Abstract
Killer cell immunoglobulin-like receptors (KIR) gene frequencies vary between populations and contribute to functional variation in immune responses to viruses, autoimmunity and reproductive success. This study describes the frequency distribution of 12 variable KIR genes and their HLA-C ligands in two Iranian populations who have lived for many generations in different environments: the Azerbaijanis at high altitude and the Jonobi people at sea level. The results are compared with those published for other human populations and a large group of English Caucasians. Differences were seen in KIR and HLA-C group frequencies, in linkage disequilibrium and inhibitory/activating KIR ratios between the groups. Similarities with geographically close populations in the frequencies of the KIR A and B haplotypes and KIR AA genotype reflected their common ancestry. The extreme variability of the KIR gene family and their HLA-C ligands is highlighted and their importance in defining differences between geographically and culturally isolated communities subject to different environmental pressures who come from the same ethnic grouping.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Natural killer (NK) cells are essential components of the innate immune system and act in the first line of defence against infection, play a role in tumour surveillance and are involved in placental development. Functionally, they are controlled by activating and inhibitory receptors (Colucci et al. 2003; Lanier 2008; McQueen and Parham 2002) and in humans the Killer cell Immunoglobulin-like Receptors (KIR) are of central importance in distinguishing normal healthy self cells from diseased or allogeneic targets. HLA class I molecules are ligands for some of the KIR and can trigger signals to inhibit effector function of the NK cells or conversely to stimulate killing or cytokine production (Colonna et al. 2000; Long et al. 2001). The KIR genes and the genes for their HLA class I ligands reside on different chromosomes and probably constitute the most diverse loci in the human genome. Indeed, the KIR region is clearly undergoing rapid evolution and selection and only the most closely related individuals will share identical KIR genotypes (Khakoo et al. 2000; Parham 2005; Shilling et al. 2002). The diversity of the KIR region is achieved by both polymorphism at individual KIR gene loci as well as variable gene content. The framework KIR genes (3DL3, 2DL4 and 3DL2) are present in nearly all individuals. Thirteen additional KIR genes have been characterised encoding five inhibitory receptors (3DL1, 2DL1–3 and 2DL5) and six activating KIR (2DS1–5, 3DS1). There are also two pseudogenes (2DP1 and 3DP1) that encode no cell surface receptors. Although KIR genotypes can vary markedly, these can be simplified into two broad categories comprised of A and/or B haplotypes. The simpler A haplotype has 7 KIR genes mainly coding for the inhibitory type (3DL1–3, 2DL1 and 2DL3) and a single KIR gene, 2DS4, coding for an activating receptor. KIR2DS4 is frequently represented by alleles with a frame-shifting deletion that abrogates the activating function of the receptor. The B haplotype has a more variable content with additional KIR genes (2DS1–3, 2DS5, 3DS1, 2DL2 and 2DL5) chiefly coding for activating receptors. In addition, the B haplotype can be divided into two halves, which are centromeric or telomeric to the centrally placed framework locus, 2DL4 (Yawata et al. 2002). These two halves were deduced from linkage disequilibrium studies and appear to be under differing selection pressures (Yawata et al. 2006). Similarly the numerous HLA-C alleles fall into one of two categories, C1 and C2, as recognized by the NK cell through the KIR receptors.
A number of studies of human populations world-wide have shown differences in KIR frequencies and haplotypes correlating with ethnicity, migration routes out of Africa and relative population isolation (Denis et al. 2005; Gendzekhadze et al. 2006; Kulkarni et al. 2008; Norman et al. 2002; Rajalingam et al. 2008; Rayes et al. 2008; Single et al. 2007). In the Middle East, the KIR frequencies are similar to those found in Caucasian populations whilst KIR genotypes in Indians or Pakistanis show a tendency towards an increase in KIR B haplotypes. Interestingly, the Parsis who migrated from Persia to North India over 1,000 years ago differ from their North Indian neighbours but show close similarities to KIR genotypes in Maharashtrians who settled in North India in ancient times and have their own language (Kulkarni et al. 2008). Until recently there had been no data available for Iranian populations which left a gap, geographically speaking, between the near-East (Mahfouz et al. 2006; Rayes et al. 2008) and Pakistan and India (Kulkarni et al. 2008; Norman et al. 2002; Rajalingam et al. 2002) in reported KIR variation.
We have now genotyped samples from two distinct Iranian populations the Azerbaijanis and the Jonobi people and compared our results with those populations that are geographical neighbours as well as an English Caucasian group. The Azerbaijanis have lived for many generations at high altitude (above 2,400 m) in East Azerbaijan and Ardabil provinces. The Jonobi people from Hormozgan province and Balochistan live at sea level along the shores of the Persian Gulf and Oman Sea (Fig. 1). In epidemiological studies, KIR and HLA ligand combinations have been shown to be important in the pathogenesis of certain viral infections such as HIV and HCV (Khakoo et al. 2004; Martin et al. 2002), in autoimmune diseases (Gardiner 2008; Nelson et al. 2004) and in reproductive fitness (Hiby et al. 2004, 2008; Parham 2004). We therefore typed our two Iranian populations for the12 KIR genes characterised by their variable presence and also identified the HLA-C groups, KIR ligands C1 and C2, carried by each individual. The Iranian results were compared to our Caucasian population and to other geographically close populations described in the literature.
Ethnographic regions of Iran showing the two populations genotyped for KIR and HLA-C groups. Borders in the figure roughly correspond to cultural regions based on ethnicity and language. From Ashrafian-Bonab et al. 2007. Azerbaijanis traditionally live at >2,400 m, the Jonobi live at sea level in the Hormozgan and Baluchistan provinces
Materials and methods
Study subjects
Blood samples were collected from unrelated adult males from two isolated Iranian communities as described (Ashrafian-Bonab et al. 2007). Marrying out of these communities is not common practice and they are very different in terms of culture and language. Samples were randomly collected from consenting volunteers to ensure an unbiased sampling strategy. Ethnic information was obtained on the place of birth of the subject and the place of birth of their parents and grandparents (Ashrafian-Bonab et al. 2007). One hundred samples from both populations were analysed in this study. Ethical approval was obtained from the Iranian Ministry of Health and the Iranian Cultural Heritage Organization.
Blood samples were taken from 584 healthy English Caucasian primiparous women and their male partners. Informed written consent and Local Ethics Committees' approval were obtained.
DNA isolation and genotyping
DNA from the Iranian samples was extracted using the PAXgene blood DNA kit (Preanalytix, GmbH, Germany). Genomic DNA from the Caucasian cohort was isolated from 5 ml of blood using the QIAamp DNA Maxi Blood Kit (Qiagen, Crawley, UK). PCR-SSP was used to amplify the genomic DNA for presence or absence of the 12 variable KIR genes coding for the inhibitory KIR, 2DL1, 2DL2, 2DL3, 2DL5 and 3DL1 and the activating KIR, 2DS1, 2DS2, 2DS3, 2DS4, 2DS5, 3DS1 and the pseudogene, 2DP1. KIR2DS4 alleles were also identified as being of the full-length type or having the 22bp deletion that would negate cell-surface expression, sometimes referred to as KIR1D. Two pairs of PCR primers were selected for each gene or allele to give relatively short amplicons of 100 to 800bp as previously described (Hiby et al. 2004). HLA-C was also genotyped as group C1 and/or group C2 as recognised by NK cells through the KIR receptors. The method used to define these groups by PCR-SSP was as previously described (Hiby et al. 2008).
Data analysis and statistical methods
Carrier frequency was calculated by direct counting of individuals possessing at least one copy of the gene (individuals heterozygous for a KIR gene cannot be distinguished from those that are homozygous without family members, who were not available for this study). KIR gene frequency estimates were calculated using the equation \( {\text{KGF}} = {\text{1}} - \surd \left( {{\text{1}} - {\text{f}}} \right) \) where f is the carrier frequency. Frequency of A and B haplotypes was deduced by defining the A haplotype as having the variable genes 2DL1-2DL3-3DL1-2DS4 only. The presence of the B haplotype would then be recognised by detection of any of the other KIR genes. Fisher's exact test was used to calculate p values for the comparison of carrier frequencies. LD coefficients for two-locus associations were calculated for unrelated Azerbaijani and Jonobi individuals according to Imanishi et al. (1991). The statistical significance of LD was calculated using Fisher's exact test. KIR2DL2/2DL3 and KIR3DL1/3DS1 allelic pairs and the HLA-C groups, C1 and C2, were found to be in Hardy–Weinberg equilibrium in all three populations.
Results and discussion
KIR and HLA-C group carrier frequencies
Only those KIR genes (n = 12) that were known to have variable occurrence were typed and thus a measure of gene content could be obtained for each individual. The framework genes, KIR3DL3, 3DL2, and 2DL4 as well as the pseudogene 3DP1were assumed to be present in all individuals as seen for most other populations. Carrier and estimated gene frequencies are given in Table 1 together with KIR frequencies from geographically close populations taken from the literature and from our English Caucasian cohort. The prevalence of the most common KIR AA genotype and that of the HLA-C2 group is also given.
Notably, there were higher frequencies of a number of KIR; both those coding for inhibitory receptors, KIR2DL1 and KIR3DL1 and activating receptors, KIR2DS1, 2DS4 and 3DS1 in the Azerbaijanis compared with the Jonobi population. The genes coding for activating KIR, 2DS1 and 3DS1 are in strong linkage disequilibrium to each other and have been implicated in resistance to infection and in reproductive success (Hiby et al. 2008; Martin et al. 2002; Qi et al. 2006). The KIR2DS4 allele coding for the full-length cell surface receptor was present at equal frequency in the two populations (carrier frequency being 29% in Azerbaijanis, 28% in Jonobis, not shown), the deleted form was more frequent in the Azerbaijanis. Expression of inhibitory KIR has been shown to be important in determining the responsiveness of NK cells to stimuli (Anfossi et al. 2006; Yawata et al. 2008).
Overall the Jonobi population more closely resembled the more westerly Palestinian and Lebanese people than did the Azerbaijanis but neither had such high frequencies of the activating KIR genes as seen in the more easterly populations of the Karachi South Asians and the Parsis.
Notably KIR2DS3 was present at high frequency in both Iranian populations (42% carriers) compared to our Caucasian population (30% carriers). This relatively high frequency of 2DS3 was also observed for neighbouring population groups from Palestine (Norman et al. 2001) to the Parsis in India (Kulkarni et al. 2008).
The KIR A and B haplotypes were deduced from the individual KIR gene frequencies as described. Overall there was a balance in the frequencies of the A and B haplotypes as has been reported in all populations analysed (Rajalingam et al. 2008). The Jonobi people had 56% A haplotypes and the Azerbaijanis, 53%. Our Caucasian population had 59% A haplotypes to 41% B haplotypes. From the ratio of frequencies of inhibitory (2DL1, 2DL2, 2DL3, 3DL1) to activating (2DS1, 2DS2, 2DS4 full length, 3DS1) KIR genes reported to have HLA class I ligands the Azerbaijanis are deviated more towards activation (ratio inhibitory to activating KIR 1.85/1) than the Jonobi people (ratio 2.01/1). The Caucasians gave a ratio with these KIR very similar to the Jonobi population of 2.04/1.
In addition there was a significantly higher frequency of the KIR ligand HLA-C2 in the Jonobis as compared to the Azerbaijanis (p value, 0.006) and the Caucasians (p < 0.001) with a notably high carrier frequency of 57% (Table 1). The C2 frequency is also high in Parsis (47%). In addition, the data from our three populations fits well with the model of Single et al. (2007) who depicted a negative correlation between KIR2DS1 and its ligand HLA-C2 that provides good evidence for their co-evolution (Fig. 2).
Correlation between KIR2DS1 and C-group 2 frequencies. Results for our populations are superimposed upon the global distribution. Each coloured circle represents a distinct population, with the colours identifying the geographic origin of the populations. The solid line represents the estimated regression line (Single et al. 2007).The Iranian and Caucasian population data reported here fits well with the study of Kulkarni et al. (2008) The activating KIR2DS1 shows a negative correlation with its HLA-C2 ligand although this did not reach significance. Boxed letters denote our populations: C Caucasian, A Azerbaijani and J Jonobi. For comparison, NI North Indian, M Maharashtrian and P Parsis are also shown within the diamonds
Since writing this report, two publications on other Iranian populations have been released (Ashouri et al. 2009 and Tajik et al. 2009). One reported on the general Iranian population selected from a number of cities (Tajik et al. 2009) and the second includes an Azeri population that was not selected on living at high altitude and indeed does not resemble our own Azerbaijani population in terms of the KIR frequencies (Ashouri et al. 2009). The possible confounding effect of racial admixture was suggested for this latter population by the authors.
KIR genotype profiles
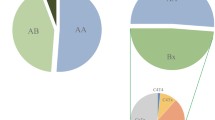
The Azerbaijanis and Jonobi people shared 16 KIR genotype profiles and 15 of these were also seen in the Caucasian population (Fig. 3). The KIR AA genotype was the most frequent in all three populations. The Azerbaijanis had seven profiles that were not found in the Jonobi people (five of these were seen in the Caucasians). In addition, the Jonobi people possessed 12 profiles not found in the Azerbaijanis and only four of these were seen in the Caucasians. The English Caucasian population had an additional 18 KIR profiles that were not seen in either Iranian population and these were uncommon genotypes present in one to four individuals only. The number of distinct profiles in the Azerbaijani, Jonobi and Caucasians numbered 23, 28 and 43, respectively. This would support the view that the Iranian populations were from relatively isolated populations as seen with the Parsis (19 profiles, Kulkarni et al. 2008) and in contrast to the genetically diverse North Indians (47 profiles, Rajalingam et al. 2002) and our Caucasian population (Shifman et al. 2003).
KIR A and B haplotypes
Unusual KIR genotypes, with all the KIR A haplotype genes but with single additional genes of the B haplotype were observed. In the Azerbaijani population, two individuals possessed KIR2DS1 with the A haplotype genes but no other B haplotype KIR. This has been observed in other populations at low frequency (Kulkarni et al. 2008). Another Azerbaijani individual had an additional KIR2DS2 with the KIR A haplotype genes as reported in several Indian populations (Rajalingam et al. 2008). In addition a single Jonobi individual had the unusual combination of KIR AA genotype genes with a KIR2DL5gene. This has not been reported before.
The next most common genotype in the Azerbaijanis after KIR AA comprised both an A haplotype and a B haplotype which was characterised by presence of the, mainly activating, telomeric group of KIR genes: 3DS1-2DL5-2DS5-2DS1 (profile 2 shared, Fig. 3). This was in contrast to the second most common in the Jonobi which had the A haplotype loci together with the centromeric B haplotype genes 2DL2 and 2DS2 (profile 4 shared, Fig. 3).
Linkage disequilibrium
Comparing linkage disequilibrium between pairs of KIR genes in the two Iranian populations reflected their different spread of KIR genotypes (Fig. 4). The Jonobi sample had 29 pairs of genes in significant positive or negative LD compared with the Azerbijani with 22 and this can be compared with the endogamous Parsis population with 39 pairs. The higher frequency of the KIR AA genotype in the Jonobi people was reflected in a more marked distinction between the LD values of the haplotype A and B KIR genes seen for this population. There were 13 pairs of genes that gave discordant results: eight pairs were in significant negative LD only in the Jonobi (3DS1-3DL1, 2DL5-3DL1, 2DS5-3DL1, 2DS1-3DL1, 2DS4-3DS1, 2DS4-2DL5, 2DS4-2DS5 and 2DS4-2DS1), whereas two pairs were in significant positive LD (2DP1-2DL3 and 2DL1-2DL3). Only three KIR gene pairs in the Azerbijani showed significance differences not found in the Jonobi with 2DL3-2DL5, 2DL3-2DS5 and 2DL3-2DS1 being in significant negative LD. Like the Parsis the Azerbijanis also showed an unusual lack of LD between KIR2DL3 and KIR2DL1 which are closely linked on the A haplotype and are reported to be in strong LD in all other published populations. The KIR2DL1*004 allele that resides on the B haplotype is not in LD with KIR2DL3 so this may explain the results obtained if the Azerbaijanis possess an unusual high frequency of the B haplotype 2DL1 alleles. KIR2DL1 allele typing would be required to confirm this.
Summary
The two Iranian populations showed distinctive differences in KIR and HLA-C group carrier and genotype frequencies. Comparison with other genotyped populations to the west and east of Iran can now be made using this data. We also describe unusual KIR genotypes emphasising the great diversity and malleability of the KIR locus in human populations that may arise from selective pressures in response to pathogens, reproductive fitness or other factors in the environment. Our other studies indicate that KIR B telomeric genes are those lacking in Caucasian women who experience problems during pregnancy. Of particular interest in this regard are pregnancy and reproduction at high altitude: birth weight decreases with altitude and the risk of foetal growth restriction and pre-eclampsia are increased (Julian et al. 2007; Zamudio 2007). Furthermore, these observations are likely due directly to hypoxia at altitude, rather than economic status (Giussani et al. 2001). Importantly, those with high altitude ancestry are partially protected from this reduction in fetal growth compared with those who have settled in the area more recently (Julian et al. 2007). When comparing these two Iranian populations for the two polymorphic gene families the increased incidence of KIR B telomeric genes in the Azerbaijanis compared to the Jonobis is interesting because the incidence of hypertension in pregnancy and recurrent miscarriage is increased in these Iranians living at high altitude in data supplied by the Iranian Ministry of Health (Table 2). Whether differences in KIR can be ascribed to contrasting environments, especially adaptations to living at high and low altitude, can only be determined as data on other similar populations increases.
References
Anfossi N, Andre P, Guia S, Falk CS, Roetynck S, Stewart CA et al (2006) Human NK cell education by inhibitory receptors for MHC class I. Immunity 25:331–342. doi:10.1016/j.immuni.2006.06.013
Ashouri E, Farjadian S, Reed EF, Ghaderi A, Rajalingam R (2009) KIR gene content diversity in four Iranian populations. Immunogenetics 61:483–492. doi:10.1007/s00251-009-0378-7
Ashrafian-Bonab M, Lawson Handley LJ, Balloux F (2007) Is urbanization scrambling the genetic structure of human populations? A case study. Heredity 98:151–156. doi:10.1038/sj.hdy.6800918
Colonna M, Moretta A, Vely F, Vivier E (2000) A high resolution view of NK-cell receptors: structure and function. Immunol Today 21:428–431. doi:10.1016/S0167-5699(00)01697-2
Colucci F, Caliguri MA, Di Santo JP (2003) What does it take to make a natural killer. Nat Rev Immunol 3:413–425. doi:10.1038/nri1088
Denis L, Sivula J, Gourraud PA, Kerdudou N, Chout R, Ricard C, Moisan JP, Gagne K, Partanen J, Bignon JD (2005) Genetic diversity of KIR natural killer cell markers in populations from France, Guadeloupe, Finland, Senegal and Reunion. Tissue Antigens 66:267–276. doi:10.1111/j.1399-0039.2005.00473.x
Gardiner CM (2008) Killer cell immunoglobulin-like receptors on NK cells: the how, where and why. Int J Immunogenet 35:1–8. doi:10.1111/j.1744-313X.2007.00739.x
Gendzekhadze K, Norman PJ, Abi-Rached L, Layrisse Z, Parham P (2006) High KIR diversity in Amerindians is maintained using few gene-content haplotypes. Immunogenetics 58:474–480. doi:10.1007/s00251-006-0108-3
Giussani DA, Phillips PS, Anstee S, Barker DJ (2001) Effects of altitude versus economic status on birth weight and body shape at birth. Pediatr Res 49:490–494. PMID:11264431
Hiby SE, Walker JJ, O'Shaughnessy KM, Redman CW, Carrington M, Trowsdale J, Moffett A (2004) Combinations of maternal KIR and fetal HLA-C genes influence the risk of preeclampsia and reproductive success. J Exp Med 200:957–965. doi:10.1084/jem.20041214
Hiby SE, Regan L, Lo W, Farrell L, Carrington M, Moffett A (2008) Association of maternal killer-cell immunoglobulin-like receptors and parental HLA-C genotypes with recurrent miscarriage. Hum Reprod 23:972–976. doi:10.1093/humrep/den011
Imanishi T, Akaza T, Kimura A, Tokunaga K, Gojobori T (1991) Estimation of allele and haplotype frequencies for HLA and complement loci. In: Tsuji K, Aizawa M, Sasazuki T (eds) HLA: 1991 Proceedings of the eleventh International Histocompatibility Workshop and Conference, vol 1. Oxford University Press, New York, 6–13 November 1991
Julian CG, Vargas E, Armaza JF, Wilson MJ, Niermeyer S, Moore LG (2007) High-altitude ancestry protects against hypoxia-associated reductions in fetal growth. Arch Dis Child Fetal Neonatal Ed 92:372–377. doi:10.1136/adc.2006.109579
Khakoo SI, Rajalingam R, Shum BP, Weidenbach K, Flodin L, Muir DG, Canavez F, Cooper SL, Valiante NM, Lanier LL, Parham P (2000) Rapid evolution of NK receptor systems demonstrated by comparison of chimpanzees and humans. Immunity 12:687–698
Khakoo SI, Thio CL, Martin MP, Brooks CR, Gao X, Astemborski J et al (2004) HLA and NK cell inhibitory receptor genes in resolving hepatitis C virus infection. Science 305:872–874. doi:10.1126/science.1097670
Kulkarni S, Single MMP, Rajalingam R, Badwe R, Joshi N, Carrington M (2008) Comparison of the rapidly evolving KIR locus in Parsis and natives of India. Immunogenetics 60:121–129. doi:10.1007/s00251-008-0279-1
Lanier L (2008) Up on the tightrope: natural killer cell activation and inhibition. Nat Immunol 9:495–502. doi:10.1038/ni1581
Long EO, Barber DF, Burshtyn DN, Faure M, Peterson M, Rajagopalan S, Renard V, Sandusky M, Stebbins CC, Wagtmann N, Watzl C (2001) Inhibition of natural killer cell activation signals by killer cell immunoglobulin-like receptors (CD158). Immunol Rev 181:223–233. doi:10.1034/j.1600-065X.2001.1810119.x
Mahfouz R, Rayes R, Mahfoud Z, Bazarbachi A, Zaatari G (2006) Distribution of killer cell immunoglobulin-like receptors genotypes in the Lebanese population. Tissue Antigens 68:66–71. doi:10.1111/j.1399-0039.2006.00605.x
Martin MP, Gao X, Lee JH, Nelson GW, Detels R, Goedert JJ et al (2002) Epistatic interaction between KIR3DS1 and HLA-B delays progression to Aids. Nat Genet 31:429–434. doi:10.1038/ng934
McQueen KL, Parham P (2002) Variable receptors controlling activation and inhibition of NK cells. Curr Opin Immunol 14:615–621
Nelson GW, Martin MP, Gladman D, Wade J, Trowsdale J, Carrington M (2004) Cutting edge: heterozygote advantage in autoimmune disease: hierarchy of protection/susceptibility conferred by HLA and killer Ig-like receptor combinations in psoriatic arthritis. J Immunol 173:4273–4276
Norman PJ, Stephens HAF, Verity DH, Chandanayingyong D, Vaughan RW (2001) Distribution of natural killer cell immunoglobulin-like receptor sequences in three ethnic groups. Immunogenetics 52:195–205. doi:10.1038/sj.gene.6363836
Norman PJ, Carrington CVF, Byng M, Maxwell LD, Curran MD, Stephens HAF et al (2002) Natural killer cell immunoglobulin-like receptor (KIR) locus profiles in African and South Asian populations. Genes Immun 3:86–95. doi:10.1038/sj.gene.6363836
Parham P (2004) NK cells and trophoblasts: partners in pregnancy. J Exp Med 200:951–955. doi:10.1084/jem.20041783
Parham P (2005) MHC class I molecules and KIRs in human history, health and survival. Nat Rev Immunol 5:201–214. doi:10.1038/nri1570
Qi Y, Mp M, Gao X, Jacobson L, Goedert JJ, Buchbinder S, Kirk GD, O'Brien SJ, Trowsdale J, Carrington M (2006) KIR/HLA pleiotropism: protection against both HIV and opportunistic infections. PloS Pathog 2:e79. doi:10.1371/journal.ppat.0020079
Rajalingam R, Krausa P, Shilling HG, Stein JB, Balamurugan A, McGinnis MD, Cheng NW, Mehra NK, Parham P (2002) Distinctive KIR and HLA diversity in a panel of north Indian Hindus. Immunogenetics 53:10009–1019. doi:10.1007/s00251-001-0425-5
Rajalingam R, Du Z, Meenagh A, Luo L, Kavitha VJ, Pavithra-Arulvani R et al (2008) Distinct diversity of KIR genes in three southern Indian populations: comparison with world populations revealed a link between KIR gene content and pre-historic human migrations. Immunogenetics 60:207–217. doi:10.1007/s00251-008-0286-2
Rayes R, Bazarbachi A, Khazen G, Sabbagh A, Zaatari G, Mahfouz R (2008) Natural killer cell immunoglobulin-like receptors (KIR) genotypes in two Arab populations: will KIR become a genetic landmark between nations? Mol Biol Rep 35:225–229. doi:10.1007/s11033-007-9074-6
Single RM, Martin MP, Gao X, Meyer D, Yeager M, Kidd JR, Kidd KK, Carrington M (2007) Global diversity and evidence for coevolution of KIR and HLA. Nat Genet 39:1114–1119. doi:10.1038/ng2077
Shifman S, Kuypers J, Kokoris M, Yakir B, Darvasi A (2003) Linkage disequilibrium patterns of the human genome across populations. Hum Mol Genet 12:771–776
Shilling HG, Guethlein LA, Cheng NW, Gardiner CM, Rodriguez R, Tyan D, Parham P (2002) Allelic polymorphism synergizes with variable gene content to individualize human KIR genotype. J Immunol 168:2307–2315
Tajik N, Shahsavar F, Mousavi T, Radjabzadeh MF (2009) Distribution of KIR genes in the Iranian population. Tissue Antigens 74:22–31. doi:10.1111/j.1399-0039.2009.01263.x
Yawata M, Yawata N, Abi-Rached L, Parham P (2002) Variation within the human killer cell immunoglobulin-like receptor (KIR) gene family. Crit Rev Immunol 22:463–482
Yawata M, Yawata N, Draghi M, Little AM, Partheniou F, Parham P (2006) Roles for HLA and KIR polymorphisms in natural killer cell repertoire selection and modulation of effector function. J Exp Med 203:633–645. doi:10.1084/jem.20051884
Yawata M, Yawata N, Draghi M, Partheniou F, Little AM, Parham P (2008) MHC class I-specific inhibitory receptors and their ligands structure diverse human NK-cell repertoires toward a balance of missing self-response. Blood 112:2369–2380. doi:10.1182/blood-2008-03-143727
Zamudio S (2007) High-altitude hypoxia and preeclampsia. Front Biosc 12:2967–2977
Acknowledgements
We would like to thank all the dedicated clinical and scientific staff who helped us to acquire the DNA samples used for this study and many thanks to the consenting subjects. The work was funded by the Wellcome Trust and the British Heart Foundation in the UK. This project has been funded in whole or in part with federal funds from the US National Cancer Institute (National Institutes of Health) under contract N01-CO-12400. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the US Government.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
An erratum to this article can be found at http://dx.doi.org/10.1007/s00251-010-0436-1
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Hiby, S.E., Ashrafian-Bonab, M., Farrell, L. et al. Distribution of killer cell immunoglobulin-like receptors (KIR) and their HLA-C ligands in two Iranian populations. Immunogenetics 62, 65–73 (2010). https://doi.org/10.1007/s00251-009-0408-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00251-009-0408-5