Abstract
The extreme variability at the Killer cell Immunoglobulin-like Receptor (KIR) locus along with that of the genes encoding their ligands, HLA class I, appears to modulate risk for viral, autoimmune, and malignant diseases, and reproductive failure. Differences in KIR gene and haplotype frequencies across world populations may reflect some combination of ancestral genotypes, locale-specific selection pressures, and genetic drift. We genotyped unrelated healthy Parsis and Maharashtrian Hindus, neighboring peoples from Western India. These two populations showed remarkable similarity in KIR gene frequencies despite their distinct ethnic background and the fairly recent migration of Parsis to Western India from Persia around 900 A.D. One clear exception is KIR3DS1, which is found at a significantly higher frequency in the Parsis than in the Maharashtrians, previously characterized North Indians, and most other world populations. The high KIR3DS1 frequency of Parsis corresponds with a low frequency of its putative HLA-B ligand group, an inverse correlation that has been observed previously across other world populations. Thus, KIR3DS1 frequency in Parsis may be a remnant of their distinct ancestral Persian origin. KIR gene frequencies and profiles of the Parsis and Maharashtrians were more similar to one another than they were to North Indians, suggesting a potential effect of local environmental factors on KIR evolution and/or some degree of admixture between Parsis and populations from Western India. Overall, these data support other studies indicating the rapid evolution of the KIR locus and the apparent dependency of this evolution on the loci encoding HLA class I ligands.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Natural killer (NK) cells are critical components of the innate immune system and they are controlled by many activating and inhibitory receptors (Colucci et al. 2002; Lanier 2005), including members of the killer cell immunoglobulin-like receptor (KIR) family. The KIR region exhibits an extensive degree of diversity, which it achieves through a combination of variable gene content and allelic polymorphism (Carrington and Norman 2003). Population studies using genomic DNA to determine the presence or absence of each KIR gene have demonstrated that across individuals, KIR gene content can vary widely. Evaluation of differences in nucleotide sequences have documented an additional dimension of KIR diversity achieved through allelic polymorphism (Shilling et al. 2002). Indeed, estimates of the extent of KIR genotype diversity within the population suggest that <0.24% of unrelated individuals can expect to have identical KIR genotypes (Shilling et al. 2002).
While gene content varies across humans, the framework genes 3DL3 and 3DL2 at either terminus of the region and 2DL4 in the middle of the locus are consistently observed in all individuals (Wilson et al. 2000). KIR haplotypes can be divided into two basic groups, A and B, both of which are found in most populations (Denis et al. 2005; Hsu et al. 2002; Jiang et al. 2005; Norman et al. 2002; Rajalingam et al. 2002; Toneva et al. 2001; Uhrberg et al. 1997; Velickovic et al. 2006; Whang et al. 2005; Yawata et al. 2002) but sometimes at very different frequencies. Haplotype A, which is estimated to be as frequent as 47–59% within Caucasian populations, comprises a common complement of KIR genes: 3DL3, −2DL3, −2DL1, −2DL4, −3DL1, −2DS4, and −3DL2. In contrast, haplotype B varies in gene content and, in general, contains more activating receptor genes (2DS and 3DS genes) than the A haplotype (Carrington and Norman 2003). Frequencies of each gene can vary remarkably across world populations, which might reflect differential selection pressures as well as persistence of ancestral genotypes. KIR and HLA ligand combinations also appear to be important in pathogenesis of several autoimmune diseases, cancers, viral infections, and reproductive fitness (Carrington and Martin 2006; Parham 2005).
The study of KIR gene and profile (the gamut of KIR genes present in a given individual irrespective of copy number) variability across distinct populations might predict the extant diseases to which a given population may be particularly resistant or susceptible. In India, KIR genotypes have been reported only in North Indian Hindus (Rajalingam et al. 2002). The present study describes KIR genotypes in Maharashtrians and Parsis who have been geographical neighbors for several centuries, but who are of distinct ethnic backgrounds; Parsis being of Persian descent and the Maharashtrians being natives of India. The Parsis have maintained their ancestral, cultural, and religious practices, with a strong tendency to marry only within the Parsi Zoroastrian community. We tested for the presence/absence of 15 KIR loci in unrelated, healthy Parsi (n = 145) and Maharashtrian (n = 139) women and compared characteristics of the Parsi KIR locus to that of Maharashtrians and the previously reported North Indians (Rajalingam et al. 2002). The study of Parsis is of particular importance, as this community is facing dwindling numbers and high incidence of cancer, neurodegenerative diseases, autoimmune diseases, and low birth rates (Desai 2004).
Materials and methods
Study subjects
Blood was collected from 145 Parsis (Zoroastrian) from Mumbai (Maharashtra, India) and 139 Maharashtrians (Marathi-speaking Hindus) residing in Mumbai with their informed consent. All individuals were healthy and unrelated. Peripheral blood lymphocytes (PBLs) were separated on Ficoll Hypaque mixture (Boyum 1968).
DNA extraction
DNA was extracted from 5 × 106 PBLs using phenol–chloroform-based extraction followed by precipitation of DNA with ethanol. DNA was washed in 70% ethanol and then dissolved in TE buffer (Wallace 1987).
Polymerase chain reaction (PCR)
Genomic DNA was amplified for presence or absence of the following genes: 2DL1, 2DL2, 2DL3, 2DL4, 2DL5, 2DS1, 2DS2, 2DS3, 2DS4, 2DS5, 3DL1, 3DL2, 3DL3, 3DS1, and 2DP1. Genotyping was performed using PCR amplification with two pairs of locus-specific primers (PCR-SSP) as previously described (Martin and Carrington 2007). Amplification control primers that amplify a 796-bp fragment of the third intron of DRB1 were also included in each PCR to confirm robust PCR. Amplification was performed in a volume of 5 μl containing 200 μM dNTP, 500 nM primer, 1.5 mM MgCl2, 20 mM Tris–HCl, pH 8.4, 50 mM KCl, and 0.5 U of Platinum Taq DNA polymerase (Invitrogen). Cycling was performed as follows: 2 min at 94°C; five cycles of 94°C for 5 s, 65°C for 15 s, 72°C for 30 s; 21 cycles of 94°C for 5 s, 60°C for 15 s, 72°C for 30 s; four cycles of 94°C for 5 s, 55°C for 1°min, 72°C for 2 min; and a final extension step of 10 min at 72°C. PCR products were electrophoresed in 3% agarose gels containing ethidium bromide, and predicted size products were visualized under ultraviolet light.
Statistical analysis
LD coefficients for two-locus associations were calculated from unrelated Parsi and Maharashtrian individuals according to Imanishi et al. (1991). The statistical significance of LD was calculated using Fisher’s exact test. p values for the comparison of carrier frequencies were corrected using Bonferroni’s method based on the number of genes compared. Figure 2 presents three panels from a study of activating KIR and their corresponding ligands (Single et al. 2007). Results for the Parsis, Maharastrians, and North Indians are superimposed upon the global distribution. Each colored circle represents a distinct population, with the colors identifying the geographic origin of the populations. The solid line represents the estimated regression line from the Single et al. study.
Results
The comparison of highly polymorphic genomic regions between the Parsis and Maharashtrians serves as a unique opportunity to test for signatures of locale-specific selection vs. ancestral ties over a fairly short period of time. The Parsis of Mumbai, sampled here, migrated to this region from Gujarat (the region of their original immigration into India) in the late seventeenth century and have maintained their ancestral genetic roots by avoiding marriage outside of the Parsi community and disallowing conversions to the faith. Thus, similarity between the two groups in frequencies of KIR genes might suggest selection for these genes, whereas distinctions may be better explained by ancestry. KIR3DL2, 3DL3, 2DL4 were present in all individuals from both populations, a characteristic that is common across the populations that have been typed to date (Wilson et al. 2000). KIR2DL1 and 3DL1 were also very common in both Parsis and Maharashtrians; Table 1). The frequencies of all other KIR were comparable in the two populations, with the exception of KIR2DS1 and KIR3DS1, the latter of which was significantly higher in Parsis than in Maharashtrians after correction for multiple tests (p < 0.001; Table 1). Thus, in general, the Parsis and Maharashtrians showed remarkably similar KIR gene frequencies except for two activating KIR that are in strong linkage disequilibrium with one another (Cook et al. 2003; Norman et al. 2001).
The North Indians genotyped previously for KIR (Rajalingam et al. 2002) were, in general, more distinct from Parsis and Maharashtrians than the latter two were from one another (Table 1 and Fig. 1), again, with the exception of activating 3DS1 and 2DS1. In particular, the frequencies of 2DL1 and 2DL3 were significantly higher in Parsis and Maharashtrians than in the North Indians, although the Parsi–North Indian comparison was not significant after adjusting for multiple tests.
KIR genotype profiles and their frequencies in Parsis, Maharashtrians, and North Indians. A total of 19 distinct profiles were observed in Parsis and 24 in Maharashtrians. The frequencies for these profiles are compared with those of the 47 distinct profiles in North Indians reported by Rajalingam et al. (2002). Presence (shaded boxes) and absence (white boxes) is indicated for 12 KIR loci. The framework genes 3DL3, 3DL2, and 2DL4 were present in each individual and are not shown in the figure. 2DP1 has also been excluded because it was not typed in the North Indians. The genotype profiles are organized by the degree to which they are shared among the three populations
KIR gene profiles represent a listing of all genes present in a given individual, without regard to copy number, and serve as an alternative molecular genetic characteristic when segregation (i.e., haplotypic) analysis is not an option. KIR profiles were compared among the subjects for whom all fifteen KIR genes were typed unambiguously (137 Parsi women and 118 Maharashtrian women). The number of distinct profiles in Parsis, Maharashtrians, and North Indians was 19, 24, and 47, respectively, supporting cohesiveness of the Parsi community and a very genetically diverse North Indian population (Fig. 1 and Rajalingam et al. 2002). The most common profile by far, in both the Parsis and Maharashtrians, was indicative of homozygosity for the A haplotype: 3DL3–2DL3–2DL1–2DL4–3DL1–2DS4–3DL2 (profile #17; 24.1%, and 23.7%, respectively). This profile was observed at a frequency of only 5.6% in the North Indians, whose KIR profiles were exceptionally diverse and evenly distributed in general (Rajalingam et al. 2002). The second most common profile in Parsis, 3DL3–2DS2–2DL2–2DL3–2DL1–2DL4–3DL1–3DS1–2DL5–2DS3–2DS5–2DS1–2DS4–3DL2 (profile #15), contained all 15 KIR genes (f = 17.5%), and this profile was also common in the Maharashtrians (11.9%), but was missing from the North Indian sample (Table 2).
The profile 3DL3–2DS2–2DL2–2DL3–2DL1–2DL4–3DL1–2DL5–2DS3–2DS4–3DL2 (profile #18), was observed at an even greater frequency in the Maharashtrians (13.6%), but was observed in only three Parsis (2.2%). This was the most common profile in North Indians (6.9%) and it represents the only clear case in which the Parsis were more similar to the North Indians than to the Maharashtrians. This is the second most common KIR genotype in populations of African origin, including African American, Afro-Caribbean, Guadeloupe Caribbean, and Senegalese (11–17%) (Denis et al. 2005; Du et al. 2007; Norman et al. 2002). KIR3DS1 is missing in this profile and the low frequency of the profile in the Parsis parallels the high frequency of KIR3DS1 in the Parsis relative to that in the other two Indian populations. There was one individual with the profile 3DL3–2DL3–2DL1–2DL4–3DL1–3DS1–2DS4–3DL2 (profile #7), which is identical to the A haplotype except for the presence of 3DS1. This profile has been observed at a frequency of 6% in French, 4.2% in Basque, and 1–2% in Han Chinese, Hispanics, American Caucasians, Finnish, Guadeloupe Caribbean and a population from the island of Reunion (Denis et al. 2005; Du et al. 2007; Jiang et al. 2005; Santin et al. 2006).
LD between pairs of KIR genes in each of the three populations also reflected the endogamy among the Parsis relative to the North Indians. In particular, 38 pairs of KIR genes were in significant positive or negative LD in the Parsi sample, 31 in the Maharashtrian sample, and only 19 in the North Indian sample (Tables 3 and 4; LD pairs that include 2DP1 were excluded from Table 4 because it was not typed in the North Indians). Further, only one of the significant LD pairs in the Maharastrians was unique to that population. There were eight pairs of KIR genes that showed significant LD in the Parsis but not in the Maharashtrians or North Indians (2DL2–3DS1, 2DL3–2DS4, 2DS2–3DS1, 2DS3–2DS1, 2DS3–2DS5 in positive LD; 2DL3–2DS1, 2DL3–2DS5, 2DL3–3DS1 in negative LD). Five of these pairs involved the 2DS1 or 3DS1 genes, which had quite different carrier frequencies in the two populations and can be attributed to genotypes carrying both 3DS1 and 2DS1 (particularly profiles 21, 22, 23, and 26), which are highly represented in the Parsis compared to the other two groups. Interestingly, in the Parsi population, KIR3DS1 was in significant LD with both KIR2DL2 and KIR2DL3, which segregate as alleles of a single locus. This situation was unique to the Parsis and, as far as we know, has not been observed in other populations. The lack of significant LD between KIR2DL3 and KIR2DL1 in the Parsis was also notable, as these genes are closely linked and in strong LD across other populations that have been tested including the Maharashtrians and the North Indians. Like the KIR gene frequency and the KIR profile frequency results, the LD analysis underscores a closer genetic relationship between the Parsis and Maharashtrians than either of these two groups to the North Indians, with the exception of the KIR3DS1 analyses, which clearly distinguish the Parsis.
Discussion
The Parsis of India are of Persian descent (Nanavutty 1997), settling in the Indian province of Gujarat around 900 A.D. and eventually moving to the state of Maharashtra. They follow the Zoroastrianism faith, which was once the official religion of the Persian Empire over 2,000 years ago. Although the Parsis have integrated themselves into Indian society, they do not marry outside of the Parsi community and conversions to the faith are strictly unacceptable. Thus, they have remained a closely knit, endogamous community, although there is evidence of admixture with native Indian groups. The Maharashtrians are natives of the Western Maharashtra region since ancient times, distinguished mainly by the language they speak, Marathi. They are far less homogeneous genetically than their Parsi neighbors and constitute a mixture of populations from the North (North Indians) and South (Dravidians) of India. The North Indians are a particularly heterogeneous group derived from the four northern states of Punjab, Haryana, Utter Pradesh, and Delhi. They are distinguished from the Parsis and Maharashtrians by geographical location, linguistics, cultural, and social activities. Further, since Delhi was the capital city of Mogul and British emperors, North India experienced massive racial migrations and admixture, which is likely to have contributed to the high KIR diversity in the North Indians detected previously (Rajalingam et al. 2002).
In this study, we have compared variation across the KIR locus in a sample of Parsis and Maharashtrians, and contrasted these results with published data available for 72 individuals from North India (Rajalingam et al. 2002). The KIR gene frequencies in Parsis did not differ significantly from those in the Maharashtrians except for KIR3DS1 and KIR2DS1, only the former of which remained significant after correction for multiple tests. However, there were 24 distinct KIR profiles identified in the Maharashtrians and only 19 in the Parsis in spite of the larger Parsi sample size (n = 137 for Parsis; n = 119 for Maharashtrians), emphasizing the limited information provided by gene frequency alone in distinguishing populations at the KIR locus. LD values between pairs of KIR genes also supported greater homogeneity at the KIR locus in the Parsis as compared with the Maharashtrians. In general, however, the Parsis and Maharashtrians do show strong genetic similarity at the KIR locus, a phenomenon that is most clearly substantiated by the observation that KIR profiles distinct to one group or the other were observed in that population only one to three times, with the exception of a profile that was observed eight times in the Maharashtrians and was missing in the Parsis. Interestingly, this latter profile was missing KIR3DS1, a gene found at high frequency in the Parsis. KIR gene profiles and LD analysis emphasize the striking diversity of the KIR locus in North Indians relative to both the Parsis and Maharashtrians. Overall, the results suggest that there is greater variability at the KIR locus in Maharashtrians as compared to Parsis, but that these two populations are more similar to one another than either one is to the North Indians.
KIR frequency data from an Iranian sample has not been reported to date, so it was not possible to determine whether the Parsi KIR frequencies are more similar to Maharashtrians or the descendants of the Persian ancestors from whom both the Parsis and Iranians stem. KIR frequency data from a sample of 120 Arabs, which represents the closest group to the Iranians geographically that has been typed so far (Mahfouz et al. 2006), suggests that Parsis are more similar to Maharashtrians than they are to Arabs. Notably, however, the Parsis have a significantly higher frequency of KIR3DS1 (62%) than do the Maharashtrians (37%), North Indians (39%), and the Arab sample (36%; Table 1 and Mahfouz et al. 2006; Rajalingam et al. 2002).
A study of Y chromosome markers has indicated that the Parsis are genetically closer to Iranians than they are to their geographical neighbors in Pakistan (Qamar et al. 2002), the other primary residence of the extant Parsi population. Conversely, a more recent study of mitochondrial DNA (matrilineal as opposed to the patrilineal Y chromosome markers) showed that Parsis display greater genetic similarity with Gujaratis in India than they do with Iranians (Quintana-Murci et al. 2004). Thus, male ancestors of the extant Parsi population may have admixed with Gujarati females, perhaps early after their migration to India and to a degree that led to the replacement of Persian mitochondrial DNA with that of Indian origin (Quintana-Murci et al. 2004). If so, nuclear DNA of present-day Parsis would be expected to be an admixture of ancestral Parsi and Western Indian background.
Based on HLA allele frequencies, the Parsis living in Maharashtra are more closely related to Parsis living in Karachi, Pakistan than to their geographical Indian neighbors (unpublished data, S. Kulkarni, S. Chhaya, R. Badwe and N. Joshi). In fact, amongst the various world populations analyzed, the Parsis were the only group that was phylogenetically separated from its geographic cluster. Such a separation is unlikely to be observed using a KIR dataset since Parsis are not easily distinguished from the Maharashtrians with regard to the KIR locus. However, the difference in KIR3DS1 frequency among the Parsis, which is the only significant distinction between the Parsis and Maharashtrians at the KIR gene complex, may actually be related to the difference in their HLA-B frequencies relative to other Indian populations. We have recently reported an inverse correlation between activating KIR gene frequencies and that of their HLA class I ligand (or putative ligand) group, especially for KIR3DS1 and HLA-B Bw4-80I (Single et al. 2007). Interestingly, the gene frequency of the Bw4-80I grouping was low in Parsis (11.1%) compared to Maharashtrians and North Indians (21.5% and 29.5%, respectively), corresponding with the higher frequency of KIR3DS1 in Parsis (62.2%) relative to the other two native Indian groups (37% and 39%, respectively; Fig. 2). A similar pattern was observed for KIR2DS1 and its ligand group, HLA-C group 2, particularly between Parsis and North Indians, although the inverse correlation for this receptor–ligand pair across all three groups was not as striking as that for KIR3DS1 and Bw4-80I. In general, these data reflect those involving worldwide frequency correlations between activating KIR and their HLA ligand groupings reported previously (Single et al. 2007) and the high frequency of KIR3DS1 in the Parsis may be a function of the low frequency of HLA-B Bw4-80I in this population.
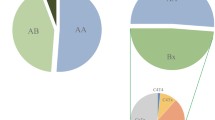
KIR and HLA ligand frequencies—correlations in a global context. Results for the Parsis (P), Maharashtrians (M), and North Indians (NI) are superimposed upon the global distribution reported in Single et al. (2007). There is a strong negative correlation between the carrier frequencies for activating KIR3DS1 and gene frequencies for its putative HLA Bw4 ligand group (a). This correlation is even stronger for the subset of Bw4-80I alleles (b). Differences between the Parsis and the other two populations are most evident for this subset of Bw4 alleles. The activating KIR2DS1 also shows a negative correlation with its C-group2 ligands (c). Each colored circle represents a distinct population, with the color corresponding to the geographic region, and the solid line represents the estimated regression line from Single et al. (2007)
HLA class I does not evolve as quickly as the KIR locus (Hao and Nei 2005; Khakoo and Carrington 2006; Khakoo et al. 2000; Vilches and Parham 2002), so it is possible that the KIR3DS1 locus in the Parsi population is under pressure to retain the high frequency observed in the founding Parsi ancestors such that an appropriate balance between KIR3DS1 and its putative HLA ligand group will be maintained. Although the frequency of KIR3DS1 is significantly higher in the Parsis relative to the Maharashtrians, the frequency of individuals with both KIR3DS1 and HLA-B Bw4-80I is much closer in the two populations (10.2% in the Parsis and 18.2% in the Maharashtrians). Further analyses of additional, historically related populations will be of great interest in elucidating the genetic evolution of the Parsi people since their migration east to India and Pakistan.
Abbreviations
- HLA:
-
human leukocyte antigen
- NK:
-
natural killer
- KIR:
-
killer immunoglobulin-like receptor
- LD:
-
linkage disequilibrium
References
Boyum A (1968) Separation of leukocytes from blood and bone marrow. Introduction. Scand J Clin Lab Invest Suppl 97:7
Carrington M, Norman RJ (2003) The KIR gene cluster. National library of medicine (US), National Centre for Biotechnology Information, Bethesda, MD
Carrington M, Martin MP (2006) The impact of variation at the KIR gene cluster on human disease. Curr Top Microbiol Immunol 298:225–257
Colucci F, Di Santo JP, Leibson PJ (2002) Natural killer cell activation in mice and men: different triggers for similar weapons? Nat Immunol 3:807–813
Cook MA, Moss PA, Briggs DC (2003) The distribution of 13 killer-cell immunoglobulin-like receptor loci in UK blood donors from three ethnic groups. Eur J Immunogenet 30:213–221
Denis L, Sivula J, Gourraud PA, Kerdudou N, Chout R, Ricard C, Moisan JP, Gagne K, Partanen J, Bignon JD (2005) Genetic diversity of KIR natural killer cell markers in populations from France, Guadeloupe, Finland, Senegal and Reunion. Tissue Antigens 66:267–276
Desai AS (2004) Addressing population concerns and community response. FEZANA J 17:31
Du Z, Gjertson DW, Reed EF, Rajalingam R (2007) Receptor–ligand analyses define minimal killer cell Ig-like receptor (KIR) in humans. Immunogenetics 59:1–15
Hao L, Nei M (2005) Rapid expansion of killer cell immunoglobulin-like receptor genes in primates and their coevolution with MHC Class I genes. Gene 347:149–159
Hsu KC, Liu XR, Selvakumar A, Mickelson E, O’Reilly RJ, Dupont B (2002) Killer Ig-like receptor haplotype analysis by gene content: evidence for genomic diversity with a minimum of six basic framework haplotypes, each with multiple subsets. J Immunol 169:5118–5129
Imanishi T, Azaka T, Kimura A et al (1991) Estimation of allele and haplotype frequencies for HLA and complement loci. In: Tsuji K, Aizawa M, Sasazuki T (eds.) Vol 1, HLA:1991 Proceedings of the Eleventh International Histocompatibility Workshop and Conference. Oxford
Jiang K, Zhu FM, Lv QF, Yan LX (2005) Distribution of killer cell immunoglobulin-like receptor genes in the Chinese Han population. Tissue Antigens 65:556–563
Khakoo SI, Carrington M (2006) KIR and disease: a model system or system of models? Immunol Rev 214:186–201
Khakoo SI, Rajalingam R, Shum BP, Weidenbach K, Flodin L, Muir DG, Canavez F, Cooper SL, Valiante NM, Lanier LL, Parham P (2000) Rapid evolution of NK cell receptor systems demonstrated by comparison of chimpanzees and humans. Immunity 12:687–698
Lanier LL (2005) NK cell recognition. Annual Review of Immunology 23:225–274
Mahfouz R, Rayes R, Mahfoud Z, Bazarbachi A, Zaatari G (2006) Distribution of killer cell immunoglobulin-like receptors genotypes in the Lebanese population. Tissue Antigens 68:66–71
Martin MP, Carrington M (2007) KIR Locus Polymorphisms: Genotyping and Disease Association Analysis. In: Ewbank J, Vivier E (eds) Methods in molecular biology: Innate immunity. Humana, Totowa, NJ, pp 49–64
Nanavutty P (1997) The Parsis. National Book Trust, New Delhi
Norman PJ, Stephens HA, Verity DH, Chandanayingyong D, Vaughan RW (2001) Distribution of natural killer cell immunoglobulin-like receptor sequences in three ethnic groups. Immunogenetics 52:195–205
Norman PJ, Carrington CV, Byng M, Maxwell LD, Curran MD, Stephens HA, Chandanayingyong D, Verity DH, Hameed K, Ramdath DD, Vaughan RW (2002) Natural killer cell immunoglobulin-like receptor (KIR) locus profiles in African and South Asian populations. Genes and Immun 3:86–95
Parham P (2005) MHC class I molecules and KIRs in human history, health and survival. Nat Rev Immunol 5:201–214
Qamar R, Ayub Q, Mohyuddin A, Helgason A, Mazhar K, Mansoor A, Zerjal T, Tyler-Smith C, Mehdi SQ (2002) Y-chromosomal DNA variation in Pakistan. Am J Hum Genet 70:1107–1124
Quintana-Murci L, Chaix R, Wells RS, Behar DM, Sayar H, Scozzari R, Rengo C, Al-Zahery N, Semino O, Santachiara-Benerecetti AS, Coppa A, Ayub Q, Mohyuddin A, Tyler-Smith C, Qasim Mehdi S, Torroni A, McElreavey K (2004) Where west meets east: the complex mtDNA landscape of the southwest and Central Asian corridor. Am J Hum Genet 74:827–845
Rajalingam R, Krausa P, Shilling HG, Stein JB, Balamurugan A, McGinnis MD, Cheng NW, Mehra NK, Parham P (2002) Distinctive KIR and HLA diversity in a panel of north Indian Hindus. Immunogenetics 53:1009–1019
Santin I, de Nanclares GP, Calvo B, Gaafar A, Castano L, Bilbao JR (2006) Killer cell immunoglobulin-like receptor (KIR) genes in the Basque population: association study of KIR gene contents with type 1 diabetes mellitus. Hum Immunol 67:118–124
Shilling HG, Guethlein LA, Cheng NW, Gardiner CM, Rodriguez R, Tyan D, Parham P (2002) Allelic polymorphism synergizes with variable gene content to individualize human KIR genotype. J Immunol 168:2307–2315
Single RM, Martin MP, Gao X, Meyer D, Yeager M, Kidd JR, Kidd KK, Carrington M (2007) Global diversity and evidence for coevolution of KIR and HLA. Nat Genet 39:1114–1119
Toneva M, Lepage V, Lafay G, Dulphy N, Busson M, Lester S, Vu-Trien A, Michaylova A, Naumova E, McCluskey J, Charron D (2001) Genomic diversity of natural killer cell receptor genes in three populations. Tissue Antigens 57:358–362
Uhrberg M, Valiante NM, Shum BP, Shilling HG, Lienert-Weidenbach K, Corliss B, Tyan D, Lanier LL, Parham P (1997) Human diversity in killer cell inhibitory receptor genes. Immunity 7:753–763
Velickovic M, Velickovic Z, Dunckley H (2006) Diversity of killer cell immunoglobulin-like receptor genes in Pacific Islands populations. Immunogenetics 58:523–532
Vilches C, Parham P (2002) KIR: diverse, rapidly evolving receptors of innate and adaptive immunity. Annu Rev Immunol 20:217–251
Wallace DM (1987) General methods for isolating and characterizing nucleic acids: Large- and small-scale phenol extractions. Methods Enzymol 152:33–44
Whang DH, Park H, Yoon JA, Park MH (2005) Haplotype analysis of killer cell immunoglobulin-like receptor genes in 77 Korean families. Hum Immunol 66:146–154
Wilson MJ, Torkar M, Haude A, Milne S, Jones T, Sheer D, Beck S, Trowsdale J (2000) Plasticity in the organization and sequences of human KIR/ILT gene families. Proc Natl Acad Sci U S A 97:4778–4783
Yawata M, Yawata N, McQueen KL, Cheng NW, Guethlein LA, Rajalingam R, Shilling HG, Parham P (2002) Predominance of group A KIR haplotypes in Japanese associated with diverse NK cell repertoires of KIR expression. Immunogenetics 54:543–550
Acknowledgements
The authors would like to thank Dr. K. A. Dinshaw, Director, Tata Memorial Centre, Mumbai, Dr. R. Sarin, Director, ACTREC, Navi Mumbai, Dr. B.B. Yeole, Director and Mrs. Lata Shirsat, Medical Social Worker of the Bombay Cancer Registry, Indian Cancer Society, Mumbai, India for their assistance in contacting and recruiting study subjects and Mrs. K. Sadhana, Statistician at Cancer Research Institute, ACTREC, Navi Mumbai, India for helpful discussions. We also wish to thank Dr. Ying Qi, NCI-Frederick for statistical support. This work was supported by a grant from the Lady Tata Memorial Trust. This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract N01-CO-12400. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This Research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Kulkarni, S., Single, R.M., Martin, M.P. et al. Comparison of the rapidly evolving KIR locus in Parsis and natives of India. Immunogenetics 60, 121–129 (2008). https://doi.org/10.1007/s00251-008-0279-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00251-008-0279-1