Abstract
We report the identification of a single major chromosomal region controlling natural killer (NK) cell-like activity in rainbow trout (Oncorhynchus mykiss). A genetic map based on 484 AFLP and 39 microsatellite genotypes from 106 doubled haploid fish was constructed. These fish were produced by androgenesis from a hybrid of two clonal lines divergent in NK-like activity. NK-like activities for 75 of the doubled haploids were quantified by an in vitro chromium release assay utilizing 51Cr-labeled YAC-1 target cells. Composite interval mapping revealed a single major quantitative trait locus (QTL) associated with NK-like activity in this rainbow trout model. Genetic mapping revealed this QTL to also be unlinked to: fragmented MHC class I and MHC class II regions, the leukocyte receptor cluster, the natural killer cell enhancement factor (NKEF) gene, the RAG-1 gene, and two QTL associated with resistance to infectious pancreatic necrosis virus in rainbow trout. Collectively, these results extend the utility of rainbow trout as an immunological model and are consistent with the idea that a single chromosomal region homologous to the natural killer cell complex (NKC) located on syntenic portions of mouse chromosome (Chr) 6, human Chr 12, and rat Chr 4 may exist in a lower vertebrate model.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
It is becoming increasingly evident that the innate immune system plays a major role in the overall health of both higher and lower vertebrates (Jones 2001). In mammals, natural killer (NK) cells are considered main effectors of the innate immune response. NK cells survey host tissues, eliminating foreign, virally infected, cancerous or otherwise compromised cells by cell-mediated lysis. Because suitable markers for NK cells have yet to be identified in lower vertebrates, it is unknown whether lower vertebrates contain homologues of mammalian NK cells. NK-like cells have been identified in amphibians (Ghoneum et al. 1990; Watson and Horton 1996) and fishes (Evans et al. 1984; Cleland and Sonstegard 1987; Faisal et al. 1989). Within fish, NK-like cells are often referred to as nonspecific cytotoxic cells (NCC) and are presumed to protect against bacteria, protozoan parasites, viruses and tumor growth (Jaso-Friedmann et al. 2001). The presence of functionally equivalent immune cells in fishes and higher vertebrates provides an interesting framework in which the evolution of innate immune responses can be studied.
It is plausible that extant differences in fish NK-like cells and mammalian NK cells may have paralleled the evolution of these organisms. In this regard, NK-like and NK cells may have diverged during the Silurian epoch some 400 million years ago to provide the same function in osteichythans and tetrapods, respectively. Because B and T cells appear conserved between mammals and fishes (Berstein et al. 1997), genetic differences in NK and NK-like cells may represent a major divergence in the evolution of the vertebrate immune system. Discerning the genetics controlling NK-like cells in a lower vertebrate model may therefore provide information useful for divulging the phylogenetics and evolution of innate immune responses in vertebrates.
In mammals, NK cells eliminate bacteria, parasites, virally infected cells and tumor cells through a complex repertoire of inhibitory and activating receptors present on the surface of NK cells (Diefenbach and Raulet 2001; Lanier 2001; Ryan et al. 2001; Sawicki et al. 2001). The existence of multiple NK receptors requiring simultaneous engagement ensures the accuracy of target cell recognition while protecting normal somatic cells from NK cell mediated lysis (Cerwenka and Lanier 2001). While activating receptors trigger a NK cell to release perforin or other proteases into a target cell (Brown et al. 2001; Moretta et al. 2002; Pinkoski and Green 2002) inhibitory receptors bind self-MHC to restrain the cytolytic capacities of the NK cell (Miller 2001; Warren et al. 2001). The presence of NK inhibitory receptors supports the “missing-self hypothesis” as NK cells survey the cells of the body for normal expression of self-MHC I (Ljunggren and Karre 1990). When confronted with aberrant MHC, NK cells are activated to initiate cell-mediated killing to confer protection against bacteria, virally infected cells and tumor growth (Kim et al. 2000).
To date, NK receptors are poorly understood in all animals below the level of mammals (Yoder et al. 2002a). Even within mammals, major differences in NK receptors have been found which necessitate further explanation. In mammals, three distinct types of inhibitory receptors have been identified: the Ly49 receptor family, the killer cell immunoglobulin-like receptors (KIR), and the CD94/NKG2 receptors (Boyington et al. 2001; Mandelboim and Porgador 2001). An extensive Ly49 receptor family exists in mice yet only one Ly49 receptor, Ly49L, has been found in humans (McQueen et al. 2002). Conversely, KIR receptors are present in humans and other primates yet appear to be absent in rodents (Karlhofer et al. 1992; Smith et al. 2001).
The existence of functionally equivalent receptors resembling lectins in mice (Ly49) and immunoglobulins in humans (KIR) appears to represent convergent evolutionary pathways. In contrast, CD94/NKG2 receptors appear to be conserved throughout several mammalian species (humans, chimpanzees, cattle, pigs, rats, mice), indicating the possibility of a shared evolutionary history (Borrego et al. 1998; Braud et al. 1998; Lee et al. 1998; Vance et al. 1998; Shum et al. 2002; Ravetch and Sun 2003). Most recently, CD94/NKG2 homologues have been identified in the urochorodate Botryllus schlosseri (Khalturin et al. 2003) and cichlid fish Paralabidochromis chilotes (Sato et al. 2003). The existence of NK receptors in primates, rodents, a bony fish and a urochordate suggests that NK receptors arose early in the evolutionary history of vertebrates.
Genetic mapping studies with traditional mammalian model systems (humans, mice and rats) have revealed NK cell activity to be largely regulated by a tight cluster of inhibitory and activating receptor genes located within a single chromosomal region called the natural killer cell complex (NKC). These NKC genes are located on syntenic portions of the mouse chromosome (Chr) 6 (Yokoyama et al. 1991), human Chr 12 (Renedo et al. 1997) and rat Chr 4 (Dissen et al. 1996). In comparison to the mammalian NKC, little is known concerning the genetic regulation of NK-like cells in fishes and other lower vertebrates (Ravtech and Lanier 2000; Litman et al. 2001; Fischer et al. 2003). To our knowledge, the CD94/NKG2 sequence identified in the cichlid (Sato et al. 2003) represents the only gene currently identified in a lower vertebrate that shares homology with the mammalian NKC. A putative activating receptor, NCCRP-1, has been reported in catfish (Jaso-Friedmann et al. 2002) although this receptor does not share homology with any genes currently identified within the mammalian NK complex. As such, it remains unclear whether a complex of genes comparable to the NKC of the mouse, rat and human genomes exists in fishes or any other lower vertebrates.
In this paper we report that a single major chromosomal region controls NK-like activity in rainbow trout. We identified this quantitative trait locus (QTL) within a doubled haploid population produced by androgenesis from an F1 hybrid of two clonal lines divergent in NK-like activity. Utilizing genetic mapping, we also show this QTL to be unlinked to either the major histocompatibility complex (MHC) or leukocyte receptor complex (LRC) in rainbow trout. These results are consistent with the idea that a region analogous to the NKC identified on syntenic portions of mouse Chromosome 6, human Chromosome 12, and rat Chromosome 4 may exist in rainbow trout. Moreover, the clonal rainbow trout lines and genetic map described herein provide a unique framework in which the complexity underlying NK-like cells in a lower vertebrate model can be further characterized.
Materials and methods
OSU and HC clonal lines of rainbow trout
Two clonal lines of rainbow trout (OSU and HC) were used in this study. The OSU clonal line was derived from a single fish obtained from a domesticated Oregon State University hatchery strain (OSU) and the HC clonal line was propagated from a single fish obtained from a hatchery strain from Hot Creek, California (HC). To produce clonal lines, outbred eggs were irradiated with 60Cobalt gamma radiation to destroy the maternal nuclear genome of the eggs. Irradiated eggs were then fertilized with sperm from either the founding HC or OSU fish to stimulate the development of haploid zygotes. Diploidy within these fertilized eggs was then achieved by applying a heat shock treatment to prevent the first cell cleavage. Resultant fish were either sexually XX or YY and homozygous at all loci. To produce lines that were genetically identical a second round of androgenesis or gynogenesis was then performed (Parsons and Thorgaard 1985; Scheerer et al. 1986). These clonal lines represent the basis of several ongoing projects in our laboratory aimed at discerning the genetic framework of complex phenotypes in rainbow trout (Robison et al. 2001; Nichols et al. 2003). Clonal lines of fish are considered analogous to the inbred mice used extensively in biomedical research (Ristow et al. 1998). The isogenicity of both the OSU and HC clonal lines has been confirmed by DNA fingerprint analysis (Young et al. 1996; Robison et al. 1999) and acceptance of skin grafts (Ristow et al. 1996).
OSU×HC doubled haploid rainbow trout
Individuals from the OSU (all female XX) and HC (all male YY) clonal lines were crossed to generate an all male (XY) F1 generation. Sperm from OSU×HC F1 hybrids was then used to produce a population of OSU×HC doubled haploids by androgenesis (Young et al. 1998). Briefly, outbred eggs were irradiated and fertilized with normal sperm from an OSU×HC F1 hybrid to create haploid zygotes. A heat shock treatment was subsequently applied to block the first mitotic division and this treatment resulted in the production of doubled haploid individuals. Doubled haploid progeny are genetically different from one another due to recombination events occurring during the production of gametes by the OSU×HC F1 hybrid.
Rainbow trout mapping progeny
Two separate OSU×HC doubled haploid populations (n=20 and n=86) were generated to yield the 106 fish utilized as the basis for constructing an OSU×HC genetic map. Genomic DNA was isolated from fin clips using the PureGene Genomic DNA purification kit (Gentra Systems, Minneapolis). Individuals in the n=86 OSU×HC doubled haploid population were PIT-tagged (BioMark, Boise, Idaho) so they could be identified in replicate testing of NK-like activity.
Rainbow trout tested for NK-like activity
The 86 PIT-tagged OSU×HC doubled haploid fish were raised in a semi-closed recirculating freshwater system at the Washington State University Hatchery facility. The fish were initially maintained in a single circular tank and to avoid overcrowding were randomly separated into two circular tanks within the same recirculating system at approximately 2 years of age. At 2.5 years of age, fish were tested for NK-like activity. A total of 75 fish were tested in triplicate in periods spanning 3 weeks during the summer season.
Phenotypic testing of NK-like activity
Fish were anesthetized and blood was collected by cardiac puncture. The NK-like phenotype was measured as described in Ristow et al. (2000). Briefly, NK-like cells were isolated from whole blood by centrifugation on Histopaque and Percoll gradients. NK-like cells from the OSU×HC doubled haploids were plated in 96-well cell culture plates at 106 cells/100 μl media/well in triplicate for each of three bleeds. 51Cr-labeled YAC-1 target cells were added to each cell set at a concentration of 104 cells/100 μl. In addition, six wells containing 104 target cells/100 μl/well and 100 μl of 2% sodium dodecyl sulfate (total release) or 100 μl of media (spontaneous release) were plated as controls. Following an 18 h incubation at 20°C, plates were centrifuged and 100 μl of supernatant of each well was measured for release of 51Cr using a gamma counter. NK-like cytotoxicity was calculated as: % cytotoxicity = (observed release−spontaneous release)/(total release−spontaneous release)×100.
AFLP genetic markers
The 106 doubled haploid fish were genotyped at 484 polymorphic amplified fragment length polymorphic (AFLP) markers following the Perkin Elmer Applied Biosystems AFLP mapping protocol for genomes. AFLP reactions were carried out as described in Vos et al. (1995) and as modified by Robison et al. (2001). Briefly, genomic DNA was digested using the restriction enzymes EcoRI and MseI prior to the ligation of specific adaptors. Selective amplification was carried out using standard AFLP EcoRI and MseI adaptor primers containing +3 selective bases. A total of 31 +3 EcoRI and MseI primer combinations were analyzed and AFLP fragments were visualized on polyacrylamide gels using Cy5-labeled EcoRI primers on a Molecular Dynamics Storm imaging system. AFLP loci were named in accordance to the selective nucleotides of the EcoRI primer followed by those of the MseI primer, the order of appearance on the gel, the size in base pairs, and the parental source of the band (OSU or HC).
Microsatellites
Microsatellites previously developed for salmonids were utilized to establish synteny between our OSU×HC genetic map and previously published OSU×Arlee (Young et al. 1998; updated Nichols et al. 2003b) and outbred (Sakamoto et al. 2000; Ozaki et al. 2001) rainbow trout maps. Primer sequences used to amplify microsatellites were obtained from sources listed in Table 1. Microsatellites were amplified using 50 ng of genomic template DNA with PCR conditions optimized for each microsatellite marker. In situations where size differences between the microsatellites amplified within the OSU and HC controls were large, microsatellites amplified in the doubled haploid progeny were scored on 2% agarose gels. Otherwise, microsatellites were run on polyacrylamide gels using Hex or Fam fluorescent labeled forward primers on an ABI system and scored using Genotyper software.
Genetic map construction
An OSU×HC genetic map based on AFLP and microsatellite genotypes of the doubled haploid progeny was constructed. This map was generated using the PC versions of Mapmaker 2.0 and Mapmaker/EXP 3.0 software (Lander et al. 1987) and the Mapmaker II program designed for the Macintosh (Dr. Scott Tingey, Dupont Experimental Station, Wilmington, Delaware). Linkage groups were established at a minimum LOD score of 3.0 and a maximum theta of 0.40 using the Kosambi map function as described in Nichols et al. (2003b).
Mapping of NKEF and MHC loci
The natural killer cell enhancement factor gene (NKEF) was genotyped and mapped on the OSU×HC map using the ABI SnaPshot dideoxynucleotide terminating protocol (Applied Biosystems International, USA). Primer sequences utilized to amplify the NKEF locus were designed to detect single nucleotide polymorphisms between the OSU and HC NKEF sequences reported in Zhang et al. (2001). Two MHC I loci (MHCIa and LMP2) and two MHC II loci (DAB and DAA) were previously mapped on the OSU×HC map as detailed in Phillips et al. (2003).
Sex phenotype
The OSU×HC doubled haploids used in the mapping panel were killed at approximately 4 years of age, at which time all were considered sexually mature. Upon sacrifice, the abdomens of each fish were cut open to expose the internal viscera. The sex of each OSU×HC doubled haploid fish was determined by a visual observation of the gonads and the male or female phenotype was assigned based on the respective presence of testis or ovaries. Phenotypic sex was mapped as a binary trait on the OSU×HC genetic map.
QTL analysis
Composite interval mapping was used to scan the OSU×HC genetic map for evidence of QTL influencing NK-like cytotoxicity in rainbow trout. This analysis was performed using Model 6 of the Windows QTL Cartographer computer program (Copyright 2001, Program in Statistical Genetics North Carolina State University) with a window size of 5.0 cM and five other markers used as cofactors in the model. Logarithm of odds (LOD) scores for the main effects were computed and significance thresholds for NK-like cytotoxicity were determined by a series of permutation tests at the 5% level (Churchill and Doerge 1994) with 1,000 replicates. Genetic markers exhibiting LOD scores greater than the calculated threshold value were then identified. Such markers are statistically correlated with regions of the rainbow trout genome associated with controlling NK-like cytotoxicity at or above the P<0.05 level of significance.
Results
Phenotypic variation of NK-like activity
The histogram shown in Fig. 1 depicts the range of NK-like activities found within both the OSU×HC doubled haploid progeny and the OSU and HC clonal lines. The phenotypic values of the 75 doubled haploids segregated into two clusters using Ward’s minimum variance cluster analysis in SAS, indicating the possibility of two discrete phenotypic categories for the NK-like trait. Average NK-like cytotoxicity values of nine OSU and HC control fish were used to produce a classification function using the DISCRIM procedure in SAS. This function was utilized to classify the 75 OSU×HC doubled haploids in two populations: 39 classified as having NK-like activity comparable to the OSU parental line while 36 classified as having NK-like activity comparable to the HC line. The mean values of fish classified as comparable to the OSU line ranged from −0.90 to 21.39% while the 36 fish classified to be comparable to the HC line ranged from 24.13 to 57.60%. Two distinct phenotypes, high and low NK-like cytotoxicity, in approximately equal proportions in the OSU and HC parental lines, and 36 high and 39 low classifications in the OSU×HC doubled haploid progeny was consistent with, but did not prove that, a single gene or genetic region controlling NK-like cytotoxicity exists in our rainbow trout model.
Genetic linkage map
A total of 523 genetic markers (484 AFLP and 39 microsatellites) were genotyped in order to construct the OSU×HC genetic map. Linkage analysis assigned 330 AFLP markers and 39 microsatellites to 29 major linkage groups and 5 smaller groups (Fig. 2). One hundred and fifty four of the AFLP markers genotyped remained unlinked at a LOD of 3.0 and a maximum θ of 0.40. Because fish within the OSU and HC clonal lines each have 60 chromosomes, 30 major linkage groups (corresponding to the haploid chromosome number) are expected within an OSU×HC population. In this respect, the constructed genetic map represents sound coverage of the OSU×HC genome. Furthermore, the 2,872.0 cM size of the OSU×HC map is consistent with the estimated 2,627.5 cM minimum size of the rainbow trout genome (Young et al. 1998).
OSU×HC Genetic map consisting of 34 linkage groups (LG). Labels indicate centimorgan distances and marker names. AFLP markers were named in accordance to the nucleotides of the EcoRI and MseI primers, order of appearance on gels, base pair size and parental source of band. Syntenies with OSU×Arlee LG are indicated in parentheses with Roman numerals. Genetic markers encompassing QTL controlling NK-like activity are outlined on LG 31
Regions of the rainbow trout genome associated with NK-like cytotoxicity
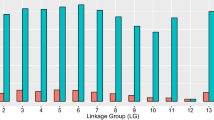
A series of permutation tests at the 5% level with 1,000 replicates resulted in a LOD threshold value for NK-like cytotoxicity of 4.33. A total of 11 AFLP markers and two microsatellites had LOD scores exceeding this calculated threshold value (Fig. 3). Therefore, these 13 markers are statistically correlated with genetic regions associated with controlling NK-like cytotoxicity. Interestingly, these 13 markers are all located on linkage group 31 (Figs. 2, 3). This clustering of markers reveals a single major chromosomal region or QTL spanning genetic distance of 25.7 cM associated with NK-like cytotoxicity. The haploid genome of rainbow trout being 2.4×109 bp (Young et al. 1998) and the 2,872 cM size of the OSU×HC map gives a tentative estimate of approximately 836 kb per cM. This estimate is tentative as recombination rates are not consistent across all chromosomal regions (Sakamoto et al. 2000) and the OSU×HC genetic map may not span the entire genome. Nevertheless, the clustering of AFLP markers within the QTL suggests that a relatively large physical distance is associated with NK-like activity in rainbow trout. This QTL also explains 63.4% of the variation in the cytotoxicity phenotype observed within the doubled haploid progeny (Table 2) indicating this chromosomal region has a major effect on the NK-like cytotoxicity phenotype.
Logarithm of odds (LOD) plot of the OSU×HC linkage group exhibiting NK-like QTL. Genetic distances are represented on the x-axis and the y-axis shows LOD scores. The dashed line indicates the 4.33 threshold value determined through a series of permutation tests at the P=0.05 level of significance. The small triangles represent positions of genetic markers on LG 31
Synteny of the OSU×HC map with other published rainbow trout maps
Mapping of common microsatellites facilitated direct alignment of 28 linkage groups between the OSU×HC and published OSU×Arlee (Nichols et al. 2003b) genetic maps. Because 29 genes are mapped on the OSU×Arlee map, this alignment enabled the inference of the chromosomal locations of these genes on the OSU×HC map. Furthermore, as common microsatellites have been used to establish synteny between the OSU×Arlee (Nichols et al. 2003b) and outbred (Sakamoto et al. 2000; Ozaki et al. 2001) rainbow trout maps, we can also infer the synteny of linkage groups on the OSU×HC map with those of the outbred rainbow trout maps (Table 3). Synteny with other published maps is important for understanding the chromosomal associations of genes and QTLs identified in other rainbow trout crosses. For example, utilizing synteny we can demonstrate the QTL associated with NK-like activity is unlinked to the leukocyte receptor cluster (LRC) (Yoder et al. 2002a), MHC class I and II loci (Phillips et al. 2003), RAG-1 gene (Nichols et al. 2003b) and two QTL associated with resistance to infectious pancreatic necrosis virus in rainbow trout (Ozaki et al. 2001).
Discussion
This is the first report concerning genetic mapping of NK-like activity in any lower vertebrate model. Utilizing cloned fish, doubled haploid progeny, in vitro immunological assays and PCR-based genetic mapping, we identified a single major QTL associated with NK-like cytotoxicity in rainbow trout. These results are consistent with the idea that a single major chromosomal region analogous to the NKC on syntenic portions of human Chromosome 12, mouse Chromosome 6 and rat Chromosome 4 may exist in a lower vertebrate model.
Our lower vertebrate model, rainbow trout (Oncorhynchus mykiss), constitutes an important species to both aquaculture and sport-fishing industries worldwide. Rainbow trout are members of the taxonomic family Salmonidae and, as salmonids, belong to one of the most primitive groups of bony fish existing today. Salmonids evolved as tetraploids by duplicating the genome of a diploid ancestor and their present genomes are in the process of rediploidization (Allendorf and Thorgaard 1984). This ancestral duplication of chromosome number and genetic fluctuation allow salmonids to tolerate a variety of chromosome set manipulation procedures which are currently not feasible or possible with traditional mammalian disease model systems.
For example, chromosome manipulations using androgenesis (Parsons and Thorgaard 1985) are used to create homozygous clonal lines of rainbow trout in only two generations. In comparison, typical inbreeding programs with mice often take up to 20 generations to produce offspring that are considered to be genetically identical. In a related approach, haploid gametes from an F1 fish can be subjected to heat shock treatments to double the chromosome number, which instantly results in progeny that are homozygous at all loci. The ability to genetically manipulate gametes to produce doubled haploid individuals is a technique currently restricted to certain plants and fishes (Lynch and Walsh 1998). This approach is amenable for genetic mapping experiments because the power of detecting QTL in experimental crosses is increased using doubled haploid designs (Carbonell et al. 1993; Martinez et al. 2002).
The combination of divergent NK-like phenotypes within the OSU and HC clonal lines and the ability to create OSU×HC doubled haploid fish provided us with a unique animal model to examine the genetic framework underlying NK-like cytotoxicity. Utilizing this model, we combined genetic mapping with phenotypic analysis to identify a single major chromosomal region controlling NK-like activity in rainbow trout. Our evidence for the existence of this genetic region is strong as this QTL accounts for 63.4% of the total variation in NK-like phenotypes observed within the doubled haploid progeny (Table 2). The additive effects of this QTL were also positive indicating that the allele(s) causing an increase in NK-like activity were inherited from the HC parent (Table 2). This finding is consistent with the increased NK-like cytotoxicity observed among individuals within the HC clonal line (Fig. 1).
Detection of a single QTL associated with NK-like cytotoxicity suggests several immediate hypotheses for NK-like cytotoxicity in this lower vertebrate model. One hypothesis is that within rainbow trout a single gene exhibiting a Mendelian inheritance pattern may control the phenotypic divergence of NK-like cytotoxicity between the OSU and HC lines. This result is consistent with suggestions that NK-like activation in channel catfish is dependent on a single receptor referred to as NCCRP-1 (Jaso-Friedmann et al. 2002). A gene carrying a mutation for NCCRP-1 in the OSU line might explain the apparent loss of function in NK-like cytotoxicity associated with the homozygous OSU genotype. To date, our attempts to amplify NCCRP-1 from cDNA derived from NK-like cells have proven unsuccessful in rainbow trout. Similarly, we have been unable to amplify a CD94/NKG2 gene in the rainbow trout using degenerate primers based on human, cichlid and urochordate sequences.
It is also possible that the OSU line might exhibit a mutant allele for another NK-like receptor, co-receptor or molecule important in either the adhesion or the signaling cascade associated with cell-mediated lysis. Because the F1 OSU×HC generation exhibits NK-like cytotoxicity comparable with the HC line (Evenhuis and Ristow, unpublished results), it appears that a loss-of-function allele could be attributed to an activating receptor in the OSU line or an inhibitory receptor in the HC line. However, because many OSU×HC doubled haploid individuals exhibited NK-like cytotoxicity (Fig. 1) at levels intermediate to the OSU and HC controls, it appears unlikely that a single gene could fully account for this variation.
It is also plausible that NK-like cytotoxicity in rainbow trout is controlled by a series of genes clustered in a single chromosomal region on linkage group 31. Such a cluster of genes would tend to be inherited together, and this scenario would also explain the high level of NK-like cytotoxicity present in the F1 OSU×HC generation and the variation of NK-like cytotoxicity observed within the OSU×HC doubled haploid individuals. Given the existence of NK gene complexes within the genomes of more extensively studied mammalian models, it appears reasonable to suggest that lower vertebrates might also possess such a genetic complex consisting of several closely linked genes.
The genetic mapping of four MHC genes in the OSU×HC cross (Phillips et al. 2003) revealed that MHC I and MHC II complexes on linkage groups 16 and 29 are unlinked to the QTL associated with NK-like cytotoxicity on linkage group 31. Furthermore, utilizing the synteny established between our OSU×HC and other maps we can demonstrate that the identified QTL is unlinked to the leukocyte receptor cluster (LRC) mapped to an OSU×Arlee linkage group (Yoder et al. 2002a) syntenic to OSU×HC linkage group 21 (Table 3). Similarly, through inferred synteny with an outbred rainbow trout map, the QTL controlling NK-like cytotoxicity is unlinked with two QTLs (inferred to be on OSU×HC linkage groups 3 and 24) that have been associated with resistance to infectious pancreatic necrosis virus in rainbow trout (Ozaki et al. 2001). In the present study, we also mapped the NKEF gene to linkage group 23 revealing that this gene is unlinked to the QTL controlling NK-like cytotoxicity. Collectively, these results establish a chromosomal framework of several important immune-related loci in rainbow trout. Moreover, these results and syntenies demonstrate that NK-like cytotoxicity, similar to the NK complex of mammals, is unlinked to both the MHC and LRC complexes.
Identification of the actual genes controlling the differences in NK-like cytotoxicity in rainbow trout is complicated by several factors including the large genetic size (in cM) of the identified QTL, possible allelic polymorphism for unrelated genes in this region, and the paucity of NK candidate genes identified in fishes. These factors exclude a direct sequencing approach for identifying the actual genes underlying the identified QTL. In efforts to overcome these obstacles, we have recently produced a new population of fish using an OSU×(OSU×HC) backcross design. These fish and subsequent backcrosses will enable genetic and phenotypic monitoring to occur at each generation for the retention of genetic markers associated with increased NK-like cytotoxicity. These populations should also enable us to conduct finer mapping on the genetic region controlling NK-like cytotoxicity in rainbow trout.
In summary, we report the identification of a single chromosomal region controlling NK-like activity differences in a rainbow trout model. This QTL was identified through the use of cloned fish, doubled haploid progeny, in vitro immunological assays and genetic mapping techniques. The OSU×HC genetic map and identified QTL presented within this paper provide a genetic framework that should facilitate further characterization of NK-like activity in rainbow trout. Given the phylogenetic distance between rainbow trout and the more traditionally studied mammalian disease models, we believe our lower vertebrate model may also prove valuable for identifying NK-related genes that may not be present in higher vertebrates. Collectively, the results presented herein are consistent with the idea that a genetic region analogous to the NKC located on syntenic portions of human Chromosome 12, mouse Chromosome 6 and the rat Chromosome 4 may also exist in lower vertebrates.
References
Allendorf F, Thorgaard G (1984) Tetraploidy and the evolution of salmonid fishes. In: Turner BJ (ed) Evolutionary genetics of fishes. Plenum, New York, pp 1–46
Berstein R, Schluter S, Marchalonis J (1997) Immunity. In: Evans DH (ed) The physiology of fishes. CRC, London, pp 215–242
Borrego F, Ulbrecht M, Weiss E, Coligan J, Brooks A (1998) Recognition of human histocompatibility leukocyte antigen HLA-E complexed with HLA class I signal sequence-derived peptides by CD94/NKG2 confers protection from natural killer cell mediated lysis. J Exp Med 187:813–818
Boyington J, Brooks A, Sun P (2001) Structure of killer cell immunoglobulin-like receptors and their recognition of the class I MHC molecules. Immunol Rev 181:66–78
Braud V, Allan D, O’Callaghan C, Soderstrom K, Phillips J, Lanier L, McMichael A (1998) HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature 391:795–799
Brown M, Dokun A, Heusel J, Smith H, Beckman D, Blattenberger E, Dubbelde C, Stone L, Scalzo A, Yokoyama W (2001) Vital involvement of a natural killer cell activation receptor in resistance to viral infection. Science 292:934–937
Carbonell E, Asins J, Baselga M, Balansard E, Gerig T (1993) Power studies in the estimation of genetic parameters and the localization of quantitative trait loci for backcross and doubled haploid populations. Theor Appl Genet 86:411–416
Cerwenka A, Lanier L (2001) Ligands for natural killer cell receptors: redundancy or specificity. Immunol Rev 181:158–169
Churchill G, Doerge R (1994) Empirical threshold values for quantitative trait mapping. Genetics 138:963–971
Cleland G, Sonstegard R (1987) Natural killer cell activity in rainbow trout: effect of dietary exposure to aroclor 1254 and mirex. Can J Fish Aquat Sci 44:636–638
Diefenbach A, Raulet D (2001) Strategies for target cell recognition by natural killer cells. Immunol Rev 181:170–184
Dissen E, Ryan J, Seaman W, Fossum S (1996) An autosomal dominant locus, Nka mapping to the Ly-49 region of a rat natural killer (NK) gene complex controls NK cell lysis of allogeneic lymphocytes. J Exp Med 183:2197–2207
Evans D, Hogan K, Graves S, Carlson R, Floyd E, Dawe D (1984) Nonspecific cytotoxic cells in fish (Ictalurus punctatus). Dev Comp Immunol 8:599–610
Faisal M, Ahmed I, Peters G, Cooper E (1989) Natural cytotoxicity of tilapia leukocytes. Dis Aquatic Org 7:17–22
Fischer U, Utke K, Ototake M, Diskstra M, Kollner B (2003) Adaptive cell-mediated cytotoxicity against allogenic targets by CD8-positive lymphocytes of rainbow trout (Oncorhynchus mykiss). Dev Comp Immunol 27:323–337
Ghoneum M, Cooper E, Sadek I. (1990) Variability of natural killer cell activity in anuran amphibians. Dev Comp Immunol 14:359–365
Jackson R, Ferguson M, Danzmann R, Fishback A, Ihssen P, O’Connell M, Crease T (1998) Identification of two QTL influencing upper temperature tolerance in three rainbow trout (Oncorhynchus mykiss) half-sib families. Heredity 80:143–151
Jaso-Friedmann L, Leary J, Evans D (2001) The nonspecific cytotoxic cell receptor (NCCRP-1): molecular organization and signaling properties. Dev Comp Immunol 25:701–711
Jaso-Friedmann L, Peterson D, Gonzalez D, Evans D (2002) The antigen receptor (NCCRP-1) on catfish and zebrafish nonspecific cytotoxic cells belongs to a new gene family characterized by an F-box associated domain. J Mol Evol 54:386–395
Jones S (2001) The occurrence and mechanisms of innate immunity against parasites in fish. Dev Comp Immunol 25:841-852
Karlhofer F, Ribaudo R, Yokoyama W (1992) MHC class I alloantigen specificity of Ly49+ IL-2 activated natural killer cells. Nature 358:66–70
Khalturin K, Becker M, Rinkevich B, Bosch T (2003) Urochordates and the origin of natural killer cells: identification of a CD94/NKRP1-related receptor in blood cells of Botryllus. Proc Natl Acad Sci USA 100:622–627
Khoo S, Ozaki A, Sakamoto T, Okamoto N (2000) Four highly polymorphic dinucleotide microsatellites in rainbow trout (Oncorhynchus mykiss). Anim Genet 31:68–79
Kim S, Iizuka K, Aguila H, Weissman I, Yokoyama W (2000) In vivo natural killer cell activities revealed by natural killer cell-deficient mice. Proc Natl Acad Sci USA 97:2731–2736
Lander E, Green P, Abrahamson J, Barlow A, Daly M (1987) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1:174–181
Lanier L (2001) Face off - the interplay between activating and inhibitory immune responses. Curr Opin Immunol 13:326–331
Lee N, Llano M, Carretero M, Ishitani A, Navarro F, Lopez-Botet M, Geraghty D (1998) HLA-E is a major ligand for the natural killer inhibitory receptor CD94/NKG2A. Proc Natl Acad Sci USA 95:5199–5204
Litman G, Hawke N, Yoder J (2001) Novel immune-type receptor genes. Immunol Rev 181:250–259
Ljunggren H, Karre K (1990) In search of the “missing self”: MHC molecules and NK cell recognition. Immunol Today 11:237–244
Lynch M, Walsh B (1998) Genetics and analysis of quantitative traits. Sinauer, Sunderland, Mass. p 401
Mandelboim O, Porgador A (2001) Molecules in focus: NKp46. Inter J Biochem Cell Biol 33:1147–1150
Martinez V, Hill W, Knott S (2002) On the use of doubled haploids for detecting QTL in outbred populations. Heredity 88:423–431
McQueen K, Wilhelm B, Harden K, Mager D (2002) Evolution of NK receptors: a single Ly49 and multiple KIR genes in the cow. Eur J Immunol 32:810–817
Miller J (2001) The biology of natural killer cells in cancer, infection and pregnancy. Exp Hematol 29:1157–1168
Moretta A, Bottino C, Mingari M, Biassoni R, Moretta L (2002) What is a natural killer cell? Nat Immunol 3:6–8
Morris D, Richard K, Wright J (1996) Microsatellites from rainbow trout (Oncorhynchus mykiss) and their use for genetic study of salmonids. Can J Fish Aquat Sci 53:120–126
Naish K, Park L (2002) Linkage relationships for 35 new microsatellite loci in Chinook salmon Oncorhynchus tschawytscha. Anim Genet 33:312–327
Nichols K, Wheeler P, Thorgaard G (2003a) Quantitative trait loci analyses for meristic traits in Oncorhynchus mykiss. Environ Biol Fishes (in press)
Nichols K, Young W, Danzman R, Robison B, Rexroad C, Noakes M, Phillips R, Bentzen P, Spies I, Knudsen K, Allendorf F, Cunningham B, Brunelli J, Zhang H, Ristow S, Drew R, Brown K, Wheeler P, Thorgaard G (2003b) A consolidated linkage map for rainbow trout. Anim Genet 34:102–115
Olsen J, Bentzen P, Seeb J (1998) Characterization of seven microsatellite loci derived from pink salmon. Mol Ecol 7:1087–1089
O’Reilly P, Hamilton L, McConell S, Wright J (1996) Rapid analysis of genetic variation in Atlantic salmon (Salmo salar) by PCR multiplexing of dinucleotide and tetranucleotide microsatellites. Can J Fish Aquat Sci 53:2292–2298
Ozaki A, Sakamoto T, Khoo S, Nakamura K, Coimbra M, Akutsu T, Okamoto N (2001) Quantitative trait loci (QTLs) associated with resistance/susceptibility to infectious pancreatic necrosis virus (IPNV) in rainbow trout (Oncorhynchus mykiss). Mol Genet Genomics 265:23–31
Parsons J, Thorgaard G (1985) Production of androgenetic diploid rainbow trout. J Heredity 76:177–181
Phillips R, Zimmerman A, Noakes M, Palti Y, Morash M, Eiben L, Ristow R, Thorgaard G, Hansen J (2003) Physical and genetic mapping of the rainbow trout major histocompatibility regions. Immunogenetics 55:561–569
Pinkoski M, Green D (2002) Lymphocyte apoptosis: refining the paths to perdition. Curr Opin Hematol 9:43–49
Ravetch J, Lanier L (2000) Immune inhibitory receptors. Science 290:84–89
Ravetch S, Sun P (2003) Structure and function of natural killer cell surface receptors. Annu Rev Biophys Biomol Struct 32:93–114
Renedo M, Arce I, Rodriguez A, Carretero M, Lanier L, Lopez-Botet M, Fernandez-Ruiz E (1997) The human natural killer gene complex is located on chromosome 12p12-p13. Immunogenetics 46:307–311
Rexroad C, Coleman R, Martin A, Hershberger W, Killefer J (2001) Thirty-five polymorphic microsatellite markers for rainbow trout (Oncorhynchus mykiss). Anim Genet 32:316–331
Rexroad C, Coleman R, Gustafson A, Hershberger W, Killefer J (2002a) Development of rainbow trout microsatellite markers from repeat enriched libraries. Mar Biotechnol 3:12–16
Rexroad C, Coleman R, Hershberger W, Killefer J (2002b) Eighteen polymorphic microsatellite markers for rainbow trout (Oncorhynchus mykiss). Anim Genet 33:72–84
Ristow S, de Avila J, Baldwin T, Wheeler P, Thorgaard G (1996) Acceptance of skin grafts by isogenic rainbow trout (Oncorhynchus mykiss). Amer J Vet Res 57:1576–1579
Ristow S, Grabowski L, Ostberg C, Robison B, Thorgaard G (1998) Development of long-term cell lines from homozygous clones of rainbow trout. J Aquat Anim Health 10:75–82
Ristow S, Evans D, Jaso-Friedmann L (2000) Analyzing nonspecific cytotoxic cells in fish. In: Campbell K, Colonna M (eds) Methods in molecular biology, vol 121. Natural killer cell protocols: cellular and molecular methods. Humana, Totowa, N.J., pp 347–357
Robison B, Wheeler P, Sundin K, Sikka P, Thorgaard G (2001) Composite interval mapping reveals a major locus influencing embryonic development rate in rainbow trout (Oncorhynchus mykiss). J Hered 92:16–22
Ryan J, Naper C, Hayashi S, Daws M (2001) Physiologic functions of activating natural killer (NK) complex-encoded receptors on NK cells. Immunol Rev 181:126–137
Sakamoto T, Danzmann, Gharbi K, Howard P, Ozaki A, Khoo S, Woram R, Okamoto N, Ferguson M, Holm L, Guyomard R, Hoyheim B (2000) A microsatellite linkage map of rainbow trout (Oncorhynchus mykiss) characterized by large sex-specific differences in recombination rates. Genetics 103:1331–1345
Sato A, Mayer W, Overpath P, Klein J (2003) Genes encoding putative natural killer cell C-type lectin receptors in teleostean fishes. Proc Natl Acad Sci USA 100:7779–7784
Sawicki M, Dimasi N, Natarajan K, Wang J, Margulies D, Mariuzza R (2001) Structural basis of MHC class I recognition by natural killer cell receptors. Immunol Rev 181:52–65
Scheerer P, Thorgaard G, Allendorf G, Knudsen K (1986) Androgenetic rainbow trout produced from inbred and outbred sperm sources show similar survival. Aquaculture 57:289–298
Scribner K, Gust J, Fields R (1996) Isolation and characterization of novel salmon microsatellite loci: cross-species amplification and population genetic applications. Can J Fish Aquat Sci 53:833–841
Shum B, Flodin L, Muir D, Rajalingam R, Khakoo S, Cleland S, Guethlein L, Uhrberg M, Parham P (2002) Conservation and variation in human and common chimpanzee CD94 and NKG2 genes. J Immunol 168:240–252
Small M, Beacham T, Withler R, Nelson R (1998) Discriminating coho salmon (Oncorhynchus kisutch) populations within the Fraser river, British Columbia, using microsatellite DNA markers. Mol Ecol 7:141–155
Smith H, Idris A, Yokoyama W (2001) Murine natural killer cell activation receptors. Immunol Rev 181:115–125
Vance R, Kraft J, Altman J, Jensen P, Raulet D (1998) CD94/NKG2A is a natural killer cell receptor for the non-classical major histocompatibility complex (MHC) class I molecule Qa-1. J Exp Med 188:1841–1848
Vos P, Hogers R, Bleeker M, Reijans M, Vandelee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23:4407–4414
Warren H, Campbell A, Waldron J, Lanier L. (2001) Biphasic response of NK cells expressing both activating and inhibitory killer Ig-like receptors. Int Immunol 13:1043–1052
Watson L, Horton J (1996) NK-like activity against allogenic tumor cells demonstrated in the spleen of control and thymectomized Xenopus. Immunol Cell Biol 74:365–373
Yoder J, Mueller M, Nichols K, Ristow S, Thorgaard G, Ota T, Litman G (2002a) Cloning novel immune-type inhibitory receptors from the rainbow trout Oncorhynchus mykiss. Immunogenetics 54:662–670
Yoder J, Nielsen M, Amemiya C, Litman G (2002b) Zebrafish as an immunological model system. Microbes Infect 4:1469–1478
Yokoyama W, Ryan J, Hunter J, Smith H, Stark M, Seaman W (1991) cDNA cloning of mouse NKR-P1 and genetic linkage with Ly49: identification of a natural killer cell gene complex on mouse chromosome 6. J Immunol 147:3229–3236
Young W, Wheeler P, Fields R, Thorgaard G (1996) DNA fingerprinting confirms isogenicity of androgenetically derived rainbow trout lines. J Hered 87:77–81
Young W, Wheeler P, Coryell V, Keim P, Thorgaard G (1998) A detailed linkage map of rainbow trout produced using doubled haploids. Genetics 148:839–850
Zhang H, Evenhuis J, Thorgaard G, Ristow S (2001) Cloning, characterization and genomic structure of the natural killer cell enhancement factor (NKEF)-like gene from homozygous clones of rainbow trout (Oncorhynchus mykiss). Dev Comp Immunol 25:25–35
Acknowledgements
The authors thank Richard Dixon, Paul Wheeler and Steve Patton for their help rearing and sampling the fish used in this study. We also thank Richard Alldredge and Krista Nichols for their respective assistance with the statistical analysis of the NK-like cytotoxicity data and the QTL analysis data. We are grateful to Paul Bentzen, Bjorn Hoyheim, Yniv Palti, Todd Seamons and Takashi Sakamoto for providing primer sequence information for several of the microsatellites used in this study. This research was funded in part by a USDA grant awarded to both Sandra Ristow and Gary Thorgaard (USDA-CSREES 2001-34452-10326).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zimmerman, A.M., Evenhuis, J.P., Thorgaard, G.H. et al. A single major chromosomal region controls natural killer cell-like activity in rainbow trout. Immunogenetics 55, 825–835 (2004). https://doi.org/10.1007/s00251-004-0645-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00251-004-0645-6