Abstract
Natural killer (NK) cell receptors belong to two unrelated, but functionally analogous gene families: the immunoglobulin superfamily, situated in the leukocyte receptor complex (LRC) and the C-type lectin superfamily, located in the natural killer complex (NKC). Here, we describe the largest NK receptor gene expansion seen to date. We identified 213 putative C-type lectin NK receptor homologs in the genome of the platypus. Many have arisen as the result of a lineage-specific expansion. Orthologs of OLR1, CD69, KLRE, CLEC12B, and CLEC16p genes were also identified. The NKC is split into at least two regions of the genome: 34 genes map to chromosome 7, two map to a small autosome, and the remainder are unanchored in the current genome assembly. No NK receptor genes from the LRC were identified. The massive C-type lectin expansion and lack of Ig-domain-containing NK receptors represents the most extreme polarization of NK receptors found to date. We have used this new data from platypus to trace the possible evolutionary history of the NK receptor clusters.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Phylogenetically, the platypus holds an important position as one of the earliest offshoots in the radiation of mammals. Monotremes diverged from therian mammals approximately 163–186 million years ago (MYA; Messer et al. 1998), and thus provide an invaluable evolutionary link between mammals and more distantly placed vertebrates, such as birds (310 MYA) and fish (450 MYA; Kumar and Hedges 1998). This makes the platypus genome an important addition to resources for studying the expansion and contraction of gene families in mammals and will provide insight into the gene repertoire of the ancestral mammal. In particular, characterization of immune gene clusters in this extraordinary mammal will shed light on the complex evolutionary dynamics of immunity.
Natural killer (NK) cells form an integral component of the innate immune system, known as natural cytotoxicity. They express cell surface receptors which recognize MHC class I or class I-like molecules on the surface of cells and, through activating and inhibitory signals, are able to destroy cells infected with viruses or intracellular parasites, as well as tumor cells (DeFranco et al. 2007). Two NK receptor clusters have been characterized in all mammals studied to date, including a marsupial, Monodelphis domestica (Belov et al. 2007). These two clusters, the leukocyte receptor complex (LRC) and the natural killer complex (NKC) contain two extensive gene families, the immunoglobulin superfamily (IgSF) and the C-type lectin superfamily (CLSF), respectively. The repertoires of NK receptor genes vary greatly between therian species, with certain species preferentially using one gene family over the other (Kelley et al. 2005). For instance, humans preferentially use LRC-encoded KIR genes (killer cell immunoglobulin-like receptor) as their main NK receptors, while mice use NKC encoded Klra (killer cell lectin-like receptor) genes for the same function (Kelley et al. 2005). A summary of NK receptor gene numbers in humans, mice, chicken, and several fish are shown in Table 1.
The variation in gene number and genomic organization of IgSF domains in LRCs is astounding. The human LRC is located on chromosome 19q13.4 and contains around 30 Ig superfamily genes, of these, 15 are KIR genes, 13 are leukocyte Ig-like receptors (LILRs), and two are leukocyte-associated Ig-like receptors (LAIRs; Kelley et al. 2005). KIRs possess two or three Ig domains, LILRs have two or four domains (Nikolaidis et al. 2005) and LAIRs have one Ig domain. The mouse LRC contains ten paired Ig-like receptors (PIRs), which have six extracellular Ig domains and one LAIR (Kelley et al. 2005). They do not have any LILRs or KIRs within their LRC, although two KIRs are located on the X chromosome (Kelley et al. 2005). The opossum LRC, found on chromosome 4, contains 154 Ig domains within 81 putative opossum MAIR (marsupial immunoglobulin-like receptor) genes (Belov et al. 2007). These genes are clearly related to the LRC genes of eutherian mammals, but direct orthologs are not found.
Genes containing Ig-like domains are also found in the extended region of the LRC, where a family of sialic-acid-binding immunoglobulin-like lectin (SIGLEC) genes contains an amino terminal V-set immunoglobulin domain and a variable number of C2-set immunoglobulin domains. These molecules regulate the action of the innate and adaptive immune systems through glycan recognition (Crocker et al. 2007). A single SIGLEC gene is found in chickens (Angata et al. 2007). It is not found within the LRC.
Fourteen different classes of CLSF proteins have been defined, with NKC genes belonging to groups II and V (Hao et al. 2006). In mammals, group V receptors are expressed on NK cells, macrophages and dendritic cells (Zelensky and Gready 2005), while group II receptors are not expressed on NK cells, but are found on the surface of dendritic cells and macrophages (Zelensky and Gready 2005). The overall structure of group II and group V receptors is very similar, as both contain a single extracellular C-type lectin domain and a transmembrane region. However, the group II receptors utilize a Ca2+-dependent carbohydrate recognition domain for pathogen recognition and cell–cell interaction, while the group V receptors recognize protein ligands independent of Ca2+ (Panagos et al. 2006).
The human NKC is located on chromosome 12p13.1, spans 2.8 Mb and contains 15 C-type lectin receptor genes, eight of which have direct NK cell receptor involvement (Hao et al. 2006). The NKC is expanded in the mouse and rat genomes. The mouse NKC contains 16 Klra (Ly49) genes distributed over 8.7 Mb, whereas the rat has 36 Klra genes over 10.3 Mb (Hao et al. 2006; Kelley et al. 2005). An NKC, containing nine C-type lectin genes, but no ortholog of Klra is present in opossum (Belov et al. 2007).
The characterization of NK receptor clusters in non-mammals is not as comprehensive as that of mammals (Sambrook and Beck 2007). The chicken LRC is located on microchromosome 31 (Viertlboeck et al. 2005). This region shares conserved synteny with the human LRC on chromosome 19. The chicken LRC contains 103 chicken Ig-like receptors (CHIRs) which consist of either one or two Ig-like domains (Laun et al. 2006). A single cluster containing C-type lectin-domain-containing NK receptors is not found in chicken. Instead, four C-type lectin-like receptor genes with NKC function are found within the chicken MHC on chromosome 16 (Rogers et al. 2003, 2005) and two C-type lectin-like receptor genes are found on chromosome 1 in a region that shares conserved synteny with the NKC in mammals (Chiang et al. 2007). However, these genes are not closely linked (Chiang et al. 2007). In addition, database searches have identified 13–17 genes containing C-type lectin domains which have not yet been mapped or assembled into the genome (Hao et al. 2006). One of these (chicken lec11) belongs to group II, while the rest belong to group V (Hao et al. 2006). Ig-like and C-type lectin receptors sharing similarity with mammalian NK receptors have also been identified in bony fish. NITRs (novel immune-type receptors) found in zebrafish possess V-type rather than C2 type Ig domains, yet they are clustered in a region of the genome which shares conserved synteny with the LRC of humans and mice (Yoder et al. 2001). The zebrafish NITR cluster is highly complex, containing 36 NITR genes plus one to three C-type lectin receptors within the same locus (Yoder et al. 2004). A single cluster of killer cell C-type lectin receptors (KLR complex) belonging to the C-type lectin family have been identified in a teleost fish, Oreochromis niloticus (Kikuno et al. 2004). None of these genes are orthologous to mammalian NKC genes, yet they are clearly paralogous, with duplications in the two clusters occurring after the divergence of the teleost and mammalian lineages. The organization of these genes is more compact than seen in humans, with approximately one KLR locus per 18 kb. Whether these domains belong to group II or group V is still a subject of debate (Kikuno et al. 2004; Panagos et al. 2006; Sato et al. 2003).
NK receptors may be inhibitory or activating. The presence of immunoreceptor tyrosine-based inhibitory motifs (ITIMs) in the cytoplasmic tail of the receptor inhibits NK lysis by decreasing activation (Kelley et al. 2005). In contrast, activating NK receptors do not typically contain ITIMs, instead encoding charged residues in their transmembrane domain which trigger activation through the recruitment of immunoreceptor tyrosine-based activation motif (ITAM)-containing adaptor molecules, such as DAP12 and DAP10 (Blery et al. 2000).
The first monotreme genome has recently been sequenced by the Genome Sequencing Center of Washington University in St Louis (Warren et al. 2008). DNA from a single female platypus, “Glennie”, from Glenrock Station, New South Wales, was sequenced using whole-genome shotgun methods. The draft assembly was produced from ∼6× coverage of reads. 177,028 contigs and 61,239 ultracontigs greater than 2 kb were produced (Contig N50 = 12 kb; Ultracontig N50 = 967 kb). High G + C content (45.5%) combined with a high density of interspersed repeats contributed to significant fragmentation levels in the assembly. Here, we describe the identification of NK receptor-like genes in the newly assembled platypus genome, adapting search strategies to account for the fragmentary assembly.
Materials and methods
All genomic analyses were performed using the Ornithorhynchus_anatinus_5.0.1 assembly from the Genome Sequencing Center of Washington University in St Louis.
Genome searches and gene prediction
To generate an LRC profile hidden Markov model (HMM), Ig domains were extracted from characterized LRC genes using SMART (http://smart.embl-heidelberg.de/). The NKC profile HMM was generated using the first exon of NKC C-type lectin genes. Genome searches with these profiles were performed using HMMER 2.3.3 with default parameters. All HMMER hits with E values less than 10 were retained as putative NKC/LRC genes.
Gene predictions were performed using GenomeScan (http://genes.mit.edu/genomescan.html). HMMER hits were padded by 5 kb and the sequence extracted. HMMER hits that did not result in any predicted genes were considered putative pseudogenes and removed from further analysis. To increase the accuracy of our gene predictions, we incorporated protein homology information into the predictions using a set of vertebrate NKC genes.
In the case of LRC genes, where it is common for a gene to contain multiple Ig domains, the raw HMMER hits were used in a blastcl3 search against the NCBI ‘nr’ nucleotide database. Top hits were chained to produce putative genes based on the genomic distance between hits (Belov et al. 2007).
NKC and LRC HMMER hits were queried against SWISSPROT using TBLASTN 2.2.11 (Altschul et al. 1997) and their top BLAST hits were examined. HMMER hits where the best BLAST result (smallest E value) did not map to NKC/LRC genes were removed from the analysis.
Transmembrane domains were predicted using TMHMM web server (V. 2.0) (http://www.cbs.dtu.dk/services/TMHMM/).
Southern blot
Genomic DNA was extracted from platypus spleen using the DNeasy blood and tissue kit (Qiagen). Ethanol precipitations were carried out to increase DNA concentration. Ten micrograms of spleen DNA was digested with HindIII and run on a 1% TAE agarose gel overnight at 30 mA. The gel was soaked for 45 min in denaturation buffer and 2 × 20 min in neutralization buffer. The gel was transferred onto Hybond-N+ membrane (Amersham, GE Healthcare) using the capillary blotting protocol according to manufacturer’s instructions (Amersham). The following day, the Hybond-N+ membrane was air dried for 15 min and oven dried at 80°C for 2 h. The probe was amplified from genomic DNA using the Ultra437_670 primers and PCR conditions described below. DNA was purified from the PCR products using the UltraClean kit (MoBio). The Random Primed DNA Labeling kit (Roche) was used to label the probe with 32P-dCTP (3,000 Ci mmol−1). The membrane was prehybridized at 60°C with Rapid-hyb Buffer (Amersham). The probe was added and hybridized at 60°C overnight. The membrane was washed by 3 × 5 min with 2× SSC, 2× 30 min with 2× SSC/0.1%SDS, 1× 30 min with 1× SSC/0.1%SDS and exposed to X-ray film at −80°C for 10 days.
cDNA sequencing
Primers were designed from platypus C-type lectin gene predictions on Contig7463 and Ultra437:671382-681519. A platypus spleen cDNA library was used as a template for PCR. The forward library vector primer (λgt10f) was coupled with the reverse gene specific primer and vice versa (i.e., forward gene specific primer and λgt10r) to ensure amplification of the 3′ and 5′ ends of the C-type lectin genes. Amplifications were performed in 25 μL containing 1× AccuPrime PCR Buffer I (Invitrogen), 4 mM Mg2SO4, 1 mM forward and reverse primer and 1 unit AccuPrime Taq polymerase (Invitrogen). The PCR amplification steps were as follows: initial denaturation at 94°C for 1 min, then 35 cycles of 94°C for 30 s, 62°C for 30 s and 68°C for 1 min. PCR products were cut from the gel and the DNA purified using the UltraClean kit (MiBio) following the protocol. The DNA was cloned into plasmids using the pGEM®-T easy vector system (Promega). Purification of plasmid DNA from overnight cultures of E. coli in LB medium was carried out using QIAprep® Spin Miniprep Kit (Qiagen). The DNA was sent to the Australian Genome Research Facility (AGRF) for sequencing.
Phylogenetic analysis
Protein sequences were aligned using MUSCLE v3.6 (Edgar 2004). Phylogenetic trees were constructed with using MEGA 4.0 (Kumar et al. 2004) using the neighbor-joining (NJ) algorithm, with pairwise deletion of gaps and under the p distance model with 1,000 bootstraps. Individual C-type lectin domains from human, mouse, chicken, and fish sequences were used. Full-length Ig-containing protein sequences were used in the LRC phylogenetic tree. Sequences for phylogenetic analysis included closely related proteins not belonging to the NKC or LRC. GenBank accession numbers are provided in Electronic Supplementary Material Table 2. Representatives of all 14 groups of C-type lectins were used in our phylogenetic analysis.
Test for positive selection
Selection tests were carried out using PAML (Yang 1997) on putative platypus NKC genes. C-type lectin domains from genes predicted by GenomeScan were extracted using SMART. Gene predictions for C-type lectin genes were incomplete, with approximately 90 bp in the carboxyl-end of the domains not predicted for around 25% of all platypus sequences. Hence, a reduced region containing 240 bp was used in our selection analysis.
DNA sequences were aligned using RevTrans 1.4 (Wernersson and Pedersen 2003). The likelihood of the data under the selection site model M2 was compared to the model of neutral selection using the likelihood ratio test with two degrees of freedom.
Homology modeling of the platypus C-type lectin gene (contig3450.1) was carried out with the program Swiss-model (Guex and Peitsch 1997). The template used to predict the platypus gene structure was Klra3 (1p1zD) from the protein data bank. PyMOL (DeLano 2002) was used to visualize residues under positive selection and identify binding sites.
FISH mapping
Fluorescence in situ hybridization (FISH) was used to localize genes to chromosomes. Platypus BACs were prepared using the Wizard miniprep kit (Promega). BAC DNA was labeled by nick translation with either digoxigenin-11-dUTP or biotin-16- dUTP (Roche Diagnostics). Hybridization, signal detection, and image capture were carried out on female and/or male platypus chromosome metaphase spreads by following the protocols used by Alsop et al. (2005), with the exception that the slides were denatured for 1 min 40 s instead of 1 min 50 s and incubation times with antibodies were all 30 min in length.
Results and discussion
Using a combination of HMMER and BLAST searches, gene prediction, and phylogenetic analysis, we identified a massive expansion of type V C-type lectin domains and no classical receptors containing immunoglobulin-like domains in the genome of the platypus.
Identification of C-type lectin domains
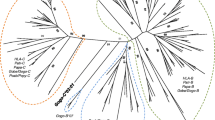
Two hundred thirteen putative C-type lectin NK receptor genes were identified and their predicted sequences are available on our website (http://bioinf.wehi.edu.au/platypus). Initial local HMMER searches using the first and most highly conserved exon of NKC C-type lectin domain led to the identification of 291 C-type lectin domains (E value <10). Sequences belonging to non-NK receptor containing C-type lectin groups I, III, VI, IX were identified using BLAST. An initial total of 266 sequences from NK receptor containing groups II and V were found and only those sequences which hit NK receptors were further analyzed using gene prediction. Six sequences did not result in any predicted genes using GenomeScan, suggesting that these were pseudogenes. NK receptor-like predictions were queried using BLAST and the best BLAST hit from each search was examined. Hits to non-NKC proteins and those with E values greater than 10−5 were removed. A final set of putative NKC genes (n = 213) comprised 38 complete peptide predictions and 175 partial predictions, 209 group V C-type lectins and only four group II C-type lectins. These sequences were incorporated into phylogenetic trees containing all 14 characterized types of C-type lectin domains from mouse. Only five platypus sequences clustered in terminal clades with non-NKC proteins with a bootstrap value less than 41%, indicating that our search strategy was largely successful in identifying true positives and removing false positives.
Accuracy of gene number estimation and gene predictions
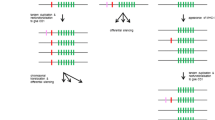
Two hundred thirteen C-type lectin NK receptors sequences were discovered in the platypus genome. However, this does not mean that the platypus has 213 NK receptor loci. Partial open-reading frames were detected for many genes. Full open-reading frames for all sequences were unavailable due to the fragmentary nature of the assembly; therefore, some genes may be pseudogenes. The sequenced animal was collected from an outbred population (Warren et al. 2008), so allelic variation is expected. If we conservatively assume that when two sequences cluster in a phylogenetic tree they are alleles of the same locus, then we have overestimated gene number by 66. Some of these clades may also represent products of homologous recombination between tandemly arrayed genes (Carrington and Cullen 2004). In either situation, it remains clear that there is a significant expansion of NKC genes in the platypus compared to any other known species. This finding is supported by multiple bands being detected using Southern blot analysis. Using a probe designed from pNKC60, we saw approximately eight bands (Fig. S1), this corresponds accordingly to six platypus NKC sequences of over 85% nucleotide identity spanning the length of this probe. The low average percent identity of the probe with all sequences, 0.40 ± 0.20 (SD), illustrates the high levels of divergence between the receptors, as evidenced in the subsequent analysis of positive selection.
To confirm the accuracy of our gene predictions, we isolated and sequenced C-type lectin genes from a spleen cDNA library. BLAT alignment of all cDNA sequences to the genome resulted in best hits to predicted sequences. Some errors in gene prediction occurred in regions flanking domains, however C-type lectin domains remain accurately predicted. cDNA sequences were identical to our predicted peptides pNKC174 (Contig36753), pNKC201 (Contig7463), and pNKC60 (Ultra437:671382-681519; Fig. S1), except for a difference in the 3′ region flanking the lectin domain in Pc2/pNKC201. Using Pc2 as a homology guide to improve the prediction, we were able to obtain a full-length gene of increased accuracy (Fig. S2). As expected, the domain-containing region of the gene is most accurately predicted due to the highly conserved nature of this sequence. Further sequencing in the flanking regions of the domain will be required to retrieve the full-length genes and will most likely reveal some prediction errors due to the short contig lengths as well as the inherent shortcomings of gene prediction. cDNA sequences are given at http://bioinf.wehi.edu.au/platypus.
Genomic organization of NK receptors in the platypus
Platypus NKC genes are found on at least two chromosomes: chromosome 7 and a small unidentified autosome. Thirty-four NKC genes in total are located on chromosome 7. Two contigs, ultra437 (32 genes; 0.95 Mb) and contig10595 (one gene; 0.03 Mb) containing 33 genes, were anchored to this chromosome using FISH (see Electronic Supplementary Data). The remaining gene (pNKC97) was localized to chromosome 7 by the assembly. Two color FISH co-localized contigs 28730 and 17200, which contain two platypus NK receptor-like genes (pNKC154 and 125, respectively) to the same small autosome (BACs KAAH-650E14a.g2 and KAAH-112E03a.b1; Fig. S5). The gene located on contig17200 was also predicted independently by REFSEQ (accession number, XP_001521201) using an alternative prediction method (GNOMON). Both peptides localizing to the small unnamed autosome were most similar to group V KLR sequences.
Four genomic clusters containing 17, 32, 8, and 35 NK receptor genes, located on ultracontigs 271, 437 (chromosome 7), 500 and 76 respectively, were identified (Fig. 1, Supplementary Table 1). The combined length of these ultracontigs is over 2.6 MB. Single genes were also found on four other ultracontigs (78, 345, 411, and 558). Due to the fragmented nature of the genome assembly, the remaining 115 peptides were found over 116 contigs and are not anchored to chromosomes.
A schematic diagram comparing the human and platypus leukocyte receptor complex and natural killer complex. An expansion of C-type lectin domain-containing genes in the NKC was identified, while classical IgSF genes of the LRC were not found. Each box represents one gene. Each platypus unit (box and line) represents one contig. All contigs are unanchored unless indicated otherwise. Lines joining genes indicate orthology based on phylogenetic tree in Fig. 2. For the platypus NKC, 8 ultracontigs, and 116 contigs are shown. Four ultracontigs and 116 contigs contain a single gene, with one contig containing two genes. Further information on gene location is available in supplementary materials Table 1
Characterization of evolutionary relationship of platypus C-type lectin domains
The evolutionary relationships between of platypus C-type lectin domains and their eutherian counterparts were determined using phylogenetic analysis and the resultant phylogenetic tree is shown in Fig. 2. A massive expansion of group V C-type lectin NKC genes occurred in the platypus. This is different to known NKC expansions in rodents where expansions occurred in both group II and V C-type lectins. In the platypus, only four putative NKC group II genes were identified. This number is comparable to the number of NKC group II genes found in human, although no direct orthologs to eutherian group II sequences were identified.
While the majority of platypus NKC genes have arisen as a result of lineage-specific expansions and do not have direct therian orthologs, putative orthologs between platypus and therian gene sequences (based on bootstrap support higher than 58%) include OLR1 (Contig13373), KLRE (Ultra437:859765-869902), dog CLEC16p (Contig12036, Contig170021), CD69 (Contig4063), and CLEC12B (Contig32313). The entire alignment including all known NKC genes from amniotes and non-NKC C-type lectin sequences is available at http://bioinf.wehi.edu.au/platypus, with separate, detailed alignments for KLRE, CLEC12B, OLR1, and CD69 in Figs. 2, 3, 4, and 5.
It is interesting to note that genes belonging to the same ultracontig intersperse throughout the phylogenetic tree (Fig. 6) and do not form discrete clades as would be expected in tandem duplications (e.g., a clade with 97% bootstrap support contains sequences from two contigs and three ultracontigs, 70–98% sequence identity). Rather, it is more likely that block duplications contributed to the genomic arrangement of these genes on ultracontigs. Alternatively, it is possible that following tandem duplications, genes were translocated to different regions of the genome.
Characterization of NK receptor-like peptides
Residues core to the C-type lectin fold are well-conserved. This included three pairs of cysteine residues involved in disulfide bridges and tryptophan residues contained within hydrophobic cores of the molecule. A Glu residue, characteristic of the domain (Zelensky and Gready 2003), is also highly conserved in the second α helix. The canonical ‘WIGL’ motif, an integrating component of the structure that is involved in the formation of all hydrophobic cores, and its variations (e.g., YIGL, WVGL, WLGL, WIGV, WIGM, WMGL, WTGL) are identified in ∼75% of all peptides (Butcher et al. 1998; Rogers et al. 2005; Zelensky and Gready 2003). Peptides missing these motifs are typically found located on contigs <1 kb, indicating that they were not predicted due to contig length.
We identified 62 sequences containing ITIMs in our predicted peptides, 33 of these peptides contain more than one ITIM motif. The variable number of ITIM motifs found in the peptides suggests differential function between characterized sequences. Two copies of the motif are required for the recruitment of the tyrosine phosphatase SHP-1 negative regulator, and a single ITIM also inhibits natural cytotoxicity responses by recruiting SHP-2 protein tyrosine phosphatase (Yusa et al. 2002). The maximum of four ITIMs were found in eight sequences, similar to the murine inhibitory molecule, PIR-B (Blery et al. 1998). Statistical analysis show that the number of ITIMs identified is significantly greater than expected by chance alone (p < 0.01) supporting the accuracy of these gene predictions. Close examination of partially predicted genes generally revealed vertebrate group II and V gene organization, with ITIMs (when present) preceding transmembrane helices, the C-type lectin domain and the 3′ terminus. Absence of this pattern including the low numbers of signal peptides identified (N = 21) can be largely attributed to short contig lengths that prevent full gene prediction and the difficulties associated with accurate computational predictions of N-termini (Bernal et al. 2007; Bina and Crowely 2001). As expected, no ITAM motifs were found in the platypus C-type lectin receptors. This is consistent with the fact that both human- and mouse-activating NK receptors are characterized by the absence of transducing elements in their intracytoplasmic domain and require ITAM-bearing adaptor proteins for activator function (Blery et al. 2000). A reduced alignment of platypus NKC sequences with eutherian NKC genes is shown in Fig. 7.
An alignment of platypus NKC sequences with eutherian NKC peptides. ITIM motifs boxed in green. Relative degrees of shading indicate level of conservation. The C-type lectin domain is marked with a line over the alignment. The transmembrane domain of CLEC4E is underlined and the positively charge residue is colored
Identification of residues under positive selection in putative NK receptor genes
We identified fourteen sites under positive selection in platypus NK receptor genes (shown in Fig. 8a and b). Four sites coincided with known contact sites between mouse Klra1 and its MHC class I ligand and are boxed in green (Fig. 8a; Tormo et al. 1999). Sites under positive selection are visualized on a projected 3-D structure in Fig. 8c. Three adjacent positively selected sites on the alignment (P > 95%) corresponded to contact sites and form an obvious surface interface and are highlighted and boxed in green. The overall ω value was 1.66. The entire alignment is available for download at http://bioinf.wehi.edu.au/platypus.
Positive selection tests indicate conservation of residues under positive selection between the platypus and mouse NK receptors. a A reduced alignment of platypus C-type lectin domains with mouse Klra1 and Klra2 gene products (whole alignment is available for download at http://bioinf.wehi.edu.au/platypus). Red asterisk indicates residues under positive selection, green plus sign indicates contact sites between mouse Klra1 and its MHC class I ligand. Green boxes indicate contact sites which are also under positive selection (1C). Conserved cysteines are highlighted. Sites identified by the Naive Empirical Bayes (NEB) analysis are shown. b A list of sites under positive selection with the probability of the site being under positive selection (ω (dn/ds) > 1) and the ω value for each site displayed. Reference residues are from contig3450. c A model of a platypus C-type lectin receptor. The sections highlighted in red indicate residues that are under positive selection. The residues in green are residues that are involved in binding as based on alignment with mouse NKC gene Klra1 and Klra2 and are also under positive selection. The green boxes relate to the green boxes in Fig. 1a, and show sites under positive selection
Contact sites between the immune receptors and ligands have been shown to be targets for pathogen evasion of the host immune response (Eagle and Trowsdale 2007; Kelley and Trowsdale 2005). As expected, these sites are under positive selection in NKC receptor genes. The similarities between the positively selected sites in platypus and the contact sites between mouse Klra1 and its MHC ligand suggests that the platypus NKC-like genes also interact with MHC class I-like ligands. In the primate KIR genes, positive selection at MHC-binding sites after duplication has led to functional divergence (Hao and Nei 2005). It is possible, therefore, that similar selective mechanisms have been responsible for the expansion of platypus NKC genes.
Identification and characterization of Ig-like domains
Exhaustive searching of the six-frame translation of the platypus genome sequence using a variety of Ig domain profile HMMs did not locate any Ig-like domain-containing NK receptor genes in the platypus genome. To check that this was not the result of our search strategy, we used the same PFAM Ig HMMER profile with chicken sequences removed to identify ∼289 CHIR-like domains in the more divergent chicken genome (E values < 1 × 10−5). We also searched for LRC genes in the platypus trace database and did not identify any. An OSCAR ortholog (contig9988, E value = 10−69) was identified, as were three putative SIGLEC genes and three Fc receptor domains (further details in Electronic Supplementary Data). No pseudogenes were identified.
Identification of Ig-like domain-containing genes like OSCAR, SIGLECs, and Fc receptors supports the stringency and level of sensitivity of our searches as these genes all contain core Ig domains and are believed to have evolved from a common ancestor (Davis et al. 2002; Dennis et al. 2000; further details in Electronic Supplementary Data).
It is impossible to determine whether the platypus genome contains an LRC region. OSCAR and SIGLEC are found in the LRC of eutherian mammals, but OSCAR is found in the MHC of the opossum (Belov et al. 2007) and has not been identified in the chicken, while CD33- and CD22-related SIGLECs map to the extended LRC in therians, but do not localize to the same chromosome as the LRC in chickens.
Evolutionary history of NK receptors
In all therian mammals studied to date, C-type lectin NK receptors are found in a single gene cluster, the natural killer complex (NKC; Hao et al. 2006; Kelley et al. 2005). The only exception is the bovine NKC, which is split over chromosomes 1 and 5 (Hao et al. 2006). Chicken C-type lectin containing NK receptors are found in three genomic regions; two in a region that shares conserved synteny with the therian NKC. The chicken also has additional genes with NKC function located outside of this region, within its MHC (chr16) and Rfp-Y region (chr16; Rogers et al. 2005). Several studies have suggested that NK receptors and MHC genes were originally localized to a single region of the genome, supporting the existence of an ancestral ‘immune supercomplex’ (Belov et al. 2006, 2007; Rogers et al. 2005; Sambrook and Beck 2007). Such an arrangement would have facilitated the co-evolution of ligands and receptors. OSCAR, a gene found in the LRC of eutherians, is located in the opossum MHC (Belov et al. 2006). The monotreme MHC is located on two pairs of sex chromosomes and appears to have evolved through a series of translocation events (Dohm et al. 2007). It is unclear as yet whether any of the putative platypus NKC genes are co-located with MHC genes or located on sex chromosomes.
However, it is clear that the platypus NKC is split over at least two chromosomes: chromosome 7 and a small unidentified autosome. Given that the chicken and platypus NKC genes do not map to a single genomic cluster, it is feasible that the single NKC region evolved after the divergence of monotremes and therians. However, at this stage, it is not possible to discount the possibility that ancestral mammals had an intact NKC, which was split in the platypus lineage. The lineage-specific expansion of NKC genes in the platypus and the split MHC (Dohm et al. 2007) support this theory. Characterization of the NK receptor clusters of the echidnas may help to answer this question.
Exhaustive searches did not uncover Ig-domain-containing receptors but did reveal IgSF members which map to a region that shares synteny with the extended LRC of eutherians. We suggest that ancestral mammals had an established LRC region which was lost in the platypus lineage due to the expansion of C-type lectin-domain-containing receptors.
Putative function of platypus NKC genes
The function of platypus NKC genes orthologous to characterized eutherian genes can be inferred through homology. CD69 is an early lymphocyte activation antigen in humans whose expression is preferentially induced by IL-3 (Cambiaggi et al. 1992; Yoshimura et al. 2002). It is conserved in all three extant mammalian lineages suggesting it plays a conserved role in mammalian immunity. KLRE lacks known signaling motifs but is able to regulate NK cell-mediated cytotoxicity by forming heterodimers with other molecules (Saether et al. 2008). CLEC16p is a pseudogene, found only in dog and related to KLRA genes (Hao et al. 2006). It is clustered with 70% bootstrap support to two platypus NKC peptides, one of which contains a full gene prediction, suggesting perhaps that a functional homolog of CLEC16p exists in the platypus. CLEC12B is an inhibitory receptor involved in myeloid cell function (Hoffmann et al. 2007), while OLR1 is a receptor to a marker protein for atherosclerosis (Sawamura et al. 1997).
Without orthology to characterized NKC genes, it is impossible to predict the function of the numerous remaining NK receptor homologs. These genes are almost exclusively group V C-type lectins. Initially, group V C-type lectin genes were thought to be restricted to NK or T cells, controlling cellular activation through recognition of MHC class I and related molecules. Recently, it has been shown that group V C-type lectin genes have a far more diverse range of functions and ligands and are expressed on a variety of myeloid cells (Pyz et al. 2006). For example, the beta-glucan receptor, Dectin-1, is expressed in a tissue-specific manner on macrophages, monocytes, dendritic cells, subsets of T cells and B cells in human and recognizes a number of fungal pathogens (Reid et al. 2004; Taylor et al. 2002; Willment et al. 2005). Some platypus NK receptors may play a key role in recognizing and responding to the fungus Mucor amphibiorum, a disease which affects Tasmanian, but not mainland platypuses (Munday et al. 1998). Additional functions attributed to group V C-type lectin genes in other species include regulation of cytokine production, involvement in antimicrobial responses, cell aggregation and adhesion, angiogenesis and inflammatory response and NK cell cytotoxicity. The range of functions is likely more broad as many of the ligands for human and mouse NK receptors have not yet been identified (Pyz et al. 2006). Alternative splicing, the number of glycosylation sites and cysteine pairs, and inhibitory motif variants all affect gene function (Pyz et al. 2006). The gene sequences described here are an important first step in characterizing the role these molecules play in the immune response of the platypus. Further studies will potentially lead to the development of immunological reagents to help conserve one of the last surviving monotreme species.
Conclusion
We have identified a massive expansion of C-type lectin NK receptor genes in the platypus lineage. These genes are found on eight ultracontigs and 116 contigs in the fragmented platypus genome assembly and have been localized to at least two chromosomes. Thirty-four NKC genes are located on chromosome 7 and 2 genes are located on an unidentified small autosome. The location of the remaining 177 genes remains to be established. No Ig-containing NK receptor genes were identified. If the genome assembly paints an accurate picture of the platypus genome, we present the most extreme case of polarization of NK receptors found to date. This study presents a first glimpse at the likely repertoire of NK receptor genes in this enigmatic mammal. Further mapping and functional studies are required to gain a comprehensive understanding of NK cell evolution and function in monotremes.
References
Alsop AE, Miethke P, Rofe R, Koina E, Sankovic N, Deakin JE, Haines H, Rapkins RW, Marshall Graves JA (2005) Characterizing the chromosomes of the Australian model marsupial Macropus eugenii (tammar wallaby). Chromosome Res 13:627–636
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Angata T, Tabuchi Y, Nakamura K, Nakamura M (2007) Siglec-15: an immune system Siglec conserved throughout vertebrate evolution. Glycobiology 17:838–846
Belov K, Deakin JE, Papenfuss AT, Baker ML, Melman SD, Siddle HV, Gouin N, Goode DL, Sargeant TJ, Robinson MD, Wakefield MJ, Mahony S, Cross JG, Benos PV, Samollow PB, Speed TP, Graves JA, Miller RD (2006) Reconstructing an ancestral mammalian immune supercomplex from a marsupial major histocompatibility complex. PLoS Biol 4:e46
Belov K, Sanderson CE, Deakin JE, Wong ES, Assange D, McColl KA, Gout A, de Bono B, Barrow AD, Speed TP, Trowsdale J, Papenfuss AT (2007) Characterization of the opossum immune genome provides insights into the evolution of the mammalian immune system. Genome Res 17:982
Bernal A, Crammer K, Hatzigeorgiou A, Pereira F (2007) Global discriminative learning for higher-accuracy computational gene prediction. PLoS Comput Biol 3:e54
Bina M, Crowely E (2001) Sequence patterns defining the 5' boundary of human genes. Biopolymers 59:347–355
Blery M, Kubagawa H, Chen CC, Vely F, Cooper MD, Vivier E (1998) The paired Ig-like receptor PIR-B is an inhibitory receptor that recruits the protein-tyrosine phosphatase SHP-1. Proc Natl Acad Sci U S A 95:2446–2451
Blery M, Olcese L, Vivier E (2000) Early signaling via inhibitory and activating NK receptors. Hum Immunol 61:51–64
Butcher S, Arney KL, Cook GP (1998) MAFA-L, an ITIM-containing receptor encoded by the human NK cell gene complex and expressed by basophils and NK cells. Eur J Immunol 28:3755–3762
Cambiaggi C, Scupoli MT, Cestari T, Gerosa F, Carra G, Tridente G, Accolla RS (1992) Constitutive expression of CD69 in interspecies T-cell hybrids and locus assignment to human chromosome 12. Immunogenetics 36:117–120
Carrington M, Cullen M (2004) Justified chauvinism: advances in defining meiotic recombination through sperm typing. Trends Genet 20:196–205
Chiang HI, Zhou H, Raudsepp T, Jesudhasan PR, Zhu JJ (2007) Chicken CD69 and CD94/NKG2-like genes in a chromosomal region syntenic to mammalian natural killer gene complex. Immunogenetics 59:603–611
Crocker PR, Paulson JC, Varki A (2007) Siglecs and their roles in the immune system. Nat Rev Immunol 7:255–266
Davis RS, Dennis G Jr, Odom MR, Gibson AW, Kimberly RP, Burrows PD, Cooper MD (2002) Fc receptor homologs: newest members of a remarkably diverse Fc receptor gene family. Immunol Rev 190:123–136
DeFranco AL, Locksley RM, Robertson M (2007) Immunity: the immune response in infectious and inflammatory disease. New Science, U.S
DeLano WL (2002) The PyMOL molecular graphics system
Dennis G Jr, Kubagawa H, Cooper MD (2000) Paired Ig-like receptor homologs in birds and mammals share a common ancestor with mammalian Fc receptors. Proc Natl Acad Sci U S A 97:13245–13250
Dohm JC, Tsend-Ayush E, Reinhardt R, Grutzner F, Himmelbauer H (2007) Disruption and pseudoautosomal localization of the major histocompatibility complex in monotremes. Genome Biol 8:R175
Eagle RA, Trowsdale J (2007) Promiscuity and the single receptor: NKG2D. Nat Rev Immunol 7:737–744
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797
Guex N, Peitsch MC (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714–2723
Hao L, Nei M (2005) Rapid expansion of killer cell immunoglobulin-like receptor genes in primates and their coevolution with MHC Class I genes. Gene 347:149–159
Hao L, Klein J, Nei M (2006) Heterogeneous but conserved natural killer receptor gene complexes in four major orders of mammals. Proc Natl Acad Sci U S A 103:3192–3197
Hoffmann SC, Schellack C, Textor S, Konold S, Schmitz D, Cerwenka A, Pflanz S, Watzl C (2007) Identification of CLEC12B, an inhibitory receptor on myeloid cells. J Biol Chem 282:22370–22375
Kelley J, Trowsdale J (2005) Features of MHC and NK gene clusters. Transpl Immunol 14:129–134
Kelley J, Walter L, Trowsdale J (2005) Comparative genomics of natural killer cell receptor gene clusters. PLoS Genet 1:129–139
Kikuno R, Sato A, Mayer WE, Shintani S, Aoki T, Klein J (2004) Clustering of C-type lectin natural killer receptor-like loci in the bony fish Oreochromis niloticus. Scand J Immunol 59:133–142
Kumar S, Hedges SB (1998) A molecular timescale for vertebrate evolution. Nature 392:917–920
Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5:150–163
Laun K, Coggill P, Palmer S, Sims S, Ning Z, Ragoussis J, Volpi E, Wilson N, Beck S, Ziegler A, Volz A (2006) The leukocyte receptor complex in chicken is characterized by massive expansion and diversification of immunoglobulin-like Loci. PLoS Genet 2:e73
Messer M, Weiss AS, Shaw DC, Westerman M (1998) Evolution of the monotremes: phylogenetic relationship to marsupials and eutherians, and estimation of divergence dates based on a-lactalbumin amino acid sequences. J Mamm Evol 5:95–105
Munday BL, Whittington RJ, Stewart NJ (1998) Disease conditions and subclinical infections of the platypus (Ornithorhynchus anatinus). Philos Trans R Soc Lond B Biol Sci 353:1093–1099
Nikolaidis N, Klein J, Nei M (2005) Origin and evolution of the Ig-like domains present in mammalian leukocyte receptors: insights from chicken, frog, and fish homologues. Immunogenetics 57:151–157
Panagos PG, Dobrinski KP, Chen X, Grant AW, Traver D, Djeu JY, Wei S, Yoder JA (2006) Immune-related, lectin-like receptors are differentially expressed in the myeloid and lymphoid lineages of zebrafish. Immunogenetics 58:31–40
Pyz E, Marshall AS, Gordon S, Brown GD (2006) C-type lectin-like receptors on myeloid cells. Ann Med 38:242–251
Reid DM, Montoya M, Taylor PR, Borrow P, Gordon S, Brown GD, Wong SY (2004) Expression of the beta-glucan receptor, Dectin-1, on murine leukocytes in situ correlates with its function in pathogen recognition and reveals potential roles in leukocyte interactions. J Leukoc Biol 76:86–94
Rogers S, Shaw I, Ross N, Nair V, Rothwell L, Kaufman J, Kaiser P (2003) Analysis of part of the chicken Rfp-Y region reveals two novel lectin genes, the first complete genomic sequence of a class I alpha-chain gene, a truncated class II beta-chain gene, and a large CR1 repeat. Immunogenetics 55:100–108
Rogers SL, Gobel TW, Viertlboeck BC, Milne S, Beck S, Kaufman J (2005) Characterization of the chicken C-type lectin-like receptors B-NK and B-lec suggests that the NK complex and the MHC share a common ancestral region. J Immunol 174:3475–3483
Saether PC, Westgaard IH, Hoelsbrekken SE, Benjamin J, Lanier LL, Fossum S, Dissen E (2008) KLRE/I1 and KLRE/I2: a novel pair of heterodimeric receptors that inversely regulate NK cell cytotoxicity. J Immunol 181:3177–3182
Sambrook JG, Beck S (2007) Evolutionary vignettes of natural killer cell receptors. Curr Opin Immunol 19:553–560
Sato A, Mayer WE, Overath P, Klein J (2003) Genes encoding putative natural killer cell C-type lectin receptors in teleostean fishes. Proc Natl Acad Sci U S A 100:7779–7784
Sawamura T, Kume N, Aoyama T, Moriwaki H, Hoshikawa H, Aiba Y, Tanaka T, Miwa S, Katsura Y, Kita T, Masaki T (1997) An endothelial receptor for oxidized low-density lipoprotein. Nature 386:73–77
Stet RJ, Hermsen T, Westphal AH, Jukes J, Engelsma M, Lidy Verburg-van Kemenade BM, Dortmans J, Aveiro J, Savelkoul HF (2005) Novel immunoglobulin-like transcripts in teleost fish encode polymorphic receptors with cytoplasmic ITAM or ITIM and a new structural Ig domain similar to the natural cytotoxicity receptor NKp44. Immunogenetics 57:77–89
Taylor PR, Brown GD, Reid DM, Willment JA, Martinez-Pomares L, Gordon S, Wong SY (2002) The beta-glucan receptor, dectin-1, is predominantly expressed on the surface of cells of the monocyte/macrophage and neutrophil lineages. J Immunol 169:3876–3882
Tormo J, Natarajan K, Margulies DH, Mariuzza RA (1999) Crystal structure of a lectin-like natural killer cell receptor bound to its MHC class I ligand. Nature 402:623–631
Viertlboeck BC, Habermann FA, Schmitt R, Groenen MA, Du Pasquier L, Gobel TW (2005) The chicken leukocyte receptor complex: a highly diverse multigene family encoding at least six structurally distinct receptor types. J Immunol 175:385–393
Warren WC, Hillier LW, Graves JAM, Birney E, Ponting CP, Grutzner F, Belov K, Miller W, Clarke L, Chinwalla AT et al (2008) Genome analysis of the platypus reveals unique signatures of evolution. Nature 453:175
Wernersson R, Pedersen AG (2003) RevTrans: multiple alignment of coding DNA from aligned amino acid sequences. Nucleic Acids Res 31:3537–3539
Willment JA, Marshall AS, Reid DM, Williams DL, Wong SY, Gordon S, Brown GD (2005) The human beta-glucan receptor is widely expressed and functionally equivalent to murine Dectin-1 on primary cells. Eur J Immunol 35:1539–1547
Yang Z (1997) PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci 13:555–556
Yoder JA, Mueller MG, Wei S, Corliss BC, Prather DM, Willis T, Litman RT, Djeu JY, Litman GW (2001) Immune-type receptor genes in zebrafish share genetic and functional properties with genes encoded by the mammalian leukocyte receptor cluster. Proc Natl Acad Sci U S A 98:6771–6776
Yoder JA, Litman RT, Mueller MG, Desai S, Dobrinski KP, Montgomery JS, Buzzeo MP, Ota T, Amemiya CT, Trede NS, Wei S, Djeu JY, Humphray S, Jekosch K, Hernandez Prada JA, Ostrov DA, Litman GW (2004) Resolution of the novel immune-type receptor gene cluster in zebrafish. Proc Natl Acad Sci U S A 101:15706–15711
Yoshimura C, Yamaguchi M, Iikura M, Izumi S, Kudo K, Nagase H, Ishii A, Walls AF, Ra C, Iwata T, Igarashi T, Yamamoto K, Hirai K (2002) Activation markers of human basophils: CD69 expression is strongly and preferentially induced by IL-3. J Allergy Clin Immunol 109:817–823
Yusa S, Catina TL, Campbell KS (2002) SHP-1- and phosphotyrosine-independent inhibitory signaling by a killer cell Ig-like receptor cytoplasmic domain in human NK cells. J Immunol 168:5047–5057
Zelensky AN, Gready JE (2003) Comparative analysis of structural properties of the C-type-lectin-like domain (CTLD). Proteins 52:466–477
Zelensky AN, Gready JE (2005) The C-type lectin-like domain superfamily. Febs J 272:6179–6217
Acknowledgments
This work was carried out as part of the Platypus Genome Project. We thank Wes Warren and his team at Washington University, St. Louis, for giving us the opportunity to work on such an exciting project. We thank Kaighan McColl for bioinformatics support. E.W. and C.S. receive scholarships from the Jean Walker Trust and the ARC Center for Kangaroo Genomics.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary materials
Below is the link to the electronic supplementary material.
ESM 1
Includes detailed information on the identification of Ig domains, specifically pertaining to the identification of OSCAR, SIGLEC and Fc receptor genes, FISH data and accession numbers for all non-platypus sequences used in phylogenetic analyses (DOC 755 kb)
Rights and permissions
About this article
Cite this article
Wong, E.S.W., Sanderson, C.E., Deakin, J.E. et al. Identification of natural killer cell receptor clusters in the platypus genome reveals an expansion of C-type lectin genes. Immunogenetics 61, 565–579 (2009). https://doi.org/10.1007/s00251-009-0386-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00251-009-0386-7