiology 150:3507–3517
Cao TB, Saier MH Jr (2001) Conjugal type IV macromolecular transfer systems of Gram-negative bacteria: organismal distribution, structural constraints and evolutionary conclusions. Microbiology147:3201–3214
Funding
This work was supported by the Fundo de Incentivo à Pesquisa e Eventos do Hospital de Clínicas de Porto Alegre, Coordenação de Aperfeiçoamento de Pessoal de Nível Superior, Fundação de Amparo à Pesquisa do Estado do Rio Grande do Sul, National Council for Scientific and Technological Development (CNPq), Ministry of Science and Technology, Brazil, and Fleury Institute.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
A. P. Z. has received honoraria for speaking engagements and consultancy from Merck, AstraZeneca, Pfizer, and United Pharmaceuticals. All other authors have no conflicts of interest to declare.
Ethical approval
The study was approved by the Ethical Committee of Hospital de Clínicas de Porto Alegre (14-0046).
Informed consent
Not applicable.
Rights and permissions
About this article
Cite this article
Magagnin, C.M., Campos, J.C., da Rocha, D.A. et al. Dissemination of blaOXA-370 is mediated by IncX plasmids and the Tn6435 transposon. Eur J Clin Microbiol Infect Dis 37, 2165–2169 (2018). https://doi.org/10.1007/s10096-018-3356-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-018-3356-x