Abstract
The availability of a contig of bacterial artificial chromosome (BAC) clones spanning the equine major histocompatibility complex (MHC) made possible a detailed analysis of horse MHC class I genes. Prior to this study, only a single horse MHC class I gene had been sequenced at the genomic level. Although many (∼60) MHC class I cDNA sequences had been determined and published, from this information, it was not possible to determine how many class I loci are expressed in horses or to assign individual sequences to allelic series. In this study, 15 MHC class I genes were identified in BAC subclones and fully sequenced. Because the BAC library donor horse had been bred for homozygosity at the MHC, these 15 genomic clones represent distinct MHC class I genes and pseudogenes and not alleles at a smaller number of loci. For five of the genes, cDNA sequences from these loci had previously been identified. Two additional expressed class I genes were discovered, bringing the known total of different equine MHC class I genes (loci) expressed as mRNA to seven. Expression of all seven loci was detected by reverse transcriptase–polymerase chain reaction in adult, fetal, and placental tissues. The remaining eight genes were designated as pseudogenes. This work resulted in moderate expansion of the horse MHC BAC contig length, and the remaining gap was shortened. The information contained in these equine MHC class I sequences will permit comparison of MHC class I genes expressed across different horse MHC haplotypes and between horses and other mammalian species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Major histocompatibility complex (MHC) class I molecules are membrane-bound glycoproteins that complex with a variety of peptides derived from intracellular degradation of endogenous and exogenous proteins. MHC class I molecules complexed with nonself peptides and β-2-microglobulin that are recognized by the T-cell receptor (TCR) on T lymphocytes initiate a specific immune response, thus providing immune surveillance against intracellular pathogens (Germain and Margulies 1993). MHC class I molecules also regulate innate immunity by serving as ligands for inhibitory receptors on natural killer (NK) cells, effecting inhibition of NK-mediated cytolytic activity (Lanier 1997).
Major histocompatibility complex class I molecules of humans and mice are often separated into two main classes: the classical, “Ia,” and the nonclassical, “Ib.” The nonclassical molecules are often distinguished by their shorter cytoplasmic tail and their low levels of polymorphism in the 5′ end (Shawar et al. 1994). The classical genes have many alleles: for HLA-A, 372; for HLA-B, 661; and for HLA-C, 190, whereas the nonclassical genes have fewer: for HLA-E, 5; for HLA-F, only 2; and for HLA-G, 15 (as of April 2005, http://www.ebi.ac.uk/imgt/hla/intro.html).
Early studies indicated the presence of at least 16, and up to 33, MHC class I genes in the equine genome (Vaiman et al. 1986; Alexander et al. 1987; Guerin et al. 1987). This estimate includes both expressed genes and pseudogenes, and the total number of bands varied between horses. The horse MHC was localized to horse chromosome 20q14–q22 by in situ hybridization (Ansari et al. 1988) and was confirmed to position 20q21 with fluorescent in situ hybridization (FISH) of bacterial artificial chromosome (BAC) clones containing MHC class I genes (Gustafson et al. 2003).
Variation in horse MHC class I genes was initially assessed serologically rather than by molecular and sequence methods. Approximately 15 distinct specificities can be discerned, and the designated nomenclature identifies them as ELA-A1, ELA-A2, ELA-A3, ..., ELA-A19 (Lazary et al. 1988). These are considered to be markers of MHC haplotypes, implying a distinctive collection of inherited alleles.
Several studies have described horse MHC class I cDNA sequences from horses of different ELA haplotypes, based on either cDNA library screening or reverse transcriptase–polymerase chain reaction (RT-PCR) experiments (Barbis et al. 1994; Ellis et al. 1995; Chung et al. 2003; McGuire et al. 2003). The full-length MHC class I gene sequences reported here may provide the genomic equivalent of some of the cDNA sequences obtained by Ellis et al. (1995) from an ELA-A3/ELA-A7 pony because of the shared ELA-A3 haplotype. Ellis and coworkers reported seven expressed MHC class I transcripts from this MHC heterozygous pony. ECMHCB1 through ECMHCB4 are considered classical MHC class I genes, and the remaining three transcripts, ECMHCA1, ECMHCC1, and ECMHCE1 are considered putative nonclassical genes (Holmes and Ellis 1999).
The present study was fueled by the combination of the genomic characterization of a classical MHC class I gene of the horse (Carpenter et al. 2001) and the construction of a BAC clone contig spanning the horse MHC (Gustafson et al. 2003). We present genomic sequence for 15 MHC class I genes of the horse from the ELA-A3 haplotype. Several correspond to previously published genes and provide a noncoding and flanking, extragenic sequence. The others are newly identified genes, some of which are expressed, and others which are pseudogenes. In total, we detected mRNA expression of seven distinct MHC class I genes from an MHC homozygous individual. Sequence variations in the promoter region regulatory elements are presented, comparing classical and nonclassical loci, as well as pseudogenes. This collection of data can be compared to MHC class I gene sequences from other equine MHC haplotypes and from other species.
Materials and methods
Animals, tissue samples, and cDNA preparation
The horses sampled for this study were from either the Equine Genetics Center herd at Cornell University or from local farms in the Ithaca, NY area. Peripheral blood lymphocytes were isolated from heparinized samples of venous jugular blood using methods described previously (Antczak et al. 1982). Conceptus tissues were recovered by nonsurgical uterine lavage from lightly sedated mares between days 33 and 34 of pregnancy. Conceptus tissues were dissected under a stereoscopic microscope (Nikon Instrument Group, Melville, NY) at 150X magnification and snap-frozen in liquid nitrogen. RNA was isolated from lymphocytes and conceptus tissues following homogenization by QIAshredder (Qiagen, Valencia, CA) as directed by the RNeasy kit (Qiagen). One microgram of RNA was treated with DNase I (Invitrogen, Carlsbad, CA) to degrade any contaminating genomic DNA. cDNA synthesis reactions were primed with oligodT (PE Biosystems, Foster City, CA) and transcribed by Moloney Murine Leukemia Virus Reverse Transcriptase (M-MLV RT, USB, Cleveland, OH).
Horse bacterial artificial chromosome library and screening
The CHORI-241 BAC library was constructed at the Children's Hospital Research Institute, Oakland, CA, using neutrophil DNA from a Thoroughbred stallion in the Cornell herd bred to be homozygous for known MHC region genes. The CHORI-241 horse BAC library was screened by hybridization with an MHC class I overgo probe as previously described (Gustafson et al. 2003). Additional overgo probes were generated from other genes in the MHC class I region, BAC end sequences, or subclones derived from BAC clones (see Table 1). The sequences were analyzed by RepeatMasker for the presence of repetitive elements (A.F.A. Smit and P. Green, unpublished data, http://repeatmasker.genome.washington.edu/cgi-bin/RepeatMasker). Overgoes were then designed from nonrepetitive sequence by Overgo Maker http://genome.wustl.edu/tools/?overgo=1-input). Overgoes were synthesized by Integrated DNA Technologies (Coralville, IA) http://www.idtdna.com).
Isolation of BAC DNA was performed using Plasmid Midi kits (Qiagen). For sequencing, 2 μg of BAC DNA was digested with BamHI at 37°C overnight. The enzyme was heat-inactivated, and the BAC DNA was sequenced with 40 pmol of primer.
Southern blot hybridization
Bacterial artificial chromosome clones from the published equine MHC class I region (Gustafson et al. 2003) and additional BAC clones identified previously in this laboratory were subjected to Southern blot hybridization with an MHC class I probe to determine the number and location of MHC class I genes.
For Southern blot analysis, 2–5 μg BAC DNA was digested with EcoRI, BamHI or HindIII restriction enzymes at 37°C overnight. The digested BAC DNA was run on a 1% Tris borate ethylenediaminetetraacetic acid (EDTA) (TBE) gel for 24–44 h at 45 V and then visualized. The BAC DNA was transferred to a nylon membrane (Hybond-N, Amersham Biosciences, Piscataway, NJ) in alkaline transfer buffer (1.5 M NaCl and 0.25 M NaOH) by capillary action overnight. The membrane was then neutralized (0.5 M EDTA, 1 M Na2HPO4, pH 7.4) and hybridized as described above.
Subcloning and sequencing of MHC class I genes from BAC clones
Restriction fragments containing horse MHC class I genes identified by Southern blot hybridization were selected for subcloning. Insert DNA was purified with the QIAEX II gel extraction kit (Qiagen). Ligation into a modified pCR2 vector was performed at a ten insert to one vector ratio with T4 Ligase (Invitrogen) and incubated overnight at 15°C. Transformation of 2–5 μl of ligation reaction into One Shot TOP10 chemically competent Escherichia coli was performed according to supplied instructions (Invitrogen). Colonies were grown at 37°C overnight and were screened by colony blot hybridization to identify those containing MHC class I genes. Methods were similar to those described above. Positive colonies were grown, and plasmids were purified with the QIAprep Spin MiniPrep Kit (Qiagen) and analyzed by restriction digest, PCR, or sequencing. Full sequencing was performed with universal vector primers M13F and M13R, MHC class I gene-specific primers, and primer walking.
Polymerase chain reaction amplification and sequencing
To determine mRNA expression for potentially expressed genes, locus-specific primers were designed (Table 1) within the hypervariable region when possible and used in RT-PCR experiments. Oligonucleotide primers were designed with Primer3 on the Web at: http://www.genome.wi.mi.edu/cgi-bin/primer/prime3.cgi and synthesized by Integrated DNA Technologies. Amplification of 50 ng cDNA or genomic DNA was performed either with Taq (Invitrogen) or Pfu DNA polymerase (Stratagene, La Jolla, CA). Amplification with Taq DNA polymerase was performed in a Robocycler 96 (Stratagene) with the following conditions: 1 cycle at 95°C for 5 min, 33 cycles of 95°C for 30 seconds, 55°C for 30 s, and 72°C for 1 min, and 1 cycle of 72°C for 10 min. Amplification with Pfu DNA polymerase was performed with the following conditions: 1 cycle at 95°C for 45 s, 33 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 2 min, and 1 cycle of 72°C for 10 min. Products were separated by electrophoresis on 1% agarose gels, stained with ethidium bromide, and visualized under a UV Transilluminator (UVP, Upland, CA).
Polymerase chain reaction products were either purified and directly sequenced or cloned. QIAquick kits (Qiagen) were used for PCR product purification. The pGEM-T Easy vector was used for TA-cloning Taq DNA polymerase products as described in the cloning kit (Promega, Madison, WI). The pCR 4Blunt-TOPO vector was used for cloning Pfu DNA polymerase products as described in the Zero Blunt TOPO PCR cloning kit (Invitrogen).
Sequencing was performed by the Biotechnology Resource Center at Cornell University, Ithaca, NY, with an ABI 3700 automated sequencer (PerkinElmer, Foster City, CA). Templates (PCR products or plasmids) were sequenced at least once in each direction. All sequences generated for this work have been submitted to GenBank.
Sequence analysis: subclone sequences, MHC class I genes, and RT-PCR products
Sequencing electropherograms were viewed with EditView 1.0.1 software from Applied Biosystems (Foster City, CA). Sequences were analyzed with VectorNTI (Invitrogen). Alignments and sequence identity tables were generated by MegAlign from the LaserGene package (DNASTAR, Inc., Madison, WI). Promoter elements were identified by comparison with the previously published horse ELA-A2 gene promoter (Carpenter et al. 2001).
Fluorescent in situ hybridization
Metaphase chromosome preparation and FISH were performed as previously described (Lear et al. 1998).
Results
Physical map of MHC class I genes
Bacterial artificial chromosome (BAC) clones from the published equine MHC class I region contig (Gustafson et al. 2003) were digested with restriction enzymes and subjected to Southern blot hybridization with an MHC class I probe to define the location and restriction fragment size of the horse MHC class I genes. Fifteen horse MHC class I genes were identified in this way, and because the donor horse of the BAC library is an ELA-A3 MHC homozygote, these genes represent 15 distinct loci rather than alleles at a smaller number of loci.
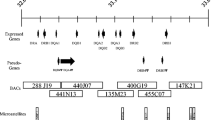
Three regions containing MHC class I genes were found, designated A, B, and C, from left to right (Fig. 1). Region A is at the BTNL2 end of the MHC class II contig and is contained within one BAC clone. This BAC clone was not included in the initial characterization of the horse MHC BAC contig but was mapped in close proximity to the BTNL2 gene by BAC end-sequence hybridization in this study, extending into the gap described by Gustafson et al. (2003). Two MHC class I genes are located in region A.
Supplemented minimal tiling path of the BAC contig of the equine MHC. Genes are shown in sequential order, as determined by hybridization. MHC class I gene clusters are shown as ovals along the top, and new BAC clones are shown as open boxes in the contig. MHC class I gene subclones are listed under the corresponding cluster. Genes are spaced equally because distance between genes has not been determined. / indicates a gap between the MHC class II and III regions. Chromosomal orientation as determined by FISH is indicated by CEN (centromeric) at the MHC class I end. Refer to Table 2 for subclone information
Region B is at the junction of the classes III and I regions, between the BAT1 and MIC genes, and is comprised of five overlapping BAC clones. Four of these clones close a previously undetected gap in the horse MHC contig. This was further supported by FISH mapping of BAC clones 41M02, 69N18, and 101I16, placing them all within the established MHC region (data not shown). Establishing this supplemental minimal tiling path was difficult because of the high prevalence of repetitive elements in BAC end sequences (54%) as identified by RepeatMasker analysis (data not shown). The major repetitive elements included long interspersed nucleotide elements (LINE), long terminal repeats (LTR), DNA/MER repeats, and short interspersed nucleotide elements (SINE). These repetitive elements are also present in the human MHC sequence, although the SINE families differ between species. Six MHC class I genes were found in this region.
Region C is in the class I region between the GNL1 and TRIM26 genes and spans four overlapping BAC clones. Seven MHC class I genes are located in this region.
Genomic structure of horse MHC class I genes
Sequences were obtained for all 15 of the identified horse MHC class I genes. MHC class I genes consist of eight exons and extend approximately 4,000 bases (Steinmetz et al. 1981; Moore et al. 1982). Exon 1 encodes the leader sequence; exons 2, 3, and 4 encode the alpha 1, 2, and 3 domains, respectively. The transmembrane-spanning domain is encoded by exon 5; exons 6, 7, and 8 encode the cytoplasmic tail. The previously identified horse MHC class I genomic sequence of the ELA-A2 gene confirmed that this genomic structure is conserved in the horse (Carpenter et al. 2001), and this sequence was used as a guide to identify intron/exon boundaries of the new class I genes. Overall, the length of each exon or intron was highly conserved among the 15 class I genes (Fig. 2).
Comparison of exon and intron lengths of horse MHC class I genes. Nucleotide length of exons, introns, and total genes listed. Expressed genes are listed on top, pseudogenes are listed below. Asterisk (*) indicates previously published cDNA sequence exists for this gene, ND indicates not detected, NS indicates not sequenced, and Underline indicates region was truncated in subclone
The coding region of the 15 new sequences were compared to cDNA or expressed sequence tag (EST) sequences deposited previously into the GenBank (Ellis et al. 1995; McGuire et al. 2003). Six of the new MHC class I genes were previously characterized at the cDNA level, and two were sequenced as ESTs (Table 2). The remaining unidentified genes were analyzed for potential expression by determining and translating the putative coding sequences. The genes can be assigned to one of two categories: (a) genes encoding a transmembrane length of 35 amino acids which are considered to be members of highly polymorphic series and therefore classical MHC class I genes, or (b) genes encoding longer transmembrane domains (36–40 amino acids) and that tend to be either monomorphic or oligomorphic and are classified as putative nonclassical MHC class I genes (Holmes and Ellis 1999). Eight MHC class I gene subclones contained a functional coding sequence with appropriate intron/exon boundaries, splicing sequences, and stop codons. Four of these encoded molecules with a transmembrane length of 35 amino acids are therefore considered classical MHC class I genes; the other four genes encoded a longer transmembrane domain and are putative nonclassical MHC class I genes.
Each subclone was assigned a number with the prefix 3, indicating the equine ELA-A3 MHC haplotype carried by the MHC homozygous horse from which the BAC library was constructed, followed by a period and a sequential number. Table 2 provides a description of each subclone.
Classical MHC class I gene subclones
Subclone 3.1 contains the genomic equivalent of the ECMHCB2 cDNA sequence (X79891). Alignment of the previously published cDNA with the exons from the genomic sequence is in 100% agreement (deduced amino acid alignment shown in Fig. 3). Subclone 3.2 provides the genomic sequence for the 8-5 transcript (AY225157). Alignment of the previously published cDNA with the exons from the genomic sequence has a 97.7% identity (deduced amino acid alignment shown in Fig. 3). Nucleotide differences between these sequences are found in exons 1 through 4. The 8-5 sequence was originally identified from a horse of the A1/A4 haplotype (McGuire et al. 2003). Subclones 3.3 and 3.4 both contained novel and potentially functional coding sequence, with a transmembrane length indicative of classical loci. All four subclones, 3.1–3.4, are located in region C (Fig. 1).
Predicted amino acid sequence alignment of expressed horse MHC class I genes from the ELA-A3 haplotype. Predicted amino acid alignment of MHC class I gene sequences determined in this study. Dots (.) indicate identity, dashes (-) indicate gaps inserted to optimize alignment, asterisks (*) indicate stop codon, and vertical bars (|) indicate exon/intron boundaries
Nonclassical MHC class I gene subclones
Subclone 3.5 contains the genomic equivalent of the ECMHCA1 cDNA sequence (X71809) and is located in region C. Alignment of the previously published cDNA with the exons from the genomic sequence is in 100% agreement (deduced amino acid alignment shown in Fig. 3). Subclone 3.6 contains the full-length gene encoding the ECMHCC1 cDNA (X79893) and is located in region B. Alignment of the previously published cDNA with the exons from the genomic sequence is also in 100% agreement (deduced amino acid alignment shown in Fig. 3). Subclone 3.7 contains the genomic sequence of the ECMHCE1 cDNA (X79894) and is also located in region B. Alignment of the previously published cDNA with the exons from the genomic sequence differs by 3% and provides exon 1 (deduced amino acid alignment shown in Fig. 3). The differences between the coding sequences occur in exons 1, 2, 3, and 7, leaving the characteristic transmembrane domain (exon 5) unchanged. In total, there are 32 nonsynonymous positions that translate into 22 amino acid changes. However, the genomic sequence identified here matches two GenBank sequences from an independent cDNA library (CD465113 and CD467042), with only one nucleotide difference. Therefore, we suggest that the new sequence is a second allele at the ECMHCE1 locus.
Pseudogenes
Pseudogenes were identified by either the presence of one or more premature stop codons, the lack of a stop codon, or lack of mRNA expression in lymphocytes and conceptus tissues. Five of the horse MHC class I genes were determined to be pseudogenes by either the presence of one or more premature stop codons or the lack of a stop codon (Table 2). Only one pseudogene subclone, 3.12, lacked entire exons. MHC class I genes contained in subclones 3.9, 3.14, and 3.15 encoded stop codons in exons before the transmembrane region (exon 5), which would presumably disrupt the conformation of any resulting proteins, and were categorized as pseudogenes without further analysis. Subclone 3.13 lacked a stop codon and provided genomic sequence for a pseudogene that is expressed as mRNA, but not protein, as described previously for the 7-7 sequence (AY225156) (McGuire et al. 2003). Subclones containing MHC class I genes that encoded premature stop codons but could produce soluble or truncated cell-surface proteins (3.10 and 3.11) were assayed for expression by RT-PCR.
Expression of MHC class I genes
Five of the eight potentially expressed horse MHC class I genes have been previously characterized at the cDNA level. To investigate the expression of all eight class I genes, locus-specific primers were used in RT-PCR experiments (Table 1). Nearly all primers were designed within the hypervariable regions for optimal specificity. We assayed cDNA from four tissues, all of the ELA-A3 haplotype: (1) adult lymphocytes from four horses, including the BAC library donor, (2) day 33 invasive trophoblast (chorionic girdle), (3) day 33 noninvasive trophoblast (allantochorion), and (4) day 33 fetus. Seven of the eight genes, 3.1 through 3.7, were expressed in all four tissues tested (adult lymphocyte gel shown in Fig. 4). RT-PCR products were purified and directly sequenced, confirming that a single gene was amplified. Further, all sequences were identical to the expected ELA-A3 coding sequences. No evidence of mRNA expression was found for the eighth gene, 3.8, in either lymphocytes or conceptus tissues. Although the full MHC class I gene sequence was not determined for the 3.8 gene, it is designated as a pseudogene in this haplotype based on the lack of mRNA expression in these tissues.
RT-PCR analysis of horse MHC class I gene mRNA expression. Peripheral blood lymphocytes express mRNA of seven distinct MHC class I genes as determined by locus-specific RT-PCR. Primers listed in Table 1. Lane 1 indicates 100-bp ladder (top band 600 b); Lane 2, 3.1 (283 b); Lane 3, 3.2 (293 b); Lane 4, 3.3 (416 b); Lane 5, 3.4 (306 b); Lane 6, 3.5 (210 b); Lane 7, 3.6 (413 b); Lane 8, 3.7 (301 b); Lane 9, 3.10 (381 b), and Lane 10, no template control
Specific primers were also designed for RT-PCR analysis of class I genes with premature stop codons that could produce soluble or truncated cell-surface proteins. The MHC class I gene contained in subclone 3.10 encoded two stop codons in the transmembrane region (exon 5). The MHC class I gene contained in subclone 3.11 encoded a stop codon in exon 6 and lacked a stop codon in exon 8, and an EST match was found in GenBank (CX603790). No mRNA expression was detected from peripheral blood lymphocytes or conceptus tissues for these genes (3.10 result shown in Fig. 4), but both primer sets produced the expected product when tested on the respective genomic subclone. Accordingly, class I genes 3.10 and 3.11 are designated as pseudogenes in this haplotype.
Previous transfection studies confirmed the protein expression of 3.1, 3.2, 3.5, and 3.6 (D.F. Antczak and S. Ellis, unpublished; Holmes and Ellis 1999; McGuire et al. 2003).
In summary, subclones 3.1 through 3.4 contain expressed classical loci, subclones 3.5 through 3.7 contain expressed putative nonclassical loci, and subclones 3.8 through 3.15 contain pseudogenes in this haplotype.
MHC class I gene regulatory element sequence analysis
Eight regulatory elements have been described in the 5′ upstream sequence of MHC class I genes, and they are conserved in multiple species (Howcroft et al. 2003; van den Elsen et al. 2004), including the horse (Carpenter et al. 2001). At least three and up to eight of these regulatory elements were present in 11 of the horse MHC class I gene subclones (Table 3). The order and spacing of the regulatory elements is conserved among all 11 genes.
High sequence conservation of the listed elements is evident for all of the expressed MHC class I loci, although a few nucleotide substitutions occur in each regulatory element. Some variations are shared among all expressed loci, such as the T to C change in the X2 box, and some are unique to a single locus. It is not possible to determine at this time whether any of these variations manifest locus specificity, as sequences from alleles of other haplotypes are necessary for this analysis. Varying levels of sequence conservation are found in these elements among the horse MHC class I pseudogenes. Pseudogenes 3.9, 3.11, and 3.15 have retained recognizable elements, and notably, the 3.11 gene is transcribed (Table 2). In contrast, the 3.14 gene only has one detectable element.
Major histocompatibiliy complex class I expression on the cell surface is dependent on stabilization from β-2-microglobulin (Zijlstra et al. 1990), thereby requiring coordinate regulation of these genes. A comparison of the regulatory elements contained in the promoter of the horse MHC class I gene ELA-A2 and the horse β-2-microglobulin is shown in Table 3, as previously published (Tallmadge et al. 2003). High sequence conservation is detected in all of the elements also found in the β-2-microglobulin promoter, with exceptions of sequence variation in the interferon-stimulated response element (ISRE), and the lack of the CCAAT box.
Finally, the sequence from the human allele HLA-A2 (L36528) is included for comparison and shows very high sequence conservation of MHC class I gene regulatory elements between these species.
Discussion
This study describes the physical map locations and sequences of 15 equine MHC class I genes. BAC clones from the published MHC class I region contig (Gustafson et al. 2003) served as the template for subcloning individual class I genes. The horse class I genes were located in three clusters: (A) at the BTNL2 end of the class II region, (B) at the junction of the class III and I regions, and (C) between the GNL1 and TRIM26 genes in the class I region. The BAC clone containing region A shortened the gap between the MHC class II and III regions, and the BAC clones placed in region B lengthened the original horse MHC BAC contig. It had been noted previously that most mammals have at least three distinct class I duplication blocks (Kulski et al. 2002). However, all three blocks are generally found within the class I region. Horse class I regions B and C correspond to the defined beta (β) and kappa (κ) duplication blocks (between BAT1 and POU5F1, and between GNL1 and TRIM26, respectively) (Kulski et al. 2000). The specific MHC class I gene content within these regions varies between human and horse. The human classical genes HLA-B and HLA-C and pseudogenes HLA-S and HLA-X are found in the β block, while the horse β block contains two putative nonclassical genes and four pseudogenes. In the κ block, one human nonclassical gene is found, HLA-E, and two pseudogenes, HLA-L and HLA-N, while in the horse, all four classical genes, one putative nonclassical gene, and two pseudogenes are found. In fact, the organization of the horse MHC class I genes is more similar to that of the pig than the human, with the exception of horse class I region A (Velten et al. 1999).
Region A is unique to the horse MHC when compared to the human or pig MHC map. Although both mice and rats encode MHC class I genes within the MHC class II genomic region (Kulski et al. 2002), the placements are not similar to that found in the horse. Horse class I region A is outside of the class II region, next to the BTNL2 gene, while the rodent class I genes map between the SACM2L and RING1 genes within the class II region. The current study describes the MHC class I physical map for the ELA-A3 haplotype only. Other ELA haplotypes may have differences in MHC class I gene content as has been reported in other species (Wroblewski et al. 1994; Ellis et al. 1999).
The horse MHC class I gene sequences characterized in this study include the genomic equivalents of previously published horse MHC class I cDNA sequences and newly identified genes and pseudogenes. This work confirms the previously published cDNA sequences and provides sequences of introns, untranslated regions, and extragenic-flanking regions. It also provides evidence for horse MHC class I mRNA expression in four tissues, indicating that at least seven different loci are expressed by horses carrying the ELA-A3 haplotype of the BAC library donor horse. We did not find a gene encoding an expressed soluble MHC class I (ESCI) gene matching the one described previously (Lew et al. 1986). The horse ESCI molecule is analogous to the mouse Q10 molecule (Lew et al. 1986), which is produced exclusively in the liver and yolk sac (David-Watine et al. 1990). Because these tissues were not included in this study, and because not all horses express soluble MHC class I molecules, it is not known whether the donor stallion expresses a soluble class I gene.
The total number of expressed MHC class I loci in horses may be higher than the seven identified thus far in the ELA-A3 haplotype. While this total of seven expressed class I loci exceeds that of human and mouse (six loci each), the rhesus macaque expresses at least nine and up to 22 class I genes (Daza-Vamenta et al. 2004). The optimal number of expressed MHC class I and II genes has been suggested to be 12, as the expression of additional MHC genes would deplete the T-cell receptor repertoire excessively (Takahata 1995).
The regulatory elements identified in the expressed MHC class I genes are likely to be functional as demonstrated by ELA-A2 reporter construct (Carpenter et al. 2001). For several class I subclones, a large amount of flanking sequence in the 5′ upstream region was obtained. This may be useful in the identification of additional regulatory elements, especially those known to be more than 1,000 bases upstream (Mavria et al. 1998). Introns have also been shown to harbor regulatory elements (Drezen et al. 1995), which could also be investigated using these subclones.
Differential regulation between the classical and putative nonclassical loci can also be pursued with the sequences provided. For example, the nonclassical HLA-G gene is regulated by different transcriptional control mechanisms than the other HLA class I genes (Gobin and van den Elsen 1999). Also, the tissue distribution of MHC class I nonclassical genes remains to be thoroughly studied in the horse. In humans, nonclassical MHC class I loci have a restricted tissue distribution and lower levels of expression, similar to the H2-Q10 gene of the mouse (David-Watine et al. 1990). In contrast, the H2-Qa2 gene in the mouse and the nonclassical genes in the pig, SLA-6, SLA-7, and SLA-8, have a wide tissue distribution at the transcriptional level (Ungchusri et al. 2001; Crew et al. 2004). Sequences for both classical and putative nonclassical loci have been detected in both horse lymphocytes and trophoblast (this study and Bacon et al. 2002), but additional somatic tissues have not yet been tested. At this time, the protein expression and function of the putative nonclassical genes of the horse is not known and warrants further investigation.
Previously, at least two classical MHC class I loci and the nonclassical loci ECMHCA1 (3.5) and ECMHCC1 (3.6) were identified in the invasive horse trophoblast of the chorionic girdle that expresses high levels of cell-surface MHC class I protein (Donaldson et al. 1990; Bacon et al. 2002). In this study, we detected two additional classical loci and the third putative nonclassical locus (ECMHCE1, 3.7) in the invasive trophoblast and all seven MHC class I genes in the noninvasive trophoblast of the allantochorion. The allantochorion trophoblast expresses only about one tenth of the mRNA level of the chorionic girdle (Bacon et al. 2002) and has little to no detectable MHC class I protein expression (Donaldson et al. 1990).
We designated MHC class I genes 3.8–3.15 as pseudogenes in this haplotype based either on inappropriate placement of stop codons, lack of mRNA expression in lymphocytes and conceptus tissues, or identity to a previously described pseudogene. However, it is possible that genes 3.8, 3.10, and 3.11 are not pseudogenes but have a restricted tissue distribution that does not encompass the tissues included in this study. This can explain the mRNA expression of 3.11 in a horse cartilage cDNA library (CX603790; J. MacLeod, University of Kentucky) and lack of expression in the tissues assayed in this study.
Common regulatory elements were detected for horse MHC class I and β-2-m genes (Enhancer A/NF-κB, ISRE, S, X1, X2, Enhancer B/Y, and TATA box), which may effect coordinate transcriptional control (Tallmadge et al. 2003). Both genes, however, have additional separate regulatory elements that might allow for disparate expression (MHC class I, NFκB2 site of Enhancer A and CCAAT; β-2-m, PAM). This information will facilitate further studies on the regulation of MHC class I and β-2-microglobulin genes in the horse.
While this study provides the sequence of 15 MHC class I genes from the ELA-A3 haplotype, further studies including multiple haplotypes are necessary to make intralocus comparisons. With additional sequence information, molecular typing methods may be devised for horse MHC class I genes. Despite the clarity of locus-specific residues in human and mouse MHC class I genes (Koller et al. 1984; Parham et al. 1989; Pullen et al. 1992; Cereb and Yang 1994; Cereb et al. 1995), such indicators have not been found in cattle or horse MHC class I genes (Ellis et al. 1995, 1999). Further, the number of loci varies among haplotypes of other species, including mice, cattle, and possibly, horse (Lew et al. 1986; Wroblewski et al. 1994; Ellis et al. 1999). The flanking extragenic sequence obtained from the subclones has the potential to serve as a unique marker, which may be useful for mapping MHC class I loci between haplotypes. A more complete understanding of the regulation of MHC class I genes may have applications in equine medicine in areas such as immunity to infectious diseases, pregnancy, autoimmunity and allergies, where the state of expression of MHC molecules is crucial.
References
Alexander AJ, Bailey E, Woodward JG (1987) Analysis of the equine lymphocyte antigen system by Southern blot hybridization. Immunogenetics 25(1):47–54
Ansari HA, Hediger R, Fries R, Stranzinger G (1988) Chromosomal localization of the major histocompatibility complex of the horse (ELA) by in situ hybridization. Immunogenetics 28(5):362–364
Antczak DF, Bright SM, Remick LH, Bauman BE (1982) Lymphocyte alloantigens of the horse. I. Serologic and genetic studies. Tissue Antigens 20(3):172–187
Bacon SJ, Ellis SA, Antczak DF (2002) Control of expression of major histocompatibility complex genes in horse trophoblast. Biol Reprod 66(6):1612–1620
Barbis DP, Maher JK, Stanek J, Klaunberg BA, Antczak DF (1994) Horse cDNA clones encoding two MHC class I genes. Immunogenetics 40(2):163
Carpenter S, Baker JM, Bacon SJ, Hopman T, Maher J, Ellis SA, Antczak DF (2001) Molecular and functional characterization of genes encoding horse MHC class I antigens. Immunogenetics 53(9):802–809
Cereb N, Yang SY (1994) The regulatory complex of HLA class I promoters exhibits locus-specific conservation with limited allelic variation. J Immunol 152(8):3873–3883
Cereb N, Maye P, Lee S, Kong Y, Yang SY (1995) Locus-specific amplification of HLA class I genes from genomic DNA: locus-specific sequences in the first and third introns of HLA-A, -B, and -C alleles. Tissue Antigens 45(1):1–11
Chung C, Leib SR, Fraser DG, Ellis SA, McGuire TC (2003) Novel classical MHC class I alleles identified in horses by sequencing clones of reverse transcription–PCR products. Eur J Immunogenet 30(6):387–396
Crew MD, Phanavanh B, Garcia-Borges CN (2004) Sequence and mRNA expression of nonclassical SLA class I genes SLA-7 and SLA-8. Immunogenetics 56(2):111–114
David-Watine B, Logeat F, Israel A, Kourilsky P (1990) Regulatory elements involved in the liver-specific expression of the mouse MHC class I Q10 gene: characterization of a new TATA-binding factor. Int Immunol 2(10):981–993
Daza-Vamenta R, Glusman G, Rowen L, Guthrie B, Geraghty DE (2004) Genetic divergence of the rhesus macaque major histocompatibility complex. Genome Res 14(8):1501–1515
Donaldson WL, Zhang CH, Oriol JG, Antczak DF (1990) Invasive equine trophoblast expresses conventional class I major histocompatibility complex antigens. Development 110(1):63–71
Drezen JM, Cohen-Tannoudji M, Pournin S, Babinet C, Morello D (1995) Developmental expression of H-2K major histocompatibility complex class I transgenes requires the presence of proximal introns. Dev Dyn 204(1):98–105
Ellis SA, Martin AJ, Holmes EC, Morrison WI (1995) At least four MHC class I genes are transcribed in the horse: phylogenetic analysis suggests an unusual evolutionary history for the MHC in this species. Eur J Immunogenet 22(3):249–260
Ellis SA, Holmes EC, Staines KA, Smith KB, Stear MJ, McKeever DJ, MacHugh ND, Morrison WI (1999) Variation in the number of expressed MHC genes in different cattle class I haplotypes. Immunogenetics 50(5–6):319–328
Germain RN, Margulies DH (1993) The biochemistry and cell biology of antigen processing and presentation. Annu Rev Immunol 11:403–450
Gobin SJ, van den Elsen PJ (1999) The regulation of HLA class I expression: is HLA-G the odd one out? Semin Cancer Biol 9(1):55–59
Guerin G, Bertaud M, Chardon P, Geffrotin C, Vaiman M, Cohen D (1987) Molecular genetic analysis of the major histocompatibility complex in an ELA typed horse family. Anim Genet 18(4):323–336
Gustafson AL, Tallmadge RL, Ramlachan N, Miller D, Bird H, Antczak DF, Raudsepp T, Chowdhary BP, Skow LC (2003) An ordered BAC contig map of the equine major histocompatibility complex. Cytogenet Genome Res 102(1–4):189–195
Holmes EC, Ellis SA (1999) Evolutionary history of MHC class I genes in the mammalian order Perissodactyla. J Mol Evol 49(3):316–324
Howcroft TK, Raval A, Weissman JD, Gegonne A, Singer DS (2003) Distinct transcriptional pathways regulate basal and activated major histocompatibility complex class I expression. Mol Cell Biol 23(10):3377–3391
Koller BH, Sidwell B, DeMars R, Orr HT (1984) Isolation of HLA locus-specific DNA probes from the 3′-untranslated region. Proc Natl Acad Sci U S A 81(16):5175–5178
Kulski JK, Gaudieri S, Dawkins RL (2000) Using alu J elements as molecular clocks to trace the evolutionary relationships between duplicated HLA class I genomic segments. J Mol Evol 50(6):510–519
Kulski JK, Shiina T, Anzai T, Kohara S, Inoko H (2002) Comparative genomic analysis of the MHC: the evolution of class I duplication blocks, diversity and complexity from shark to man. Immunol Rev 190:95–122
Lanier LL (1997) Natural killer cell receptors and MHC class I interactions. Curr Opin Immunol 9(1):126–131
Lazary S, Antczak DF, Bailey E, Bell TK, Bernoco D, Byrns G, McClure JJ (1988) Joint Report of the Fifth International Workshop on Lymphocyte Alloantigens of the Horse, Baton Rouge, Louisiana, 31 October–1 November 1987. Anim Genet 19(4):447–456
Lear TL, Breen M, Ponce de Leon FA, Coogle L, Ferguson EM, Chambers TM, Bailey E (1998) Cloning and chromosomal localization of MX1 and ETS2 to chromosome 26 of the horse (Equus caballus). Chromosome Res 6(4):333–335
Lew AM, Bailey E, Valas RB, Coligan J (1986) The gene encoding the equine soluble class I molecule is linked to the horse MHC. Immunogenetics 24(2):128–130
Mavria G, Hall KT, Jones RA, Blair GE (1998) Transcriptional regulation of MHC class I gene expression in rat oligodendrocytes. Biochem J 330(Pt 1):155–161
McGuire TC, Leib SR, Mealey RH, Fraser DG, Prieur DJ (2003) Presentation and binding affinity of equine infectious anemia virus CTL envelope and matrix protein epitopes by an expressed equine classical MHC class I molecule. J Immunol 171(4):1984–1993
Moore KW, Sher BT, Sun YH, Eakle KA, Hood L (1982) DNA sequence of a gene encoding a BALB/c mouse Ld transplantation antigen. Science 215(4533):679–682
Parham P, Lawlor DA, Lomen CE, Ennis PD (1989) Diversity and diversification of HLA-A, B, C alleles. J Immunol 142(11):3937–3950
Pullen JK, Horton RM, Cai ZL, Pease LR (1992) Structural diversity of the classical H-2 genes: K, D, and L. J Immunol 148(3):953–967
Shawar SM, Vyas JM, Rodgers JR, Rich RR (1994) Antigen presentation by major histocompatibility complex class I-B molecules. Annu Rev Immunol 12:839–880
Steinmetz M, Moore KW, Frelinger JG, Sher BT, Shen FW, Boyse EA, Hood L (1981) A pseudogene homologous to mouse transplantation antigens: transplantation antigens are encoded by eight exons that correlate with protein domains. Cell 25(3):683–692
Takahata N (1995) MHC diversity and selection. Immunol Rev 143:225–247
Tallmadge RL, Lear TL, Johnson AK, Guerin G, Millon LV, Carpenter SL, Antczak DF (2003) Characterization of the beta2-microglobulin gene of the horse. Immunogenetics 54(10):725–733
Ungchusri T, Chiang EY, Brown G, Chen M, Tabaczewski P, Timares L, Stroynowski I (2001) Widespread expression of the nonclassical class I Qa-2 antigens in hemopoietic and nonhemopoietic cells. Immunogenetics 53(6):455–467
Vaiman M, Chardon P, Cohen D (1986) DNA polymorphism in the major histocompatibility complex of man and various farm animals. Anim Genet 17(2):113–133
van den Elsen PJ, Holling TM, Kuipers HF, van der Stoep N (2004) Transcriptional regulation of antigen presentation. Curr Opin Immunol 16(1):67–75
Velten FW, Renard C, Rogel-Gaillard C, Vaiman M, Chardon P (1999) Spatial arrangement of pig MHC class I sequences. Immunogenetics 49(11–12):919–930
Wroblewski JM, Kaminsky SG, Nakamura I (1994) Bat-1 genes and the origin of multiple class I loci in the H-2D region. Immunogenetics 39(4):276–280
Zijlstra M, Bix M, Simister NE, Loring JM, Raulet DH, Jaenisch R (1990) Beta 2-microglobulin deficient mice lack CD4-8+cytolytic T cells. Nature 344(6268):742–746
Acknowledgements
This research was funded by NIH grants HD-34086, HD-15799, and HD-049545, the Harry M. Zweig Memorial Fund for Equine Research, the Dorothy Russell Havemeyer Foundation, Inc., the USDA Regional Grant Program, and the Morris Animal Foundation. This manuscript is published in connection with a project of the University of Kentucky Agricultural Experiment Station as paper number 05-14-076. The authors thank Judy Lundquist (Kentucky) for technical assistance, and Don Miller and Bettina Wagner (Cornell) for helpful discussion and assistance. All experiments described in this manuscript comply with current laws of the USA.
Author information
Authors and Affiliations
Corresponding author
Additional information
The nucleotide sequence data reported are available in the GenBank database under the accession numbers CZ552047–CZ552159, CZ559424–CZ559456, and DQ083407–DQ083421
Rights and permissions
About this article
Cite this article
Tallmadge, R.L., Lear, T.L. & Antczak, D.F. Genomic characterization of MHC class I genes of the horse. Immunogenetics 57, 763–774 (2005). https://doi.org/10.1007/s00251-005-0034-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00251-005-0034-9