Abstract
Natural killer (NK) cells affect a form of innate immunity that recognizes and eliminates cells that are infected with certain viruses or have undergone malignant transformation. In mammals, this recognition can be mediated through immunoglobulin- (Ig) and/or lectin-type NK receptors (NKRs). NKR genes in mammals range from minimally polymorphic single-copy genes to complex multigene families that exhibit high levels of haplotypic complexity and exhibit significant interspecific variation. Certain single-copy NKR genes that are present in one mammal are present as expanded multigene families in other mammals. These observations highlight NKRs as one of the most rapidly evolving eukaryotic gene families and likely reflect the influence of pathogens, especially viruses, on their evolution. Although well characterized in human and mice, cytotoxic cells that are functionally similar to NK cells have been identified in species ranging from birds to reptiles, amphibians and fish. Although numerous receptors have been identified in non-mammalian vertebrates that share structural relationships with mammalian NKRs, functionally defining these lower vertebrate molecules as NKRs is confounded by methodological and interpretive complexities. Nevertheless, several lines of evidence suggest that NK-type function or its equivalent has sustained a long evolutionary history throughout vertebrate species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The vertebrate adaptive immune system somatically rearranges genes encoding immunoglobulin (Ig) and T cell antigen receptors (TCRs) in individual lymphocyte precursors and then clonally selects and expands cell populations bearing specific surface receptors, with essentially limitless regionalized structural diversity and antigen recognition. However, these de novo responses are associated with significant temporal limitations. In marked contrast, innate immune responses, which are mediated by many different classes of molecules (e.g., Toll-like receptors (TLRs), nucleotide-binding oligomerization domain-containing proteins (NODs), and scavenger receptors) are expressed by a variety of different cell types, possess generalized specificity, and require neither a specific genetic rearrangement nor complex clonal selection. The signaling pathways that regulate the innate immune response are linked to and can directly augment adaptive immune responses. Of the many potentially injurious challenges that confront a host organism, those caused by certain viral infections or malignant transformation are of particular consequence. In both cases, an alteration in a cell may not trigger a conventional innate immune response; the time lag in effecting a protective adaptive response could be fatal.
A particularly diverse collection of molecules and corresponding signaling pathways are associated with natural killer receptor (NKR) function in mammals. The extensively characterized natural killer (NK) cells are lymphocytes that are distinct from both T and B lineages and mediate an alternative form of innate immunity, which is triggered through receptors that recognize anomalies on the surfaces of some tumor cells, e.g., changes in cell surface glycoprotiens or certain bacterial and viral molecules (particularly those deriving from herpes viruses). In addition, receptors sense modulation (e.g., down regulation) of the cell surface expression of major histocompatibility complex (MHC) I associated with certain viral infections. Upregulation of cell surface molecules, which are expressed in response to stress, serves as the basis for another mechanism of NK recognition. Other NK functions are mediated by NKT lymphocytes that recognize CD1d with a restricted repertoire of TCRs, express NKRs and, like NK cells, produce cytokines upon receptor engagement (Bendelac et al. 2007). NKT function is also mediated by non-rearranging monomorphic molecules (e.g., NKp30) that encode Ig V or C2 domains (Barrow and Trowsdale 2008).
The phylogenetic origins of NK-type immunity are of great interest and NK-like cytotoxic cells capable of effecting allogeneic killing in avians, amphibians, and fish have been described (Gobel et al. 1994; Goyos and Robert 2009; Horton et al. 1996; Rogers et al. 2008; Shen et al. 2004; Stuge et al. 1997; Yoder 2004). A prevailing general impression is that NK immunity is of ancient origin, possibly predating adaptive immunity, with its obligatory requirements of genetic rearrangement and selection of appropriate receptors in individual somatic cell lineages (Du et al. 2004; Khalturin et al. 2004; Lin et al. 2001; Parrinello et al. 1993). Although NK or NK-like cytotoxic cells have been demonstrated functionally in diverse species, the molecular mechanisms that regulate these cells are best understood in mammals. One of the challenges of identifying the receptors that regulate cytotoxicity in non-mammalian species is that despite the extensive characterization of NKR sequences and structures in mammals, the receptors mediating NK immunity as well as other forms of immune function in lower vertebrates, cannot be predicted reliably (Litman et al. 2007), e.g., TLR4, which is presumed to bind LPS in all eutherian (placental) mammals, does not appear to bind LPS in zebrafish (Sepulcre et al. 2009; Sullivan et al. 2009).
Despite the paucity of functional information relating to NK recognition in non-mammalian vertebrates, enabling of genomes in a growing number of representative species has identified individual genes and members of gene families that exhibit varying degrees of homology to mammalian innate immune receptors, including candidate NKRs. Although, the recent and rapid evolution of NKRs makes the orthology between mediators of NK function in higher and lower vertebrates less than certain, structural features and limited functional characterization of some molecules suggest that several receptor types found in lower vertebrates may mediate functions that are similar to those affected by NKRs in higher vertebrates. This review will provide a general background of NKRs in mammals but will focus primarily on various forms of receptors that exhibit genetic and structural relationships to known NKRs and are found in representative non-mammalian vertebrate species as well as discuss what, if any, inferences can be made regarding their potential functions.
Functional analyses of NK cells
Mammalian NK cells possess the capacity to directly recognize and kill tumor target cells without any prior induction period, thus presenting a primary defense during the mobilization of an adaptive immune response (Orr and Lanier 2010). Mammalian NK cells induce apoptosis in target cells by making direct contact with the target cell and killing is perforin and granzyme mediated (Voskoboinik et al. 2010). NK function in large part is defined by functional assays that have been developed and validated over the past three decades using a number of allogeneic and xenogeneic NK target cell lines. These assays quantify the ability of NK cells to induce cell death in target cells by monitoring target cell lysis or apoptosis. Unfortunately, and unlike all other hematopoietic cell types in gnathostomes, no cell type-specific markers typify NK cells in all mammalian species (Walzer et al. 2007b) and thus, basic cellular assays take on particular significance when defining NK or NK-type function in diverse species.
Diversity of mammalian NKRs
Mammalian NKRs can be classified into two groups based on the physiological function of signaling motifs. Specifically, NK function is mediated by inhibitory and activating NKRs that bind ligands on the target cell surface and mediate release of cytolytic granules. Inhibitory NKRs typically encode a cytoplasmic immunoreceptor tyrosine-based inhibition motif (ITIM; (S/I/V/L)xYxx(I/V/L)) (Ravetch and Lanier 2000). Activating NKRs typically possess a positively charged residue within the transmembrane domain that associates physically with an adaptor protein such as DAP12, which encodes a negatively charged residue within the transmembrane domain and a cytoplasmic immunoreceptor tyrosine-based activation motif (ITAM; YxxLX6–12YxxL/I) (Pitcher and van Oers 2003) (Fig. 1). Upon ligand recognition, activating NKRs initiate a signaling cascade (e.g., Syk/ZAP70 → PI3K → Rac → PAK → MEK → MAPK/ERK) that leads to the production of cytokines and chemokines and the release of cytolytic granules. In contrast, the signaling initiated by inhibitory NKRs counters that of the activating NKRs (Djeu et al. 2002; Lanier 2005). As the distribution of these signaling motifs within immune receptors is particularly broad, their utility for defining a receptor as a NKR is limited.
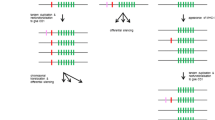
Model of mammalian natural killer receptor (NKR) signaling. NK cells display both inhibitory and activating NKRs along with adaptor proteins (e.g., DAP12 dimer). Ligand recognition by a mammalian activating NKR will induce the ITAM in DAP12 to be tyrosine phosphorylated, leading to the recruitment of the tyrosine kinases Syk and ZAP70, which in turn activates a signaling cascade that stimulates the production of cytokines and chemokines and the release of the cytolytic granules. In contrast to activating NKRs, ligand recognition by an inhibitory NKR induces its ITIM to be tyrosine phosphorylated that in turn leads to the recruitment of the tyrosine-specific SHP phosphatases: SHP-1 and SHP-2, or the phospholipid-specific phosphatase, SHIP. In mammalian NK cells, the recruitment and activation of SHP-1 and SHP-2 by inhibitory NKRs can result in decreased phosphorylation of numerous intracellular signaling proteins, including Syk and ZAP-70. Activating NKRs typically bind non-polymorphic MHC-I- or MHC-I-related stress-induced molecules and inhibitory NKRs typically bind polymorphic MHC I. Not all activating NKRs partner with DAP12 (Fig. 2)
A second basis for classifying NKRs in mammals relates to their extracellular protein domains, which correspond to different gene complexes. The leukocyte receptor complex (LRC) is located on human chromosome 19q13 and encodes multiple Ig-type receptor families that are type I transmembrane proteins including the NKR family of inhibitory and activating killer cell immunoglobulin-like receptors (KIRs; Fig. 2). The human LRC includes additional Ig-type receptors such as the leukocyte Ig-like receptors (LILRs or LIRs) that also are known as Ig-like transcripts (ILTS), and the leukocyte-associated Ig-like receptors (LAIRS) (Barrow and Trowsdale 2008). Interestingly, the number of KIR genes encoded at the LRC varies between primate species (Fig. 3 and the Electronic supplementary material (Table S1)) as well as between individuals within a species. For example, humans possess >130 unique KIR genotypes (Hollenbach et al. 2010). In contrast, KIR diversity in a population of macaques, which are geographically isolated on the island of Mauritius, exhibit limited genetic diversity: only eight common KIR haplotypes (encoding from three to six KIRs) were identified in 274 macaques representing about 1% of the population (Bimber et al. 2008). Whereas, primates typically encode multiple KIRs, the grey mouse lemur, a prosimian primate whose lineage diverged from other primate lineages (including human) nearly 50 million years ago (MYA), encodes only a single functional KIR (Averdam et al. 2009). The number of KIR genes at the LRC of non-primate mammals ranges from zero in the dog to more than four in the cow (Fig. 4 and the Electronic supplementary material (Table S1)) (Guethlein et al. 2007; Hammond et al. 2009). Mice do not encode any KIR genes at the LRC but do encode a KIR and KIR-like genes on a different chromosome. The mouse LRC also encodes paired Ig-like receptors (PIRs), which represent putative orthologs of human LILR genes (Takai 2005).
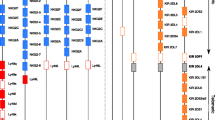
Mammalian NKRs. Representative members of the human KIR, human CD94/NKG2 and mouse Ly49 multigene families of NKRs are shown along with examples of single-copy human NKRs; NKp30, NKp44, and NKp46 (also known as natural cytotoxicity receptors). Inhibitory NKRs possess cytoplasmic immunoreceptor tyrosine-based inhibition motifs (ITIMs); activating NKRs possess a positively charged transmembrane residue (+). Activating NKRs utilize adaptor proteins with a negatively charged transmembrane residue (−) (e.g. DAP12, DAP10, FcεRIγ, or CD3ζ) to effect cytoplasmic signaling; although two DAP12, FcεRIγ or CD3ζ adaptor proteins associate with an individual activating NKR only one is shown (dimer model for DAP12 is shown in Fig. 1). The exception is KIR2DL4 which possesses an ITIM, but signals via FcεRIγ. Specific MHC I ligands have been defined for most KIR, NKG2 and Ly49 receptors. NKp30 binds the tumor cell ligand B7-H6 and NKp46 binds viral haemagglutinins (HA). NKp46 can partner with either CD3ζ or FcεRIγ. NKG2D can partner with either DAP10 or DAP12. All other NKG2 receptors hetero-dimerize with CD94. More complete descriptions of the human and mouse NKRs and their adaptors and ligands are available (Biassoni 2009; Brandt et al. 2009; Call and Wucherpfennig 2007; Mandelboim et al. 2001; Yokoyama and Plougastel 2003). CTLD C-type lectin domain, C2 C2 Ig domain, V V Ig domain, MIC MHC class-I-related molecule, ULBP cytomegalovirus UL16 binding proteins
Identification of NKRs in primates. Data are summarized from the Electronic supplementary material (Table S1). Question marks indicate genes that have yet to be reported as present or absent. Plus sign indicates that additional genes are likely present. Strike-through text indicates a pseudogene. Phylogenetic tree adapted from (Glazko and Nei 2003)
Identification of NKRs in non-primate eutherian mammals. Data are summarized from the Electronic supplementary material (Table S1). NKR data for primate species are presented in Fig. 3. Asterisk indicates that although mouse possess a KIR gene, it not encoded at the LRC. Question marks indicate genes that have yet to be reported as present or absent. Plus sign indicates that additional gene are likely present. Strike-through text indicates a pseudogene. Not all lineages are shown and species included are biased towards those with described NKRs. Phylogenetic tree adapted from (Arnason et al. 2008)
The human LRC also encodes the single-copy Ig-type natural cytotoxicity receptor gene, NKp46 (NCR1). The other single-copy Ig-type natural cytotoxicity receptors genes, NKp30 (NCR3) and NKp44 (NCR2), are encoded on chromosome 6p21 (Fig. 2) (Barrow and Trowsdale 2008; Biassoni et al. 2001). Of these receptors, NKp46, is the most conserved between mammalian species and it has been reported as a “unifying marker” for NK cells across mammalian species. A single NKp46 ortholog has been described in multiple primates, cows, pig, sheep, dog, and in multiple strains of mice. No NKp44 ortholog has been reported in mice, in which NKp30 is a pseudogene (Walzer et al. 2007a; Walzer et al. 2007b). These marked interspecific variations, which will be further elaborated upon below, underscore the likely functional redundancy within NKR systems and are predictive of difficulties that may be introduced in broader phylogenetic investigations of both NKR structure and function. No evidence for orthologs of the natural cytotoxicity receptors has been described outside of eutherian mammals.
The natural killer complex (NKC) is located on human chromosome 12 and mouse chromosome 6 and encodes multiple lectin-type receptor families that are type II transmembrane proteins and include the NKR family of inhibitory and activating CD94/NKG2 receptors (Fig. 2). CD94 forms heterodimers with various NKG2 receptors such as NKG2A, NKG2B (which is a splice variant of NKG2A), NKG2C, NKG2E, and NKG2H (which is a splice variant of NKGE); by contrast, NKG2D is a homodimer. The genomic organization of the CD94/NKG2 family is relatively constant between primate and rodent species with two exceptions, specifically: (1) NKG2CE, which is predicted to represent the precursor gene that gave rise to both NKG2C and NKG2E, is encoded in several primates and (2) three CD94 and five NKG2A/C/E-like genes have been described in the grey mouse lemur (Fig. 3 and the Electronic supplementary material (Table S1)). The number and organization of CD94/NKG2 genes in the small number of non-primate and non-rodent mammals that have been investigated varies with NKG2D being the most conserved (Fig 3 and the Electronic supplementary material (Table S1) and discussed below). In addition to the CD94/NKG2 receptors, the NKC of mouse includes the Ly49 family of lectin-type NKRs. The number of Ly49 genes encoded at the NKC varies between rodent species (Fig. 4 and the Electronic supplementary material (Table S1)) as well as between strains of mice (eight in BALB/c, 15 in C57BL/6, 19 in 129S6 and 21 in NOD). In contrast, the NKC of human and certain other primates encodes a single Ly49L pseudogene (Fig. 3 and the Electronic supplementary material (Table S1)) (Carlyle et al. 2008; Yokoyama and Plougastel 2003). The expansion of the Ly49 genes has been observed only in rodents and horse (Fig. 4 and the Electronic supplementary material (Table S1)). Despite the considerable intra- and interspecific variation in gene number and complexity, a unifying theme between the mammalian Ig-type and lectin-type NKRs is that they are expressed on NK cells, bind MHC I or proteins that share structural similarities with MHC I, and can inhibit or activate target cell killing and/or cytokine release through competing signaling pathways.
What can mammalian KIR and Ly49 genes inform us about non-mammalian NKRs?
The characterization and cataloging of genomic and cDNA sequences encoding KIRs and Ly49s from numerous mammals, including human and mouse as well as other primates, rodents, carnivores, ungulates and representative marsupial and monotreme species has provided a wealth of information about how NKRs have evolved within the relatively narrow period of vertebrate phylogeny that is represented by the mammalian radiations (Figs. 3 and 4 and the Electronic supplementary material (Table S1)). The extensive databases of receptor structures and functional specificities provide a basis for understanding some of the common features these receptors share with other immune receptors encoded by multigene families as well as many of the unique influences that shape the functional NK repertoire.
In terms of the broad evolutionary focus of this review, several points are of particular phylogenetic significance of which the disparity in numbers and complexity of KIRs relative to Ly49 is the most apparent. As alluded to above, primates typically encode multiple KIR genes at the LRC and a single, and commonly non-functional, Ly49 gene at the NKC. In contrast, mice and rats encode multiple Ly49 genes at the NKC and a single or no KIR gene at the LRC. Seals, sea lions and the grey mouse lemur possess a single, intact and presumably functional KIR gene and a single, intact and presumably functional Ly49 gene (Hammond et al. 2009). In marked contrast, the dog lacks a KIR gene and its single Ly49 lacks a conserved cysteine suggesting that its function might be altered from other Ly49 receptors (Gagnier et al. 2003; Hammond et al. 2009). Given the relatively high level of variation, the significance of any given NKR in terms of overall NK function across diverse species is uncertain.
An evaluation of the KIR genotypes in higher vertebrates led to a model in which a single ancient KIR gene duplicated ∼135 MYA (predating the radiation of placental mammals, after the genesis of the monotremes and coincident with the emergence of marsupials) (Guethlein et al. 2007; Parham et al. 2010). Depending on the lineage, one duplicated KIR gene underwent an expansion leading to multiple KIR genes in cow whereas the other duplicated KIR gene underwent an expansion leading to multiple KIR genes in primates. This model suggests that during the evolution of pig and dog either one or both duplicated KIR genes were lost. If this model is correct, it would suggest that if an expansion of KIR genes outside of mammals is observed, it would be reflective of a lineage-specific KIR expansion rather than an overriding phylogenetic trend of gain and loss of gene complexity. Balancing selection, which factors in the maintenance of HLA class I polymorphism, likely accounts for both the haplotypic and allelic complexity of KIRs (Gendzekhadze et al. 2009; Norman et al. 2007). Of the many assumptions that have been drawn from a very large data set, the studies in primates suggest what may be one unifying theme, i.e., the diversity and complexity of the KIR locus is related directly to variation and complexity in their MHC I ligands; KIRs and their MHC I ligands appear to be co-evolving (Parham et al. 2010). Given the considerable genomic information that has been acquired for species that appear in phylogeny prior to the mammals, NKRs in lower vertebrates might be expected to exhibit complex haplotypic variation and differ markedly between species. Furthermore, their overall complexity may be correlated with complexity of MHC I molecules. In this regard, it is important to note that MHC I is conserved across jawed vertebrate species (Kulski et al. 2002) and the degree of MHC I polymorphism throughout the jawed vertebrates may well be equivalent to that seen in mammals (Flajnik 2002). As such, MHC I and potentially MHC II, which also is conserved phylogenetically, remain “general” candidates for NK receptor ligands among all jawed vertebrates even though the receptor types that recognizes these molecules may be structurally dissimilar. It is critical to note that past successes in charactering mammalian NKR-MHC I interactions have relied heavily on dedicated cell lines, which are available for very few lower vertebrate systems. Furthermore, ligand searches that are based on recognition of recombinant MHC I structures can be technologically cumbersome as appropriate folding and certain expression densities are difficult to achieve using routine cell transfection strategies since effective presentation requires the presence of β2-microglobulin. Finally, binding of molecules such as CpG by KIRs (Sivori et al. 2010) underscores the importance of considering other classes of potential ligands.
Genetic searches for NK-type receptors in non-mammalian vertebrates
Given our current understanding of NK receptors in mammals and paucity of functional assays that could be applied in lower vertebrates, the most obvious, albeit potentially misleading, means to search for NK orthologs is based on nucleotide sequence identity/homology. On the simple assumption that Ig-type molecules often mediate immune-related functions, including at least certain forms of NK reactivity, we and others have experimentally targeted large families of Ig-type genes that do not represent rearranging receptors or orthologous forms of molecules that are not associated with NK function. Such approaches are fraught with technical and interpretive complications. As an example, in mammals there are many Ig gene families, e.g., Fc receptors, which bear an overall general resemblance to KIRs but effect very different functions. Likewise, certain lectin-type receptors that function as immune mediators are related to genes that exhibit unrelated functions. Overlaps in structure and signaling function preclude unequivocal assignments of orthology (e.g., the C-type lectin domains of NKRs belong to the group V family of C-type lectin proteins, but not all group V proteins are NKRs). Parallel searches that seek to identify monomorphic forms of NKRs are confounded by the very rapid changes that occur in gene families that are presumably under strong (immediate) selective pressure.
Notwithstanding these complications and noting that sequence identity does not necessarily predict functional homology, a number of approaches based largely on molecular homology have been carried out, including: low stringency library screens, degenerate primer PCR strategies and genome screening analyses, including EST scanning. The criteria that we propose to be characteristic of NKRs include: (1) the molecule must to be expressed on NK cells, (2) the receptor must play a role in differentiating between normal and abnormal (e.g., infected, transformed, or stressed) cells, (3) ligand engagement by the receptor must initiate a signaling cascade within the NK cell, and (4) the signaling cascade must influence cytotoxicity or another classic immune function such as cytokine secretion. Whereas marsupials and monotremes exhibit receptors resembling NKRs of the eutherian mammals, the vast majority of putative NKR-like molecules identified below the phylogenetic level of mammals fall short of meeting all of these conditions; however, certain molecules exhibit at least some major features of NKRs and are discussed within the context of the species in which they have been identified.
NKRs in marsupials and monotremes
Marsupials and monotremes diverged from eutherian mammals ∼145 and ∼175 MYA, respectively (Miller 2010). It might be predicted that marsupials and monotremes would encode NKRs similar to eutherian mammals; however, a paucity of KIR, Ly49 and CD94/NKG2 genes has been observed in the genomes of opossum (marsupial) and platypus (monotreme). Analyses of the platypus genome identified no KIR genes and more than 213 lectin domains that may represent NKRs; however, no clear orthologs of Ly49 or CD94/NKG2 were reported (Warren et al. 2008; Wong et al. 2009). Analyses of the opossum genome identified 124 KIR/LILR-like Ig domains, no Ly49 genes, and only a single NKG2D ortholog (Belov et al. 2007; Hao et al. 2006). Opossum NKG2D is encoded at the NKC along with eight additional C-type lectin receptors. An opossum ortholog of MIC, a NKG2D ligand, has been described (Belov et al. 2007). In contrast to KIR genes, the CD94/NKG2 family (with the exception of NKG2D), recognizes non-polymorphic, non-classical MHC I (Borrego et al. 2006) and is more highly conserved between mammalian species. The conservation of NKG2D in primates, rodents, pig, cow, dog and opossum does not fit the paradigm of a rapidly evolving NKR (as compared with KIRs and Ly49 genes) and suggests specific selective pressure to maintain its function.
CD94/NKG2 in chicken
Avian species diverged from mammalian species ∼326 MYA (Blair and Hedges 2005). More so then in other non-mammalian species, the susceptibility of avians to numerous pathogens as well as the genetic basis for susceptibility and resistance to infectious disease have been characterized extensively (Rogers et al. 2008); furthermore, unique aspects of MHC I molecules in these species potentially could be reflected in their NKRs (Kaufman et al. 1999). A single gene exhibiting homology to CD94/NKG2, which displays conserved synteny with the human and mouse NKC, is located in a region of chicken chromosome 1 (Fig. 5) (Chiang et al. 2007). Chicken CD94/NKG2 is physically linked to orthologs of multiple genes located at or near the NKC, including CD69, which represents an early activation gene that also is present in the opossum. Although chicken CD94/NKG2 and CD69 are group V C-type lectins, CD69 does not possess any obvious signaling motifs, is broadly expressed across multiple tissues and unlikely to function as a NKR. In contrast, chicken CD94/NKG2 is expressed primarily in leukocytes and lung. The cytoplasmic tail of chicken CD94/NKG2 encodes: GYTALNLRTPASDITDGYLSN, which resembles but does not fully match the ITIM (S/I/V/L)xYxxx(I/V/L) or the ITAM (YxxLX6-12 YxxL/I) consensus (Chiang et al. 2007); CD94/NKG2 may be an inhibitory, activating or dual-signaling NKR. CD94/NKG2 in chicken is related more to mammalian CD94 than to individual NKG2 family members. The presence of CD94/NKG2 below the phylogenetic level of the avians is uncertain (see below) and authentic Ly49 orthologs have not been identified outside of eutherian mammals.
Candidate avian (chicken) NKRs. Various group V lectin-type receptors that map within the NKC (CD94/NKG2), near MHC-like genes in the Rfp-Y region (17.5/Y-Lec family), and near the MHC B locus (B-NK/B-Lec) have been identified in chicken. CD94/NKG2 and B-NK are the only chicken lectin-type receptors identified thus far that possess inhibitory or activating motifs. Six representative chicken Ig-like receptors (CHIRs) are shown and reflect the diversity of 84 CHIR genes characterized to date, including inhibitory CHIR-A, activating CHIR-B and CHIR-AB, which possess putative inhibitory and activating signaling motifs; certain CHIRs lack known signaling motifs (e.g., CHIR-B6). CHIR-AB1 functions as an Fc receptor (for IgY) and along with CHIR-A2, can partner with FcεRIγ. Asterisk indicates that structures have been confirmed by analysis of EST or cDNA sequences. C-type lectin domains (CTLDs), C2 Ig domains (C2), positively charged transmembrane residues (+), cytoplasmic immunoreceptor tyrosine-based inhibition motifs (ITIMs), immunoreceptor tyrosine-based activation motifs (ITAM), immunoreceptor tyrosine-based switch motifs (ITSM), the YXXM activating motif, and endocytosis signal motif (Endo) are indicated
Other C-type lectins in all vertebrates
Additional candidate C-type lectin (group V) NKRs have been identified on chicken chromosome 16 in the B locus, which encodes the classical MHC, and in the Rfp-Y locus, which encodes non-classical MHC genes (reviewed in (Rogers et al. 2008)). A C-type lectin receptor encoded by gene 17.5 in the chicken MHC Rfp-Y genomic region shares sequence similarity with mammalian NK receptor NKRP1 (aka CD161, NKLRB1) (Bernot et al. 1994). The 17.5 gene is predicted to encode a transmembrane receptor with a single extracellular group V C-type lectin domain that lacks the features of an activating or inhibitory NKR. Y-Lec1 and Y-Lec2 represent additional group V C-type lectin receptor genes in the Rfp-Y region and are similar to 17.5 in overall structure, including the lack of signaling motifs (Fig. 5) (Rogers et al. 2003). Additional 17.5-like genes have been identified (Bernot et al. 1994). The expression of 17.5-like genes is elevated in spleen and a lymphoblastoid cell line, which is consistent with lymphocyte expression; weaker expression is detected in the thymus (Bernot et al. 1994).
Chicken B-NK (NKr) and B-Lec genes represent additional group V, C-type lectin receptors (similar to 17.5) that map to the chicken B locus (Fig. 5) (Kaufman et al. 1999). B-NK shares sequence similarity to mammalian NKRP1, encodes a cytoplasmic ITIM and is transcribed actively by a NK cell line but not by B, T and macrophage cell lines (Kaufman et al. 1999; Rogers et al. 2005). In contrast, B-Lec encodes a cytoplasmic endocytosis motif, is transcriptionally activated by PMA and is more similar to the group V C-type lectin early activation antigens CD69, LLT1, and AICL (Rogers et al. 2005). A different haplotype of B-NK and B-Lec seen in red jungle fowl includes additional C-type lectin receptor genes, suggesting that they underwent recent and rapid birth and death events. Genomic sequences from quail reveal multiple genes similar to B-NK and B-Lec as well as marked variation in gene numbers between haplotypes (Hosomichi et al. 2006).
Bony fish (Osteichthyes) include the ray finned (Actinopterygii) and the lobe finned fish (Sarcopterygii). The ray finned fish diverged from tetrapods ∼476 MYA and lobe finned fish diverged from other tetrapods ∼430 MYA (Blair and Hedges 2005). Functional assays have demonstrated that teleosts possess cytotoxic cells capable of xenogeneic and allogeneic recognition and killing (Evans et al. 1984; Fischer et al. 2006; Moss et al. 2009; Shen et al. 2004; Somamoto et al. 2004; Stuge et al. 1997). A candidate lectin NKR, termed cichlid KLR (cKLR) was identified in an EST screen of Paralabidochromis chilotes (a cichlid fish). cKLR consists of a single extracellular C-type lectin domain and a positively charged transmembrane residue. cKLR is expressed in leukocytes and pharyngeal tissues but not in 11 other tissues, including: erythrocytes, intestine, kidney, and spleen (Sato et al. 2003). At least 28 cKLR genes, of which at least half are predicted to be pseudogenes, were identified in Oreochromis niloticus, a related cichlid species. Although complete sequences of only a few cKLR genes are available, seven of eight identifiable transmembrane domains possess a charged residue suggesting a high density of activating receptors (Kikuno et al. 2004). Based on structural and sequence similarities, the cKLRs were predicted initially to encode group V C-type lectin domains, suggesting a relationship to mammalian CD94/NKG2 genes (Sato et al. 2003); however, no conserved synteny has been demonstrated between cKLRs and mammalian NKRs. CD209 (DC-SIGN) in zebrafish, medaka and fugu exhibits the highest similarity to cKLR (3e−04 to 2e−10), suggesting that the cKLR genes may be a derived feature in cichlid species (Yoder, unpublished observations). In addition, the results from a whole genome survey of the pufferfish Fugu rubripes reported no evidence for group V C-type lectins in Fugu suggesting that, perhaps, bony fish lack group V C-type lectin receptors (Zelensky and Gready 2004). A phylogenetic comparison of a cKLR lectin domain with 66 other groups II and V lectin domains indicates that cKLR is actually a group II lectin (Zelensky and Gready 2004). Additional group II lectin receptors have been described in other fish species that include inhibitory and/or activating forms (Panagos et al. 2008; Soanes et al. 2004; Soanes et al. 2008; Zhang et al. 2000) and although some of these receptors may influence NK function, they are not reviewed here.
A cDNA encoding a C-type lectin protein, termed BsCD94-1, was identified in a urochordate (Botryllus schlosseri) and reported as a CD94-related receptor. BsCD94-1 encodes a type II transmembrane protein with a single extracellular lectin domain and lacks conventional inhibitory or activating motifs. Southern blot analyses reveal polymorphic variation and the possible presence of additional homologous genes. Genomic sequence information is not available to determine if the BsCD94-1 locus (or possible gene cluster) is potentially syntenic to the mammalian NKC (Khalturin et al. 2003). Initial phylogenetic analyses suggested that BsCD94-1 is most related to mammalian CD94, and NKGD, but subsequent phylogenetic analyses with additional proteins suggest that BcCD94-1 is more similar to CLEC5A (Sato et al. 2003).
Taken together, the evidence for a single CD94/NKG2 gene within the NKC in opossum and chicken and the observation that Ly49 genes only have been identified in eutherian mammals suggests that the expansion of these genes may be specific to the mammalian lineage. No convincing evidence has been presented for a syntenic NKC in any species below chicken.
Ig-type NKRs in chicken
Although no clear orthologs of KIRs have been identified outside of mammals, multiple Ig-type receptors that share characteristics with KIRs have been identified in marsupial (discussed above), avian, amphibian and fish species. Chicken Ig-type receptors were identified in EST database searches using a mouse PIR query, leading to the identification of the chicken Ig-like receptors (CHIR) that consist of activating (CHIR-A) and inhibitory (CHIR-B) forms (Dennis et al. 2000). In addition, CHIRs encoding cytoplasmic ITSMs as well as forms with a charged transmembrane and cytoplasmic ITIM, termed CHIR-AB, have been described (Fig. 5) (Nikolaidis et al. 2005; Viertlboeck et al. 2005). The chicken CHIR locus, which is located on chromosome 32 and shares conserved synteny with the human LRC, encodes 84 genes (35 CHIR-A, 26 CHIR-B, and 23 CHIR-AB), and 46 pseudogenes (Laun et al. 2006; Lochner et al. 2010; Nikolaidis et al. 2005; Viertlboeck et al. 2005). By contrast, 103 different nucleotide sequences encoding 98 different two Ig domain CHIR proteins have been identified from an individual M11 chicken. A single R11 chicken revealed 76 different CHIR nucleotide sequences encoding 70 different proteins, of which only three are100% identical with the M11 animal (Viertlboeck et al. 2010). The CHIR gene cluster is highly polymorphic and likely is undergoing a recent and rapid evolution (Lochner et al. 2010; Viertlboeck et al. 2010).
CHIR-B2 is expressed on the surfaces of B cells and a subset of T cells. CHIR-AB1 is expressed on B cells, NK cells, and monocytes/macrophages (Viertlboeck et al. 2004; Viertlboeck et al. 2005; Viertlboeck et al. 2007). Expression of CHIRs in B cells and monocyte/macrophages more reflects a LILR than a KIR expression pattern in mammalian NK cells. CHIR-AB1 is a high affinity receptor of avian IgY (Arnon et al. 2008; Viertlboeck et al. 2007), and other CHIR-AB1-like molecules also may bind IgY at different relative affinities or mediate unrelated functions of which NK activity cannot be ruled out (Viertlboeck et al. 2009).
MHC-linked Ig superfamily V genes in Xenopus
Amphibians diverged from mammals (e.g., amniotes) ∼370 MYA (Blair and Hedges 2005). The ease of in vitro fertilization and the large and transparent nature of Xenopus laevis embryos make this species a powerful developmental model. NK-like cytotoxic cells capable of effecting allogeneic killing have been identified in Xenopus (Horton et al. 1996) and studies with thymectomized animals have defined an NK cell population that is capable of allogeneic killing (Horton et al. 2000; Horton et al. 2003; Rau et al. 2002). Inbred MHC homozygous strains and gynogenetically produced MHC-defined syngeneic clones of X. laevis provide advantages for immunological investigations (Robert and Ohta 2009). However, the genome of X. laevis is pseudo-tetraploid and confounds both genetic mapping and genome sequencing. Genomic efforts have focused on the diploid species Xenopus tropicalis (Hellsten et al. 2010).
Six Xenopus MHC-linked Ig superfamily V genes (XMIV) that are candidate Ig-type NKR genes, which include both putative activating and inhibitory forms, have been identified. Different Xenopus species possess XMIV genes that encode either one or two V domains (Ohta et al. 2006) (Fig. 6). Genes encoding XMIVs and the human natural cytotoxicity receptor NKp30 are located at the same relative position within the MHC class III region; however, there is only minimal sequence similarity between NKp30 and XMIV (Ohta et al. 2006). The expression patterns of XMIV genes have not yet been determined.
Representative candidate amphibian (Xenopus) NKRs. Xenopus MHC-linked Ig superfamily V (XMIV) are predicted to encode both inhibitory and activating forms. Xenopus Ig-like receptors (xILRs) identified thus far are predicted to exhibit activating function. Asterisk indicates that structures have been confirmed from EST or cDNA sequences. Proteins encoded by alternatively spliced transcripts are denoted with “sp”. V Ig domains (V), C2 Ig domains (C2), positively charged transmembrane residues (+), cytoplasmic immunoreceptor tyrosine-based inhibition motifs (ITIMs), immunoreceptor tyrosine-based activation motifs (ITAM), and immunoreceptor tyrosine-based switch motifs (ITSM) are indicated
Additional Ig-like receptor genes in Xenopus
Multiple cDNAs with weak identity to mammalian KIRs, LILRs, PIRs and avian CHIRs also have been identified in X. laevis and X. tropicalis. The Xenopus Ig-like receptors (xILRs) possess one to three Ig domains and all forms identified thus far are predicted to encode activating receptors (Fig. 6). Three xILR genes are expressed in skin, kidney, testes, brain, intestine, spleen, liver and muscle; the expression of a fourth xILR was detected primarily in: testes, brain, intestine and spleen. Although the xILRs share overall protein architecture with KIRs, LILRs, PIRs and CHIRs (Guselnikov et al. 2010), their expression profiles do not resemble those described for other NKRs.
Novel immune-type receptors in bony fish
Our initial efforts to detect NK-type receptors in bony fish were founded on the postulate that some form of NK recognition could be initiated by the V region of a cell membrane-bound receptor that is neither an Ig nor a TCR and would not undergo somatic diversification. Such a receptor likely would be encoded in an extended gene cluster. A strategy, based on short sequence homologies (three to four amino acids), was developed that potentially could amplify any V region from genomic DNA. Pufferfish was chosen for its relatively reduced genome content and markedly shorter intervening sequences, which facilitates sequencing of gene clusters, and minimizes the types of PCR amplification artifacts that arise from the use of short primers; the novel immune-type receptors (NITRs) were identified using this approach (Rast et al. 1995). The prototypic NITR molecule consists of an N-terminal V domain (most often containing a J homology region), an intermediate (I) domain, a transmembrane region, cytoplasmic tail with an ITIM signaling motif(s). NITRs can be depicted graphically as a fusion between: (N-terminal) a TCR-like molecule and (C-terminal) an Ig domain-containing receptor, such as a KIR, that functions in activating/inhibitory signaling (Strong et al. 1999). NITRs are members of large diversified multigene families that are ubiquitous in bony fish (Fig. 7) (Ferraresso et al. 2009; Hawke et al. 2001; Yoder et al. 2001; Yoder et al. 2004; Yoder 2009).
Representative candidate bony fish NKRs. Predicted protein structures of select NITRs from zebrafish, NILTs from trout and LITRs from catfish. Ten different NITR forms are inferred from 39 different zebrafish gene sequences. Cross-linking inhibitory NITRs down-regulate the MAPK signal transduction pathway presumably via SHP-1 and SHP-2 (not shown). Activating NITRs associate with and signal through the adaptor protein Dap12 and, in the context of human NK cells, redirect cytolysis of target cells (in vitro). Six NILTs isoforms are encoded by four genes in trout via alternative mRNA splicing. Neither a NILT ligand nor functional confirmation of NILT signaling has been reported. Certain ITIM-containing LITRs can associate with both SHP-1 and SHP-2 (not shown). Surface expression of certain activating LITRs is enhanced by cotransfection with FcεRIγ and FcεRIγ-like molecules. FcεRIγ and FcεRIγ-like proteins can be co-precipitated with activating LITRs. Although a CD3ζ-like adaptor protein can be co-precipitated with an activating LITR, its expression does not influence the surface expression of activating LITRs and is not shown. Asterisk indicates that structures have been confirmed from EST or cDNA sequences. Proteins encoded by alternatively spliced transcripts are denoted with “sp”. V Ig domains (V), I Ig domains (I), C2 Ig domains (C2), positively charged transmembrane residues (+), cytoplasmic immunoreceptor tyrosine-based inhibition motifs (ITIMs), ITIM-like sequences (itim), immunoreceptor tyrosine-based activation motifs (ITAM) and immunoreceptor tyrosine-based switch motifs (ITSM) are indicated. The classifications: LITR1-like and 2-like sequences are described in (Stafford et al. 2007)
NITRs are complex V region-containing activating and inhibitory molecules
At the gene family level, NITRs: (1) encode one (V) or two (V and I) domains, (2) lack or possess a positively charged transmembrane residue, (3) possess different numbers or lack ITIMs in the cytoplasmic region, and/or (4) lack or possess a transmembrane region (Fig. 7). The overall degree of structural variation is among the most extensive seen in any large IgSF. The positively charged transmembrane residue in NITRs, which is associated with activating function, interacts with DAP12 (Wei et al. 2007).
Annotation of NITR genes is complete for both zebrafish and the Japanese medaka (Oryzias latipes) (Desai et al. 2008; Yoder et al. 2004; Yoder et al. 2008). In zebrafish, 12 NITR gene families comprising 36 genes have been identified on chromosome 7; two other NITR gene families are represented on chromosome 14. Efforts to identify NITRs outside of bony fish using PCR-based methods, genome database screens and EST screens as of yet have not been successful. In addition, 24 families of NITRs in medaka are located in gene clusters on chromosomes 10, 18, and 21 and show a pattern of intrafamily diversification similar to that seen in pufferfish and zebrafish (Desai et al. 2008). In each of these cases, the largest gene family is arbitrarily designated NITR1 and exhibits multiple variant forms (as defined by 70% or greater sequence relatedness to other members of the same family). The availability of stable inbred lines of medaka, the lack of which represents a significant shortcoming in zebrafish immunological research, greatly facilitate genome assembly and will benefit efforts to elucidate NK function.
Only minimal sequence overlaps are evident in comparisons between the V regions of zebrafish and medaka, i.e., V families in zebrafish are no more related to those seen in medaka than the different zebrafish families are related to each other. Such species-specific expansions of various NITR types have been interpreted to reflect recent “birth” and “death” events as described above for other receptor types (Desai et al. 2008; Ferraresso et al. 2009). Similar patterns of sequence variation in the V regions of Igs and TCRs as well as in the (non-rearranging) KIR family of NK receptors have been reported (Martinez-Borra and Khakoo 2008; Ota et al. 2000; Parham 2005); all three receptor families likely have undergone a parallel process of evolution. Finally, evidence of positive Darwinian selection in the V domain of NITR genes supports the hypothesis that NITRs may function as NKRs.
The majority of NITRs function through ITIM-based inhibitory mechanisms (Yoder 2009). Nitr9 is the activating NITR present in zebrafish and is encoded by a single-copy gene that undergoes alternative splicing to generate three isoforms: Nitr9-L (the long form that encodes both V and I domains), Nitr9-S (a shorter form in which splicing deletes part of the V domain) and Nitr9SS (the shortest form in which splicing removes the entire V domain) (Wei et al. 2007). Six different adaptor molecule orthologs: Dap10, Dap12, CD3ζ, CD3ζ-like, FcεRIγ and FcεRIγ-like, have been identified in zebrafish (Yoder et al. 2007). Nitr9L partners preferentially with the zebrafish ortholog of mammalian Dap12 (Wei et al. 2007) and cytotoxicity was shown to be induced in human NK cells by transfection with zebrafish Nitr9L and Dap12 (Yoder et al. 2004). Cross-linking of the Nitr9L-Dap12 complex activates the phosphytidylinositol 3-kinase → AKT → extracellular signal-regulated kinase pathway (Wei et al. 2007). Activating NITRs appear to participate in adaptor-based pathways similar to those associated with mammalian KIR function.
NITR expression patterns
Developmental stage-specific expression of the 14 families of zebrafish NITRs has been characterized in 0 h post-fertilization embryos through 6-day post-fertilization (dpf) larvae as well as in adult tissues (Yoder et al. 2010). Larval expression (trace levels) of nitr1, nitr5, nitr7, nitr10, and nitr11 genes can be detected at 6 dpf. Only nitr3 is expressed throughout and at the highest levels during embryonic development.
NITR expression was evaluated from lymphoid and myeloid cells separated from zebrafish whole kidney marrow via flow cytometry by forward and side scatter (Traver et al. 2003; Yoder et al. 2007). All zebrafish NITRs are expressed in lymphoid but not myeloid cells (Yoder et al. 2010); a similar conclusion was reached on the basis of equivalent levels of expression of NITRs in kidney (the major site of lymphopoiesis in bony fish) in rag1 −/− and rag1 +/− zebrafish (Jima et al. 2009). NITRs are expressed in non-T, non-B lymphocytes, consistent with expression patterns of other NKRs.
Although lineage-specific antibodies to NK cell types in bony fish have not been characterized, NK-like cell lines that lack markers for neutrophils, macrophages, B cells and T cells, express perforin and multiple granzymes, possess dense cytoplasmic granules, and demonstrate allogeneic killing have been derived from clonal channel catfish (Ictalurus punctatus) (Shen et al. 2002; Shen et al. 2004; Stuge et al. 1997; Yoder 2004). The 3H9 cell line was derived by allogeneic stimulation and selection, exhibits in vitro cytotoxicity for the IG8 (stimulator) allogeneic B cell line, possesses azurophilic cytoplasmic granules, does not express Ig heavy chains or TCRαβ and is negative for staining with an anti-nonspecific cytotoxic cell (NCC) antibody. Collectively, these observations, along with general morphological characteristics of 3H9 cells, are consistent with the possibility that 3H9 is an NK-related cell lineage (Shen et al. 2002). Multiple NITRs are expressed by the 3H9 cell line (Evenhuis et al. 2007; Hawke et al. 2001) and in macrophage, T, B, and other CTL cell lines (Evenhuis et al. 2007).
Allogeneic recognition by an NITR
A cDNA library was derived from the 3H9 NK-like cell line and different NITRs were characterized (Hawke et al. 2001). An in vitro assay that coupled different NITR ectodomains to the cytoplasmic domain of CD3ζ was developed in which constructs were transfected into a mouse T cell hybridoma line (43-1) that contains a GFP transgene under the control of an NFAT-responsive promoter (Cannon et al. 2008). Of nine different catfish NITR ectodomains that were tested, only NITR11 produced a positive response with IG8 but not with 3B11, an MHC I/II disparate control B cell line for which 3H9 exhibits only minimal cytotoxicity (Shen et al. 2002). The same specificity patterns were revealed when the same group of NITR ectodomains was coupled to Fc regions (NITR V-Fc chimeras). NITR10, which is structurally related to NITR11, does not bind IG8. Using gain and loss of function mutants, NITR11 specificity for IG8 ultimately was localized to (i.e., shown to be dependent on) Asn50 (Cannon et al. 2008).
The crystal structures of NITR10 and NITR11 as well as their respective gain and loss of function mutants were solved in both native and selenomethionine forms. NITRs adopt the same dimerization mode as the antigen receptors and conserve the structural features imparted to (TCR) VαVβ and (Ig) V H V L interfaces by the highly conserved FGXG motif present in the J segment, a structural feature shared by a large number of NITRs and the V domain-containing, heterodimeric proteins that mediate adaptive immunity. The specificity of NITR11 for a determinant on the surface of the IG8 B cell line is dependent on the local electrostatic potential of Asn50 in the (Ig/TCR) CDR1 analogous loop in the V domain, referred to above (Cannon et al. 2008).
Of the many classes of Ig receptors now classified in fish, the NITRs are most appealing as potential mediators of NK-type function. NITRs are expressed by lymphocytes and NK-like cell lines (Evenhuis et al. 2007; Hawke et al. 2001; Yoder et al. 2010), a NITR has been implicated in allogeneic recognition (Cannon et al. 2008), cross-linking NITRs initiates the same signaling pathways as mammalian NKRs (Wei et al. 2007; Yoder et al. 2001) and, in the context of human NK cells, NITRs can influence cytotoxicity (Yoder et al. 2004). Specifically, the expression patterns, role in allogeneic recognition, possible cytotoxicity and signaling pathways in large part meet the criteria for NKRs. However, whether or not this gene family accounts for any, all, or some NK function remains open to questions. What is increasingly more certain is that if NK reactivity is ubiquitous throughout at least the jawed vertebrates, the real possibility exists that this function is mediated by diverse individual and multigene families of molecules, of which bony fish possess many.
Novel Ig-like transcripts in bony fish
The prototypic inhibitory (cytoplasmic ITIM) and activating (cytoplasmic ITAM) novel Ig-like transcripts (NILTs) that possess a single Ig domain and are structurally similar to TREM, CMRF35 and the natural cytotoxicity receptor NKp44 were identified first in carp (Cyprinis carpio L.) (Stet et al. 2005). NILTs with two Ig domains, a NILT with both ITAM and ITIM sequences and a NILT that is predicted to be secreted also were identified in rainbow trout (Fig. 7) (Kock and Fischer 2008; Ostergaard et al. 2009). The highest levels of NILT expression in carp and trout are in spleen, kidney, intestine and gills (Ostergaard et al. 2009; Stet et al. 2005). NILTs are highly polymorphic and 53 different NILT-like sequences were identified in an individual carp. NILTs and the zebrafish MHC I “ze” loci (Dare-ZE) are linked on chromosome 1, reminiscent of the linkage between MHC and the NKp44/TREM gene cluster on human chromosome 6 (Stet et al. 2005).
Leukocyte immune-type receptors in bony fish
The prototypic leukocyte immune-type receptors (LITRs) LITR1, LITR2 and LITR3, were identified as ESTs from channel catfish. LITR1 encodes four extracellular C2 domains and a cytoplasmic ITIM and ITIM-like sequence as well as an ITSM. LITR2 and LITR3 encode three and six extracellular C2 domains, respectively, and both possess a positively charged residue within their transmembrane domains (Stafford et al. 2006). Additional LITRs have been identified that vary in the number of extracellular Ig domains as well as in the presence of cytoplasmic ITAMs, ITIMs, and ITSMs (Fig. 7) (Stafford et al. 2007). LITRs are encoded on three different zebrafish chromosomes and are polymorphic and polygenic (Stafford et al. 2006). Certain LITRs can homo- and hetero-dimerize when transfected in cell culture (Mewes et al. 2009).
Cytoplasmic tails of LITR1 family members transfected into mammalian cells are tyrosine phosphorylated after pervanadate stimulation. Zebrafish SHP-1 and SHP-2 can be immunoprecipitated with LITR cytoplasmic domains that include an ITIM (Montgomery et al. 2008). Co-expression of recombinant activating LITRs with the catfish adaptor proteins, FcεRIγ and FcεRIγ-like, enhance the surface expression of activating LITRs. Co-immunoprecipitation studies have established the physical association of an activating LITR with the adaptor proteins FcεRIγ, FcεRIγ-like, and CDζ-like (Mewes et al. 2009).
The highest level of LITR expression is in the kidney; reduced expression is seen in the spleen and gill and much lower levels are detected in muscle, heart, liver, intestine and thymus. LITR1 and LITR2 transcripts were detected in a catfish mixed leukocyte culture, a macrophage cell line, and two cytotoxic T cell lines, but not in another T or B cell line. Only LITR1 (and not LITR2) was detected in a NK-like cell line. Alloantigen stimulation of both peripheral blood leukocytes and CTL cell lines resulted in increased LITR expression (Stafford et al. 2007); however, interpretations of such findings need to consider selective lineage expansion as a basis for increased levels of expression.
Certain LITR Ig domains are more similar to certain LRC encoded receptors (e.g. PIR, LILR, KIR as well as Xenopus XFL and chicken CHIR), whereas, others are more similar to mammalian Fc receptors (Stafford et al. 2006). Although the number of Ig domains varies between different LITRs, the two amino-terminal Ig domains are similar suggesting that these receptors may bind the same ligand (or class of ligand) (Stafford et al. 2007). In the amino-terminal Ig domain of human LILRB1 (which interacts with the α3 domain of MHC I), six of six key residues were identical or conserved between the amino-terminal Ig domains of catfish LITRs. By contrast, only two of 13 key residues in human KIR2DL1, which make contact with the α1 and α2 domains of MHC I, are conserved in the amino-terminal LITR Ig domain (Stafford et al. 2007). Computer modeling of the LITR Ig domains supports the hypothesis that the amino-terminal Ig domain of certain LITRs may bind MHC I; however, direct proof of this interaction is lacking. Interactions of receptors with MHC I do not represent an exclusive property of molecules associated with NK function.
Other NKR-like gene families in bony fish
As alluded to previously, NK activity is mediated by an exceptionally large number of different receptor types that mediate many different functions as defined by cell lineage- and/or species-specific assays. Identifying potential NK gene candidates on the basis of structural features alone at best represent reasonable guesses. Three more multigene families are significant in consideration of NK-type and other immunoregulating functions. The modular domain immune-type receptors (MDIRs) were identified in the course of an unsuccessful effort to identify NITR homologs in cartilaginous fish (Cannon et al. 2006; Cannon et al. 2010). MDIRs are type I transmembrane proteins that exhibit transmembrane and cytoplasmic tail features seen in NITRs and many other families of Ig domain-containing cell surface molecules, e.g., Fc receptors, poly Ig receptors, CD300s, TREMs, etc. MDIRs appear more abundant than any of these gene families as defined currently in mammals. In the course of characterizing MDIRs, we also identified diverse I domain-containing proteins (DICPs), which exhibit features of activating and inhibitory Ig-containing transmembrane receptors (Cannon et al. 2010). Some members of this family also appear to be secreted, as is seen with some NITRs (Yoder et al. 2004), MDIRS (Yoder, unpublished observations) and LITRs (Stafford et al. 2007). Recent findings suggest that MDIRs and DICPs exhibit some similarities in ligand binding activity (Haire, Cannon, O’Driscoll and Litman, unpublished observations). Although shared synteny can provide important clues regarding past relationships of genes and gene families (Flajnik and Kasahara 2010) and in the case of NKRs could be highly informative, it is not possible to draw any conclusions based on the current state of our analysis of NITRs, NILTs, LITRs, DICPs, and MDIRs and knowledge of mammalian genome structure.
Future directions
Our current understanding of adaptive and innate immunity underscores the potential advantages for an NK-type mechanism of recognition in all vertebrates. Although findings in a number of lower vertebrate species have identified a variety of molecules that represent potential mediators of NK-type immune functions, additional and far more detailed characterization of cells, genes and molecules is necessary before rational hypotheses regarding broad phylogenetic relationships can be formed. The lack of clear orthology between many of the candidate and verified mediators of NK function confound the reconstruction of a rational evolutionary history for NK recognition (Fig. 8). Furthermore, it is possible that non-Ig-type and non-lectin-type protein domains, which cannot be identified by sequence homology, are integral to NK function in non-mammalian species and thus cannot be recognized in sequence homology-based strategies. The take-home message from what has been observed both at the relatively early stage of work in avians, amphibians and bony fish reviewed here and the remarkably large body of work that has been conducted in mammals underscores the need to develop system-specific functional assays that can lead to the “functional” identification of NKRs and/or are suitable for screening candidate NKR molecules. Despite the inherent complexity of such investigations, there is good reason to believe that we will be able to define the important differences if not necessarily the patterns of successive acquisition of NK-type functions within vertebrate and potentially in invertebrate species.
Identification of candidate NKRs in marsupials, monotremes and non-mammalian vertebrates. NKR data for eutherian mammals is presented in Fig. 3, Fig. 4, and the Electronic supplementary material (Table S1). Opossum and platypus gene predictions are based on genome analyses only. Chicken CHIRs are structurally related to KIRs, but certain CHIRs function as Ig receptors (see text and Fig. 5). CD94/ NKG2 and B-NK are the only chicken lectin-type receptors thus far identified that possess inhibitory or activating motifs. Xenopus XMIV genes are predicted to encode both inhibitory and activating forms whereas, xILRs encode putative activating receptors (Fig. 6). NITRs, LITRs, and NILTs are multigene Ig-type receptors that possess both inhibitory and activating forms (Fig. 7). Phylogenetic trees adapted from Blair and Hedges (Blair and Hedges 2005)
References
Arnason U, Adegoke JA, Gullberg A, Harley EH, Janke A, Kullberg M (2008) Mitogenomic relationships of placental mammals and molecular estimates of their divergences. Gene 421:37–51
Arnon TI, Kaiser JT, West AP Jr, Olson R, Diskin R, Viertlboeck BC, Gobel TW, Bjorkman PJ (2008) The crystal structure of CHIR-AB1: a primordial avian classical Fc receptor. J Mol Biol 381:1012–1024
Averdam A, Petersen B, Rosner C, Neff J, Roos C, Eberle M, Aujard F, Munch C, Schempp W, Carrington M, Shiina T, Inoko H, Knaust F, Coggill P, Sehra H, Beck S, Abi-Rached L, Reinhardt R, Walter L (2009) A novel system of polymorphic and diverse NK cell receptors in primates. PLoS Genet 5:e1000688
Barrow AD, Trowsdale J (2008) The extended human leukocyte receptor complex: diverse ways of modulating immune responses. Immunol Rev 224:98–123
Belov K, Sanderson CE, Deakin JE, Wong ES, Assange D, McColl KA, Gout A, de BB B, AD STP, Trowsdale J, Papenfuss AT (2007) Characterization of the opossum immune genome provides insights into the evolution of the mammalian immune system. Genome Res 17:982–991
Bendelac A, Savage PB, Teyton L (2007) The biology of NKT cells. Ann Rev Immunol 25:297–336
Bernot A, Zoorob R, Auffray C (1994) Linkage of a new member of the lectin supergene family to chicken Mhc genes. Immunogenetics 39:221–229
Biassoni R (2009) Human natural killer receptors, co-receptors, and their ligands. Curr Protoc. Immunol Chapter 14:Unit 14.10
Biassoni R, Cantoni C, Pende D, Sivori S, Parolini S, Vitale M, Bottino C, Moretta A (2001) Human natural killer cell receptors and co-receptors. Immunol Rev 181:203–214
Bimber BN, Moreland AJ, Wiseman RW, Hughes AL, O’Connor DH (2008) Complete characterization of killer Ig-like receptor (KIR) haplotypes in Mauritian cynomolgus macaques: novel insights into nonhuman primate KIR gene content and organization. J Immunol 181:6301–6308
Blair JE, Hedges SB (2005) Molecular phylogeny and divergence times of deuterostome animals. Mol Biol Evol 22:2275–2284
Borrego F, Masilamani M, Marusina AI, Tang X, Coligan JE (2006) The CD94/NKG2 family of receptors: from molecules and cells to clinical relevance. Immunol Res 35:263–278
Brandt CS, Baratin M, Yi EC, Kennedy J, Gao Z, Fox B, Haldeman B, Ostrander CD, Kaifu T, Chabannon C, Moretta A, West R, Xu W, Vivier E, Levin SD (2009) The B7 family member B7-H6 is a tumor cell ligand for the activating natural killer cell receptor NKp30 in humans. J Exp Med 206:1495–1503
Call ME, Wucherpfennig KW (2007) Common themes in the assembly and architecture of activating immune receptors. Nat Rev Immunol 7:841–850
Cannon JP, Haire RN, Mueller MG, Litman RT, Eason DD, Tinnemore D, Amemiya CT, Ota T, Litman GW (2006) Ancient divergence of a complex family of immune-type receptor genes. Immunogenet 58:362–373
Cannon JP, Haire RN, Magis AT, Eason DD, Winfrey KN, Hernandez Prada JA, Bailey KM, Jakoncic J, Litman GW, Ostrov DA (2008) A bony fish immunological receptor of the NITR multigene family mediates allogeneic recognition. Immunity 29:228–237
Cannon JP, Dishaw LJ, Haire RN, Litman RT, Ostrov DA, Litman GW (2010) Recognition of additional roles for immunoglobulin domains in immune function. Semin Immunol 22:17–24
Carlyle JR, Mesci A, Fine JH, Chen P, Belanger S, Tai LH, Makrigiannis AP (2008) Evolution of the Ly49 and Nkrp1 recognition systems. Semin Immunol 20:321–330
Chiang HI, Zhou H, Raudsepp T, Jesudhasan PR, Zhu JJ (2007) Chicken CD69 and CD94/NKG2-like genes in a chromosomal region syntenic to mammalian natural killer gene complex. Immunogenetics 59:603–611
Dennis GJ, Kubagawa H, Cooper MD (2000) Paired Ig-like receptor homologs in birds and mammals share a common ancestor with mammalian Fc receptors. Proc Natl Acad Sci USA 97:13245–13250
Desai S, Heffelfinger AK, Orcutt TM, Litman GW, Yoder JA (2008) The medaka novel immune-type receptor (NITR) gene clusters reveal an extraordinary degree of divergence in variable domains. Evol Biol 8:177–188
Djeu JY, Jiang K, Wei S (2002) A view to a kill: signals triggering cytotoxicity. Clin Cancer Res 8:636–640
Du PL, Zucchetti I, De SR (2004) Immunoglobulin superfamily receptors in protochordates: before RAG time. Immunol Rev 198:233–248
Evans DL, Graves SS, Cobb D, Dawe DL (1984) Nonspecific cytotoxic cells in fish (Ictalurus punctatus). II. Parameters of target cell lysis and specificity. Dev Comp Immunol 8:303–312
Evenhuis J, Bengten E, Snell C, Quiniou SM, Miller NW, Wilson M (2007) Characterization of additional novel immune type receptors in channel catfish, Ictalurus punctatus. Immunogenetics 59:661–671
Ferraresso S, Kuhl H, Milan M, Ritchie DW, Secombes CJ, Reinhardt R, Bargelloni L (2009) Identification and characterisation of a novel immune-type receptor (NITR) gene cluster in the European sea bass, Dicentrarchus labrax, reveals recurrent gene expansion and diversification by positive selection. Immunogenetics 61:773–788
Fischer U, Utke K, Somamoto T, Kollner B, Ototake M, Nakanishi T (2006) Cytotoxic activities of fish leucocytes. Fish Shellfish Immunol 20:209–226
Flajnik MF (2002) Comparative analyses of immunoglobulin genes: surprises and portents. Nat Rev Immunol 2:688–698
Flajnik MF, Kasahara M (2010) Origin and evolution of the adaptive immune system: genetic events and selective pressures. Nat Rev Genet 11:47–59
Gagnier L, Wilhelm BT, Mager DL (2003) Ly49 genes in non-rodent mammals. Immunogenetics 55:109–115
Gendzekhadze K, Norman PJ, Abi-Rached L, Graef T, Moesta AK, Layrisse Z, Parham P (2009) Co-evolution of KIR2DL3 with HLA-C in a human population retaining minimal essential diversity of KIR and HLA class I ligands. Proc Natl Acad Sci U S A 106:18692–18697
Glazko GV, Nei M (2003) Estimation of divergence times for major lineages of primate species. Mol Biol Evol 20:424–434
Gobel TW, Chen CL, Shrimpf J, Grossi CE, Bernot A, Bucy RP, Auffray C, Cooper MD (1994) Characterization of avian natural killer cells and their intracellular CD3 protein complex. Eur J Immunol 24:1685–1691
Goyos A, Robert J (2009) Tumorigenesis and anti-tumor immune responses in Xenopus. Front Biosci 14:167–176
Guethlein LA, Abi-Rached L, Hammond JA, Parham P (2007) The expanded cattle KIR genes are orthologous to the conserved single-copy KIR3DX1 gene of primates. Immunogenetics 59:517–522
Guselnikov SV, Reshetnikova ES, Najakshin AM, Mechetina LV, Robert J, Av T (2010) The amphibians Xenopus laevis and Silurana tropicalis possess a family of activating KIR-related Immunoglobulin-like receptors. Dev Comp Immunol 34:308–315
Hammond JA, Guethlein LA, Abi-Rached L, Moesta AK, Parham P (2009) Evolution and survival of marine carnivores did not require a diversity of killer cell Ig-like receptors or Ly49 NK cell receptors. J Immunol 182:3618–3627
Hao L, Klein J, Nei M (2006) Heterogeneous but conserved natural killer receptor gene complexes in four major orders of mammals. Proc Natl Acad Sci U S A 103:3192–3197
Hawke NA, Yoder JA, Haire RN, Mueller MG, Litman RT, Miracle AL, Stuge T, Shen L, Miller N, Litman GW (2001) Extraordinary variation in a diversified family of immune-type receptor genes. Proc Natl Acad Sci USA 98:13832–13837
Hellsten U, Harland RM, Gilchrist MJ, Hendrix D, Jurka J, Kapitonov V, Ovcharenko I, Putnam NH, Shu S, Taher L, Blitz IL, Blumberg B, Dichmann DS, Dubchak I, Amaya E, Detter JC, Fletcher R, Gerhard DS, Goodstein D, Graves T, Grigoriev IV, Grimwood J, Kawashima T, Lindquist E, Lucas SM, Mead PE, Mitros T, Ogino H, Ohta Y, Poliakov AV, Pollet N, Robert J, Salamov A, Sater AK, Schmutz J, Terry A, Vize PD, Warren WC, Wells D, Wills A, Wilson RK, Zimmerman LB, Zorn AM, Grainger R, Grammer T, Khokha MK, Richardson PM, Rokhsar DS (2010) The genome of the Western clawed frog Xenopus tropicalis. Science 328:633–636
Hollenbach JA, Meenagh A, Sleator C, Alaez C, Bengoche M, Canossi A, Contreras G, Creary L, Evseeva I, Gorodezky C, Hardie RA, Hemming KT, Lie B, Luo M, Martinetti M, Navarette C, de Oliveira DC, Ozzella G, Pasi A, Pavlova E, Pinto S, Porto LC, Santos P, Slavcev A, Srinak D, Tavoularis S, Tonks S, Trachtenberg E, Vejbaesya S, Middleton D (2010) Report from the killer immunoglobulin-like receptor (KIR) anthropology component of the 15th International Histocompatibility Workshop: worldwide variation in the KIR loci and further evidence for the co-evolution of KIR and HLA. Tissue Antigens 76:9–17
Horton TL, Ritchie P, Watson MD, Horton JD (1996) NK-like activity against allogeneic tumour cells demonstrated in the spleen of control and thymectomized Xenopus. Immunol Cell Biol 74:365–373
Horton TL, Minter R, Stewart R, Ritchie P, Watson MD, Horton JD (2000) Xenopus NK cells identified by novel monoclonal antibodies. Eur J Immunol 30:604–613
Horton TL, Stewart R, Cohen N, Rau L, Ritchie P, Watson MD, Robert J, Horton JD (2003) Ontogeny of Xenopus NK cells in the absence of MHC class I antigens. Dev Comp Immunol 27:715–726
Hosomichi K, Shiina T, Suzuki S, Tanaka M, Shimizu S, Iwamoto S, Hara H, Yoshida Y, Kulski JK, Inoko H, Hanzawa K (2006) The major histocompatibility complex (MHC) class IIB region has greater genomic structural flexibility and diversity in the quail than the chicken. BMC Genomics 7:322
Jima DD, Shah RN, Orcutt TM, Joshi D, Law JM, Litman GW, Trede NS, Yoder JA (2009) Enhanced transcription of complement and coagulation genes in the absence of adaptive immunity. Mol Immunol 46:1505–1516
Kaufman J, Milne S, Gobel TWF, Walker BA, Jacob JP, Auffray C, Zoorob R, Beck S (1999) The chicken B locus is a minimal essential major histocompatibility complex. Nature 401:923–925
Khalturin K, Becker M, Rinkevich B, Bosch TCG (2003) Urochordates and the origin of natural killer cells: identification of a CD94/NKR-P1-related receptor in blood cells of Botryllus. Proc Natl Acad Sci USA 100:622–627
Khalturin K, Panzer Z, Cooper MD, Bosch TC (2004) Recognition strategies in the innate immune system of ancestral chordates. Mol Immunol 41:1077–1087
Kikuno R, Sato A, Mayer WE, Shintani S, Aoki T, Klein J (2004) Clustering of C-type lectin natural killer receptor-like loci in the bony fish Oreochromis niloticus. Scand J Immunol 59:133–142
Kock H, Fischer U (2008) A novel immunoglobulin-like transcript from rainbow trout with two Ig-like domains and two isoforms. Mol Immunol 45:1612–1622
Kulski JK, Shiina T, Anzai T, Kohara S, Inoko H (2002) Comparative genomic analysis of the MHC: the evolution of class I duplication blocks, diversity and complexity from shark to man. Immunol Rev 190:95–122
Lanier LL (2005) NK cell recognition. Annu Rev Immunol 23:225–274
Laun K, Coggill P, Palmer S, Sims S, Ning Z, Ragoussis J, Volpi E, Wilson N, Beck S, Ziegler A, Volz A (2006) The leukocyte receptor complex in chicken is characterized by massive expansion and diversification of immunoglobulin-like Loci. PLoS Genet 2:e73
Lin W, Zhang H, Beck G (2001) Phylogeny of natural cytotoxicity: cytotoxic activity of coelomocytes of the purple sea urchin, Arbacia punctulata. J Exp Zool 290:741–750
Litman GW, Dishaw LJ, Cannon JP, Haire RN, Rast JP (2007) Alternative mechanisms of immune receptor diversity. Curr Opin Biol 19:526–534
Lochner KM, Viertlboeck BC, Gobel TW (2010) The red jungle fowl leukocyte receptor complex contains a large, highly diverse number of chicken immunoglobulin-like receptor (CHIR) genes. Mol Immunol 47:1956–1962
Mandelboim O, Lieberman N, Lev M, Paul L, Arnon TI, Bushkin Y, Davis DM, Strominger JL, Yewdell JW, Porgador A (2001) Recognition of haemagglutinins on virus-infected cells by NKp46 activates lysis by human NK cells. Nature 409:1055–1059
Martinez-Borra J, Khakoo SI (2008) Speed and selection in the evolution of killer-cell immunoglobulin-like receptors. Int J Immunogenet 35:89–96
Mewes J, Verheijen K, Montgomery BC, Stafford JL (2009) Stimulatory catfish leukocyte immune-type receptors (IpLITRs) demonstrate a unique ability to associate with adaptor signaling proteins and participate in the formation of homo- and heterodimers. Mol Immunol 47:318–331
Miller RD (2010) Those other mammals: the immunoglobulins and T cell receptors of marsupials and monotremes. Semin Immunol 22:3–9
Montgomery BC, Mewes J, Davidson C, Burshtyn DN, Stafford JL (2008) Cell surface expression of channel catfish leukocyte immune-type receptors (IpLITRs) and recruitment of both Src homology 2 domain-containing protein tyrosine phosphatase (SHP)-1 and SHP-2. Dev Comp Immunol 33:570–582
Moss LD, Monette MM, Jaso-Friedmann L, Leary JH III, Dougan ST, Krunkosky T, Evans DL (2009) Identification of phagocytic cells, NK-like cytotoxic cell activity and the production of cellular exudates in the coelomic cavity of adult zebrafish. Dev Comp Immunol 33:1077–1087
Nikolaidis N, Makalowska I, Chalkia D, Makalowski W, Klein J (2005) Origin and evolution of the chicken leukocyte receptor complex. Proc Natl Acad Sci USA 102:4057–4062
Norman PJ, Abi-Rached L, Gendzekhadze K, Korbel D, Gleimer M, Rowley D, Bruno D, Carrington CV, Chandanayingyong D, Chang YH, Crespi C, Saruhan-Direskeneli G, Fraser PA, Hameed K, Kamkamidze G, Koram KA, Layrisse Z, Matamoros N, Mila J, Park MH, Pitchappan RM, Ramdath DD, Shiau MY, Stephens HA, Struik S, Verity DH, Vaughan RW, Tyan D, Davis RW, Riley EM, Ronaghi M, Parham P (2007) Unusual selection on the KIR3DL1/S1 natural killer cell receptor in Africans. Nat Genet 39:1092–1099
Ohta Y, Goetz W, Hossain MZ, Nonaka M, Flajnik MF (2006) Ancestral organization of the MHC revealed in the amphibian Xenopus. J Immunol 176:3674–3685
Orr MT, Lanier LL (2010) Natural killer cell education and tolerance. Cell 142:847–856
Ostergaard AE, Martin SA, Wang T, Stet RJ, Secombes CJ (2009) Rainbow trout (Oncorhynchus mykiss) possess multiple novel immunoglobulin-like transcripts containing either an ITAM or ITIMs. Dev Comp Immunol 33:525–532
Ota T, Sitnikova T, Nei M (2000) Evolution of vertebrate immunoglobulin variable gene segments. Curr Top Microbiol Immunol 248:221–245
Panagos PG, Dobrinski KP, Chen X, Grant AW, Traver D, Djeu JY, Wei S, Yoder JA (2008) Immune-related, lectin-like receptors are differentially expressed in the myeloid and lymphoid lineages of zebrafish. Immunogenet 58:31–40
Parham P (2005) MHC class I molecules and KIRs in human history, health and survival. Nat Rev Immunol 5:201–214
Parham P, Abi-Rached L, Matevosyan L, Moesta AK, Norman PJ, Older Aguilar AM, Guethlein LA (2010) Primate-specific regulation of natural killer cells. J Med Primatol 39:194–212
Parrinello N, Arizza V, Cammarata M, Parrinello DM (1993) Cytotoxic activity of Ciona intestinalis (Tunicata) hemocytes: properties of the in vitro reaction against erythrocyte targets. Dev Comp Immunol 17:19–27
Pitcher LA, van Oers NS (2003) T-cell receptor signal transmission: who gives an ITAM? Trends Immunol 24:554–560
Rast JP, Haire RN, Litman RT, Pross S, Litman GW (1995) Identification and characterization of T-cell antigen receptor related genes in phylogenetically diverse vertebrate species. Immunogenet 42:204–212
Rau L, Gantress J, Bell A, Stewart R, Horton T, Cohen N, Horton J, Robert J (2002) Identification and characterization of Xenopus CD8+ T cells expressing an NK cell-associated molecule. Eur J Immunol 32:1574–1583
Ravetch JV, Lanier LL (2000) Immune inhibitory receptors. Science 290:84–89
Robert J, Ohta Y (2009) Comparative and developmental study of the immune system in Xenopus. Dev Dyn 238:1249–1270
Rogers S, Shaw I, Ross N, Nair V, Rothwell L, Kaufman J, Kaiser P (2003) Analysis of part of the chicken Rfp-Y region reveals two novel lectin genes, the first complete genomic sequence of a class I alpha-chain gene, a truncated class II beta-chain gene, and a large CR1 repeat. Immunogenetics 55:100–108
Rogers SL, Gobel TW, Viertlboeck BC, Milne S, Beck S, Kaufman J (2005) Characterization of the chicken C-type lectin-like receptors B-NK and B-lec suggests that the NK complex and the MHC share a common ancestral region. J Immunol 174:3475–3483
Rogers SL, Viertlboeck BC, Gobel TW, Kaufman J (2008) Avian NK activities, cells and receptors. Semin Immunol 20:353–360
Sato A, Mayer WE, Overath P, Klein J (2003) Genes encoding putative natural killer cell C-type lectin receptors in teleostean fishes. Proc Natl Acad Sci USA 100:7779–7784
Sepulcre MP, Alcaraz-Perez F, Lopez-Munoz A, Roca FJ, Meseguer J, Cayuela ML, Mulero V (2009) Evolution of lipopolysaccharide (LPS) recognition and signaling: fish TLR4 does not recognize LPS and negatively regulates NF-kappaB activation. J Immunol 182:1836–1845
Shen L, Stuge TB, Zhou H, Khayat M, Barker KS, Quiniou SMA, Wilson M, Bengten E, Chinchar VG, Clem LW, Miller NW (2002) Channel catfish cytotoxic cells: a mini-review. Dev Comp Immunol 26:141–149
Shen L, Stuge TB, Bengten E, Wilson M, Chinchar VG, Naftel JP, Bernanke JM, Clem LW, Miller NW (2004) Identification and characterization of clonal NK-like cells from channel catfish (Ictalurus punctatus). Dev Comp Immunol 28:139–152
Sivori S, Falco M, Moretta L, Moretta A (2010) Extending killer Ig-like receptor function: from HLA class I recognition to sensors of microbial products. Trends Immunol 31:289–294
Soanes KH, Figuereido K, Richards RC, Mattatall NR, Ewart KV (2004) Sequence and expression of C-type lectin receptors in Atlantic salmon (Salmo salar). Immunogenet 56:572–584
Soanes KH, Ewart KV, Mattatall NR (2008) Recombinant production and characterization of the carbohydrate recognition domain from Atlantic salmon C-type lectin receptor C (SCLRC). Protein Expr Purif 59:38–46
Somamoto T, Sato A, Nakanishi T, Ototake M, Okamoto N (2004) Specific cytotoxic activity generated by mixed leucocyte culture in ginbuna crucian carp. Fish Shellfish Immunol 17:187–191
Stafford JL, Bengten E, Du Pasquier L, McIntosh RD, Quiniou SM, Clem LW, Miller NW, Wilson M (2006) A novel family of diversified immunoregulatory receptors in teleosts is homologous to both mammalian Fc receptors and molecules encoded within the leukocyte receptor complex. Immunogenet 58:758–773
Stafford JL, Bengten E, Du PL, Miller NW, Wilson M (2007) Channel catfish leukocyte immune-type receptors contain a putative MHC class I binding site. Immunogenet 59:77–91
Stet RJM, Hermsen T, Westphal AH, Jukes J, Engelsma M, Verburg-van Kemenade BML, Dortmans J, Aveiro J, Savelkoul HFJ (2005) Novel immunoglobulin-like transcripts in teleost fish encode polymorphic receptors with cytoplasmic ITAM and ITIM and a new structural Ig domain similar to the natural cytotoxicity receptor NKp44. Immunogenet 57:77–89
Strong SJ, Mueller MG, Litman RT, Hawke NA, Haire RN, Miracle AL, Rast JP, Amemiya CT, Litman GW (1999) A novel multigene family encodes diversified variable regions. Proc Natl Acad Sci USA 96:15080–15085
Stuge TB, Yoshida SH, Chinchar VG, Miller NW, Clem LW (1997) Cytotoxic activity generated from channel catfish peripheral blood leukocytes in mixed leukocyte cultures. Cell Immunol 177:154–161
Sullivan C, Charette J, Catchen J, Lage CR, Giasson G, Postlethwait JH, Millard PJ, Kim CH (2009) The gene history of zebrafish tlr4a and tlr4b is predictive of their divergent functions. J Immunol 183:5896–5908
Takai T (2005) Paired immunoglobulin-like receptors and their MHC class I recognition. Immunology 115:433–440
Traver T, Herbomel P, Patton EE, Murphy R, Yoder JA, Litman GW, Catic A, Amemiya CT, Zon LI, Trede NS (2003) The zebrafish as a model organism to study development of the immune system. Adv Immunol 81:253–330
Viertlboeck BC, Crooijmans RP, Groenen MA, Gobel TW (2004) Chicken Ig-like receptor B2, a member of a multigene family, is mainly expressed on B lymphocytes, recruits both Src homology 2 domain containing protein tyrosine phosphatase (SHP)-1 and SHP-2, and inhibits proliferation. J Immunol 173:7385–7393
Viertlboeck BC, Habermann FA, Schmitt R, Groenen MA, Du PL, Gobel TW (2005) The chicken leukocyte receptor complex: a highly diverse multigene family encoding at least six structurally distinct receptor types. J Immunol 175:385–393
Viertlboeck BC, Schweinsberg S, Hanczaruk MA, Schmitt R, Du PL, Herberg FW, Gobel TW (2007) The chicken leukocyte receptor complex encodes a primordial, activating, high-affinity IgY Fc receptor. Proc Natl Acad Sci USA 104:11718–11723
Viertlboeck BC, Schweinsberg S, Schmitt R, Herberg FW, Gobel TW (2009) The chicken leukocyte receptor complex encodes a family of different affinity FcY receptors. J Immunol 182:6985–6992
Viertlboeck BC, Gick CM, Schmitt R, Du PL, Gobel TW (2010) Complexity of expressed CHIR genes. Dev Comp Immunol 34:866–873
Voskoboinik I, Dunstone MA, Baran K, Whisstock JC, Trapani JA (2010) Perforin: structure, function, and role in human immunopathology. Immunol Rev 235:35–54
Walzer T, Blery M, Chaix J, Fuseri N, Chasson L, Robbins SH, Jaeger S, Andre P, Gauthier L, Daniel L, Chemin K, Morel Y, Dalod M, Imbert J, Pierres M, Moretta A, Romagne F, Vivier E (2007a) Identification, activation, and selective in vivo ablation of mouse NK cells via NKp46. Proc Natl Acad Sci U S A 104:3384–3389
Walzer T, Jaeger S, Chaix J, Vivier E (2007b) Natural killer cells: from CD3(−)NKp46(+) to post-genomics meta-analyses. Curr Opin Immunol 19:365–372
Warren WC, Hillier LW, Marshall Graves JA, Birney E, Ponting CP, Grutzner F, Belov K, Miller W, Clarke L, Chinwalla AT, Yang SP, Heger A, Locke DP, Miethke P, Waters PD, Veyrunes F, Fulton L, Fulton B, Graves T, Wallis J, Puente XS, Lopez-Otin C, Ordonez GR, Eichler EE, chen L, Cheng Z, Deakin JE, Alsop A, Thompson K, Kirby P, Papenfuss AT, Wakefield MJ, Olender T, Lancet D, Huttley GA, Smit AF, Pask A, Temple-Smith P, Batzer MA, Walker JA, Konkel MK, Harris RS, Whittington CM, Wong ES, Gemmell NJ, Buschiazzo E, Vargas Jentzsch IM, Merkel A, Schmitz J, Zemann A, Churakov G, Kriegs JO, Brosius J, Murchison EP, Sachidanandam R, Smith C, Hannon GJ, Tsend-Ayush E, McMillan D, Attenborough R, Rens W, Ferguson-Smith M, Lefevre CM, Sharp JA, Nicholas KR, Ray DA, Kube M, Reinhardt R, Pringle TH, Taylor J, Jones RC, Nixon B, Dacheux JL, Niwa H, Sekita Y, Huang X, Stark A, Kheradpour P, Kellis M, Flicek P, Chen Y, Webber C, Hardison R, Nelson J, Hallsworth-Pepin K, Delehaunty K, Markovic C, Minx P, Feng Y, Kremitzki C, Mitreva M, Glasscock J, Wylie T, Wohldmann P, Thiru P, Nhan MN, Pohl CS, Smith SM, Hou S, Nefedov M, de Jong PJ, Renfree MB, Mardis ER, Wilson RK (2008) Genome analysis of the platypus reveals unique signatures of evolution. Nature 453:175–183
Wei S, Zhou J, Chen X, Shah RN, Liu J, Orcutt TM, Traver D, Djeu JY, Litman GW, Yoder JA (2007) The zebrafish activating immune receptor Nitr9 signals via Dap12. Immunogenet 59:813–821
Wong ES, Sanderson CE, Deakin JE, Whittington CM, Papenfuss AT, Belov K (2009) Identification of natural killer cell receptor clusters in the platypus genome reveals an expansion of C-type lectin genes. Immunogenetics 61:565–579
Yoder JA (2004) Investigating the function, morphology and genetics of cytotoxic cells in bony fish. Comp Biochem Physiol 138:271–290
Yoder JA (2009) Form, function and phylogenetics of NITRs in bony fish. Dev Comp Immunol 33:135–144
Yoder JA, Mueller MG, Wei S, Corliss BC, Prather DM, Willis T, Litman RT, Djeu JY, Litman GW (2001) Immune-type receptor genes in zebrafish share genetic and functional properties with genes encoded by the mammalian lymphocyte receptor cluster. Proc Natl Acad Sci USA 98:6771–6776
Yoder JA, Litman RT, Mueller MG, Desai S, Dobrinski KP, Montgomery JS, Buzzeo MP, Amemiya CT, Trede NS, Wei S, Djeu JY, Humphray S, Jekosch K, Hernandez Prada JA, Ostrov DA, Litman GW (2004) Resolution of the NITR gene cluster in zebrafish. Proc Natl Acad Sci USA 101:15706–15711
Yoder JA, Orcutt TM, Traver D, Litman GW (2007) Structural characteristics of zebrafish orthologs of adaptor molecules that associate with transmembrane immune receptors. Gene 401:154–164
Yoder JA, Cannon JP, Litman RT, Murphy C, Freeman JL, Litman GW (2008) Evidence for a transposition event in a second NITR gene cluster in zebrafish. Immunogenet 60:257–265
Yoder JA, Turner PM, Wright PD, Wittamer V, Bertrand JY, Traver D, Litman GW (2010) Developmental and tissue-specific expression of NITRs. Immunogenetics 62:117–122
Yokoyama WM, Plougastel BFM (2003) Immune functions encoded by the natural killer complex. Nat Rev Immunol 3:304–316
Zelensky AN, Gready JE (2004) C-type lectin-like domains in Fugu rubripes. Genomics 5:51
Zhang H, Robison B, Thorgaard GH, Ristow SS (2000) Cloning, mapping and genomic organization of a fish C-type lectin gene from homozygous clones of rainbow trout (Oncorhynchus mykiss). Biochim Biophys Acta 1494:14–22
Acknowledgments
We thank Barb Pryor for editorial assistance and Dr. Martin Flajnik for invaluable input. The authors are supported by funds from the National Institutes of Health (R01 AI057559 to GWL and JAY and R01 AI23337 to GWL).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 98.8 kb)
Rights and permissions
About this article
Cite this article
Yoder, J.A., Litman, G.W. The phylogenetic origins of natural killer receptors and recognition: relationships, possibilities, and realities. Immunogenetics 63, 123–141 (2011). https://doi.org/10.1007/s00251-010-0506-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00251-010-0506-4