Abstract
Passerine birds have a supernumerary chromosome in their germ cells called the germline-restricted chromosome (GRC). The GRC was first discovered more than two decades ago in zebra finch but recent studies have suggested that it is likely present in all passerines, the most species rich avian order, encompassing more than half of all modern bird species. Despite its wide taxonomic distribution, studies on this chromosome are still scarce and limited to a few species. Here, we cytogenetically analyzed the GRC in five closely related estrildid finch species of the genus Lonchura. We show that the GRC varies enormously in size, ranging from a tiny micro-chromosome to one of the largest macro-chromosomes in the cell, not only among recently diverged species but also within species and sometimes even between germ cells of a single individual. In Lonchura atricapilla, we also observed variation in GRC copy number among male germ cells of a single individual. Finally, our analysis of hybrids between two Lonchura species with noticeably different GRC size directly supported maternal inheritance of the GRC. Our results reveal the extraordinarily dynamic nature of the GRC, which might be caused by frequent gains and losses of sequences on this chromosome leading to substantial differences in genetic composition of the GRC between and even within species. Such differences might theoretically contribute to reproductive isolation between species and thus accelerate the speciation rate of passerine birds compared to other bird lineages.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
It is typically assumed that, apart from some malformations like cancer tissues, multicellular organisms normally contain the same karyotype and genetic information in all their cells. However, a growing number of exceptions to this paradigm have been noted, such as cases when parts of the genome are eliminated from some or all somatic cells (Wang and Davis 2014; Dedukh and Krasikova 2022; Smith et al. 2021; Suh and Dion-Côté 2021). A striking example of such programmed DNA elimination occurs in passerine birds (Passeriformes) (Pigozzi and Solari 1998; Torgasheva et al. 2019). Here, programmed DNA elimination concerns the germline-restricted chromosome (GRC), which is eliminated from somatic cells very early during embryogenesis and maintained only in the germline (Pigozzi and Solari 1998; Goday and Pigozzi 2010). The GRC was first discovered two decades ago in zebra finch (Taeniopygia guttata) (Pigozzi and Solari 1998) and later observed in bengalese finch (Lonchura domestica) (del Priore and Pigozzi 2014). Interestingly, recent studies have suggested that the GRC is very likely present in all passerine birds (Kinsella et al. 2019; Torgasheva et al. 2019), the largest and most diverse order of modern birds with more than 6000 species (Fjeldså et al. 2020). This makes the Passeriformes the largest taxon with obligatory programmed DNA elimination. Analysis of GRC sequences in zebra finch revealed that this chromosome is surprisingly gene rich, containing more than one hundred protein coding genes (Biederman et al. 2018; Kinsella et al. 2019). Many of these genes showed ovary or testis expression, suggesting that this chromosome plays an important role in the germ cell development (Biederman et al. 2018; Kinsella et al. 2019).
Despite its wide taxonomic distribution, the cytogenetic characterization of the GRC has only been performed in a handful of species so far (Pigozzi and Solari 1998; del Priore and Pigozzi 2014; Torgasheva et al. 2019; Poignet et al. 2021). Interestingly, these studies show that the GRC varies noticeably in size among species, suggesting that we are still far from understanding its evolutionary significance and function for passerine birds. Specifically, of the 18 species analyzed so far (Torgasheva et al. 2019; Poignet et al. 2021), 10 species, including zebra finch and bengalese finch, showed a large GRC comparable in size with macro-chromosomes, while 8 species exhibited a small GRC of similar size to micro-chromosomes. However, there is no evident phylogenetic clustering of species according to GRC size, and species within the same family sometimes differ in GRC length (Torgasheva et al. 2019). This suggests rapid and independent changes to the GRC genetic content across the passerine phylogeny. Indeed, cross-species hybridizations of the whole-GRC microdissected DNA probes revealed significant changes in the GRC genetic content between species (Torgasheva et al. 2019, 2021). Such changes might theoretically contribute to reproductive isolation between species and thus accelerate the speciation rate of passerine birds (Kinsella et al. 2019). Nonetheless, except for two nightingale species (Poignet et al. 2021) and two martin species (Torgasheva et al. 2019; Malinovskaya et al. 2020), where the GRC showed similar size within each genus, GRC size has not yet been compared among closely related species in the early stages of divergence and thus, it is not clear whether changes in the GRC size and associated changes in genetic content are fast and frequent enough to potentially contribute to speciation in passerines.
Remarkable variation in size is not the only peculiarity of the GRC. This chromosome also shows atypical copy-number variation between individuals (i.e., polymorphism) and sometimes even within individuals (i.e., mosaicism). In females, the GRC is normally present in two copies that form a bivalent and recombine during meiosis like standard autosomes (Pigozzi and Solari 1998, 2005; del Priore and Pigozzi 2014; Torgasheva et al. 2019; Malinovskaya et al. 2020). However, female individuals with a single GRC copy have been occasionally found in zebra finch (Pigozzi and Solari 2005) and sand martin (Riparia riparia) (Malinovskaya et al. 2020). In addition, some females in great tit (Parus major) showed mosaicism in GRC copy number, with some oocytes carrying one GRC copy and others two GRC copies (Torgasheva et al. 2021). By contrast, the GRC in males is typically present as a single copy that behaves as univalent during the pachytene of the first meiosis, and later is eliminated from the nucleus (Pigozzi and Solari 1998, 2005; del Priore and Pigozzi 2014; Torgasheva et al. 2019). But mosaicism in GRC copy number was recently described in some males of pale martin (Riparia diluta), in which a single individual can sometimes carry one, two, and even three GRC copies in its germ cells (Malinovskaya et al. 2020).
The observation that the GRC is eliminated from the spermatocyte nucleus during the first meiotic division (Pigozzi and Solari 1998; Goday and Pigozzi 2010), later forming a micronucleus with fragmented DNA that is finally expelled from the cell (Schoenmakers et al. 2010), has led to the assumption that the GRC is inherited only through females. However, a recent study that analyzed zebra finch mitochondrial and GRC haplotypes in captive-bred populations suggested that the GRC might occasionally be inherited from males (Pei et al. 2022).
In the present study, we used classical and antibody-based cytogenetic methods to explore polymorphism in GRC size and copy-number at interspecific, intraspecific, and intra-individual levels. Specifically, we (1) examined variation in GRC size and copy number among five closely related species of estrildid finches (genus Lonchura), (2) performed extensive individual sampling to test for intra-individual mosaicism in GRC copy number and size, and finally, (3) crossed two Lonchura species with distinct GRC sizes to directly test for maternal inheritance of the GRC. Our results brought clear support for maternal inheritance of the GRC and suggested that the GRC varies enormously in size not only among recently diverged species but can also exhibit polymorphism in size within species and even between germ cells of a single individual. We also found evidence for mosaicism in GRC copy number in male germ cells of one analyzed individual. Our results suggest that the GRC is an extraordinarily rapidly evolving chromosome, which strongly contrasts with the otherwise conserved avian karyotype (Ellegren 2010), and highlight its potential role in the speciation of passerine birds.
Methods
Examined species
The GRC was examined in five species of the genus Lonchura: bengalese finch (L. striata var. domestica; hereafter L. domestica), scaly-breasted munia (L. punctulata), chestnut-breasted mannikin (L. castaneothorax), tricolored munia (L. malacca), and chestnut munia (L. atricapilla). From each species, two unrelated reproductively active males were analyzed, except for L. malacca from which only one individual was used. Additionally, two F1 hybrid males from a cross between a female L. domestica and a male L. punctulata were used to study GRC inheritance. All birds were sacrificed by cervical dislocation and their left testis was immediately dissected for the preparation of meiotic spreads. The birds were obtained from different Lonchura breeders in the Czech Republic and Netherlands and kept at the Institute of Vertebrate Biology (Studenec, Czech Republic). The work with birds followed the Czech law on protection of animals (law no. 246/1992 Sb.) and was approved by the ethical committee of the Charles University (permission no: UKPRF/28830/2021).
Meiotic spreads and immunostaining
Meiotic spreads were prepared following Peters et al. (1997) with some modifications. Briefly, the left testis was placed in hypotonic solution (30 mM Tris, 50 mM sucrose, 17 mM trisodium citrate dehydrate, 5 mM EDTA, pH = 8.2) immediately after dissection. Once there, it was cut into small pieces to release the germ cells and left in the solution for 40 min. Subsequently, the cells were re-suspended in 100 mM sucrose and dropped on slides previously covered by a thin layer of 1% paraformaldehyde fixative (PFA) and 0.15% Triton (Sigma Aldrich). The slides were put in a humid chamber for 1.5 h and washed with 1 × PBS for 2 min. The slides were then incubated for 1.5 h with one or more of the following primary antibodies: (i) rabbit polyclonal anti-SYCP3 (ab15093, Abcam), which stains the lateral element of the synaptonemal complex; (ii) human monoclonal serum (CREST, 15–234, Antibodies Incorporated) staining centromeres; (iii) mouse monoclonal anti-MLH1 (ab14206, Abcam), which stains recombination sites; and (iv) anti-H3S10p (GTX128116, GeneTex) detecting histone H3 phosphorylated at serine 10, which labels the eliminated GRC when it forms a micronucleus localized in the cytoplasm of secondary spermatocytes or young spermatids (Goday and Pigozzi 2010; Del Priore and Pigozzi 2014). Anti-SYCP3 and anti-H3S10p antibodies were diluted 1:200 in 1 × PBS with 1% blocking reagent (Roche); the rest of the antibodies were diluted 1:50. The secondary antibodies applied to detect the corresponding primary antibodies were anti-rabbit Alexa 594 (A32740, Invitrogen), anti-human Alexa-488 (A-11013, Invitrogen), and anti-mouse Alexa 488 (A-11029, Invitrogen). Each secondary antibody was diluted 1:200 in 1% blocking reagent (Roche) in 1 × PBS. The slides were air-dried and counterstained with Vectashield/DAPI (1.5 mg/ml) (Vector, Burlingame, Calif., USA) to label the chromatin. Observation of cells was done with an Olympus BX53 microscope. Pictures were taken with an Olympus DP30BW digital microscope camera using the Olympus Acquisition Software and processed using the Adobe Photoshop CS6 software.
GRC identification and size measurement
The univalent GRC was identified in pachytene spermatocytes according to diagnostic features previously reported by Pigozzi and Solari (2005) and Torgasheva et al. (2019). Specifically, (i) the GRC is stained weaker with anti-SYCP3 antibody when compared to the normal set of bivalent chromosomes, (ii) the GRC shows signal of the CREST antibody not only at the centromere, but also diffusely along the entire chromosome, and (iii) the GRC shows no signal of anti-MLH1 antibody staining recombination sites. The presence and size of the GRC was visually evaluated in 100 pachytene cells per individual in all species and hybrids, except for one individual of L. atricapilla for which a larger number of cells (288) were inspected due to mosaicism in GRC size and copy number. In a subset of cells (Table 1, Supplementary Table S1), we estimated the size of the GRC by measuring the length of its chromosomal axis coated by anti-SYCP3 antibody using ImageJ (Schneider et al. 2012).
Furthermore, we visualized the GRC in the form of an eliminated micronucleus, which is typically localized next to nuclei of secondary spermatocytes or young spermatids and is positively stained with anti-H3S10p antibody (del Priore and Pigozzi 2014). The diameter of GRC micronuclei in each individual was measured (Table 1, Supplementary Table S2) with Image J (Schneider et al. 2012).
Mapping GRC size on the Lonchura phylogeny
Since there is, to our knowledge, no published phylogeny including all the Lonchura species examined here, we built a calibrated tree with BEAST.v2.4.6 (Bouckaert et al. 2014), using zebra finch (Taeniopygia guttata) as an outgroup. We used sequence data of the mitochondrial NADH dehydrogenase 2 (ND2) locus (1042 bp) available in GenBank (AY323596.2, CM016794.1, MN991460.1, MF765864.1, MN991459.1, KP965879.1), a strict clock and the GTR + I + G nucleotide substitution model selected in jModeltTest v.2.1.10 (Darriba et al. 2012), using the Akaike Information Criteria. The tree was calibrated using the divergence times of T. guttata/L. punctulata (9.2 million years ago (Mya), 95% highest posterior density (HPD) interval: 8.6–9.7 Mya), and L. domestica/L. castaneothorax (3.5 Mya, HPD interval: 3.7–3.3 Mya), following results from Stryjewski and Sorenson (2017). We ran a total of 50 million generations sampling each 5000 generations and discarding the first 10% as burn in. The convergence of parameters was verified by checking the effective sample size (ESS values > 200) in TRACER v1.6 (Drummond and Rambaut 2007); the consensus tree was obtained from TreeAnotator (Drummond et al. 2012), and plot in FigTree v.1.4.3 (Rambaut 2012). We then mapped the GRC size of individual species to this phylogenetic tree.
Results
With no exception, the GRC was observed in each examined pachytene cell of all individuals. Surprisingly, the GRC varied dramatically in size among the Lonchura species. In L. domestica and L. castaneothorax, the GRC was a macro-chromosome, comparable in size with the largest chromosomes in the cell (Fig. 1; Supplementary Fig. S1). The mean length of the GRC SYCP3 axis was larger than 20 μm in both of these species (Table 1; Supplementary Table S1). Consistent with this observation, L. domestica and L. castaneothorax showed relatively large GRC micronuclei (Fig. 1) with a mean diameter larger than 3.7 μm (Table 1; Supplementary Table S2).
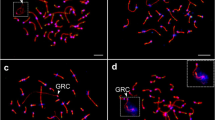
The germline-restricted chromosome (GRC) in males of four Lonchura species. Pachytene chromosomes are immunostained with anti-SYCP3 (red) and CREST (green) (a) or anti-SYCP3 (red) and anti-MLH1 (green) antibodies (b). Anti-SYCP3, CREST, and anti-MLH1 antibodies label the lateral elements of synaptonemal complexes, centromeres, and recombination sites, respectively. The arrows point to the GRC which is less intensively stained with anti-SYCP3 antibody, is diffusely labeled with CREST and shows no recombination sites. The boxes display a magnification of the GRC with reduced CREST signal for better visibility of the chromosome. The GRC micronuclei immunostained with anti-H3S10p antibody (red) next to nuclei of secondary spermatocytes, where DNA is counterstained with DAPI (blue) (c). Arrowheads point to the GRC micronucleus. In L. domestica and L. castaneothorax, the GRC is a macro-chromosome, while in L. punctulata and L. malacca, the GRC is a micro-chromosome. Scale bar: 10 μm
By contrast, in L. punctulata and L. malacca, the GRC was a tiny chromosome, comparable or smaller in size than the smallest micro-chromosome (Fig. 1; Supplementary Fig. S1). In both species, the mean GRC length was smaller than 1 μm (Table 1; Supplementary Table S1). However, unlike the larger GRCs in L. domestica and L. castaneothorax, where CREST and SYCP3 signals corresponded in size, the SYCP3 signal was smaller than the CREST signal in L. punctulata and L. malacca, which might mean that for small GRCs, the length of the SYCP3 signal slightly underestimates the real size of the GRC. The small size of the GRC in L. punctulata and L. malacca was supported by small GRC micronuclei (Fig. 1), with a mean diameter smaller than 1.5 μm (Table 1; Supplementary Table S2) in these two species.
Interestingly, in L. atricapilla we observed mosaicism in GRC size and copy number (Fig. 2; Table 2). In one of the two individuals, 57% of the pachytene spermatocytes carried a macro-GRC (Fig. 2a1), while 38% of the cells carried a micro-GRC (Fig. 2a2). The remaining 5% of cells either contained both macro- and micro-GRC (Fig. 2a3) or two micro-GRCs (Fig. 2a4). The mean length of the SYCP3 axis was 22.4 μm for the macro-GRC and 0.6 µm for the micro-GRC (Table 1; Supplementary Table S1). Consistent with this observation, both large and small GRC micronuclei were detected in this individual (Fig. 2; Table 1; Supplementary Table S2). The proportion of large and small micronuclei was similar to the ratio of macro- and micro-GRCs found in the pachytene cells (Table 2). A single large micronucleus was found next to the nucleus of a secondary spermatocyte or young spermatid in 65% of cases (Fig. 2b1), while a single small micronucleus was found in 32% (Fig. 2b2). The combination of one large and one small micronuclei, two small micronuclei, or two large micronuclei in the vicinity of a single secondary spermatocyte or spermatid were each observed in 1% of cases (Fig. 2b3–b5; Table 2). In the second individual of L. atricapilla, all pachytene spermatocytes had only a single micro-GRC and only small GRC micronuclei were observed in the later meiotic stages (Table 1).
Mosaicism in size and copy number of the germline-restricted chromosome (GRC) in one L. atricapilla male. Pachytene chromosomes immunostained with anti-SYCP3 (red) and CREST (green) antibodies labeling the lateral element of the synaptonemal complexes and centromeres, respectively (a). Most primary spermatocytes carried either a single macro-GRC (a1) or a single micro-GRC (a2), but a few cells had one macro- and one micro-GRC (a3) or two micro-GRCs (a4). Congruently, a single large GRC micronucleus (b1) or a single small GRC micronucleus (b2) was found next to nuclei of secondary spermatocytes or spermatids in most cases, but in a few cases one small and one large GRC micronucleus (b3), two small GRC micronuclei (b4) or two large GRC micronuclei (b5) were found next to the nuclei of secondary spermatocytes or spermatids. Anti-H3S10p antibody (red) labels GRC micronuclei and DAPI (blue) counterstains the chromatin (b). Scale bar: 10 μm
In contrast with the extraordinarily large variation in GRC size among the Lonchura species, all the other chromosomes showed similar sizes in all the examined species, which also presented the same diploid chromosome number (2n = 80). Distribution of the GRC size along the Lonchura phylogeny revealed that the species with large and small GRCs do not form a single clade but are rather scattered across the phylogeny (Fig. 3). This suggests that change between macro- and micro-GRC may have occurred several times in this genus. However, given the existence of polymorphism in GRC size within a species and the low number of analyzed individuals per species, it is hard to infer how many times the GRC has actually changed size.
Distribution of the macro-GRCs (black dots) and micro-GRCs (white dots) along a calibrated-tree of five Lonchura species. The Bayesian phylogeny is based on 1042 bp of the NADH dehydrogenase 2 (ND2) locus and uses zebra finch (Taeniopygia guttata) as an outgroup. Divergence times on nodes 1 and 3 were set at 9.2 Mya and 3.5 Mya, respectively, based on data from Stryjewski and Sorenson (2017). Bars at nodes indicate the 95% highest posterior density (HPD) intervals
Finally, the two examined F1 hybrid males between L. domestica (female) and L. punctulata (male) carried a macro-GRC comparable in size with the maternal GRC of L. domestica, as well as large GRC micronuclei (Fig. 4; Table 1), supporting maternal inheritance of the GRC.
The GRC in F1 hybrids between a female of L. domestica and a male of L. punctulata. The pachytene chromosomes are labeled with anti-SYCP3 (red) and CREST (green) (a) or anti-SYCP3 and anti-MLH1 (green) antibodies (b). SYCP3, CREST, and MLH1 antibodies stain the lateral element of synaptonemal complexes, centromeres, and recombination sites, respectively. The box in (a) displays a magnification of the GRC with reduced CREST signal for better visibility of the chromosome. The GRC micronucleus next to the nucleus of a secondary spermatocyte labeled with anti-H3S10p (red) antibody; DNA counterstained with DAPI (blue) (c). Arrows point to the GRC. The arrowhead points to the expelled GRC micronucleus. The hybrids exhibit a macro-GRC and large GRC micronuclei comparable in size with that of the maternal species (see Fig. 1). Scale bar: 10 μm
Discussion
The exclusion of the GRC from somatic cells and its maintenance only in the germline represents an intriguing example of programmed DNA elimination (Wang and Davis 2014; Dedukh and Krasikova 2022; Smith et al. 2021). Although the GRC might have originally been a parasitic, non-essential B chromosome (Camacho et al. 2000; Johnson Pokorná and Reifová 2021), its presence in all the passerine species studied so far suggests that it gained some important function that has prevented its loss in this taxon. This function is still unknown, but it has been suggested that the GRC might play an important role in germ cell development or even the germline determination itself (Kinsella et al. 2019). Our results showing enormous variation in GRC size among species, within species and even among germ cells of a single individual seemingly contrast with the apparent importance of this chromosome and suggest that large parts of the GRC might in fact be dispensable.
The vast majority of genes identified on the zebra finch GRC have paralogs on autosomes and sex chromosomes (Itoh et al. 2009; Kinsella et al. 2019). It thus appears that the GRC contains duplicated sequences from regular chromosomes (Kinsella et al. 2019). Interestingly, while some genes on the GRC of zebra finch seem to be quite old, others were apparently added to the GRC very recently (Kinsella et al. 2019). We thus suggest that the great variability in GRC size among species, and even among individuals of the same species may be due to rapid turnover of the GRC genetic content caused by frequent addition of sequences from other chromosomes and their subsequent loss. This idea is supported by FISH (fluorescent in situ hybridization) experiments with GRC-derived DNA probes, showing that GRC probes derived from different species hybridized to different regions of the genome (Torgasheva et al. 2019, 2021). Another reason for GRC size variability might be sequence duplications within the GRC including species-specific amplifications of repetitive sequences such as transposable elements. The large GRC of zebra finch seems to lack a high density of transposable elements, but many GRC genes are indeed duplicated within the GRC (Kinsella et al. 2019). Comparison of GRC genetic content among the Lonchura species will be needed to determine the reasons for the enormous GRC size variation.
The dynamic evolution of the GRC strongly contrasts with the rather conserved avian karyotypes (Zhang et al. 2014), which are remarkably stable in chromosome number, gross chromosome morphology and synteny, even between distant bird taxa (e.g., Galliformes and Passeriformes) (Griffin et al. 2007; Nanda et al. 2011; Zhang et al. 2014; O’Connor et al. 2019). The speed at which the GRC evolves thus appears extraordinarily fast when compared with any other chromosome, including the W chromosome, which is the fastest evolving chromosome in the somatic karyotype, due to its highly repetitive genetic content (Peona et al. 2021). Distribution of the GRC size across the Lonchura phylogeny (Fig. 3) suggests that the GRC has dramatically changed its size more than once during the last 5 million years. However, the low number of examined individuals per species prevents us from identifying possible polymorphisms in GRC size in all analyzed species and thus, we cannot estimate the real number of GRC size changes during the Lonchura radiation.
The rapid evolution of the GRC allows for the intriguing possibility that this chromosome might be involved in the establishment of reproductive isolation between passerine species. Passerines represent the most species rich avian order comprising more than half of all modern species (Fjeldså et al. 2020). Some evidence suggests that postzygotic incompatibilities causing sterility of hybrids evolve faster in passerines compared to other bird lineages (Price and Bouvier 2002), which could contribute to a higher speciation rate in this taxon. Although there is currently no empirical evidence that the GRC is associated with hybrid sterility, such a possibility is intriguing given the dynamic changes in GRC size and genetic content observed in this and previous studies (Kinsella et al. 2019; Torgasheva et al. 2019). Further studies on hybrids among different Lonchura species, which often show partial or full sterility (McCarthy 2006), could test this hypothesis. Besides postzygotic isolation, the GRC might theoretically also be involved in postmating prezygotic isolation as it could, in a species-specific manner, affect sperm or ovum characteristics, including sperm size and speed or proteins on gamete surfaces. This kind of reproductive isolation is not well understood in birds, but recent studies suggest it might be common (Cramer et al. 2016; Albrecht et al. 2019).
The GRC has been assumed to only be passed to the zygote through females. This assumption was mainly based on cytogenetic evidence of GRC elimination from spermatocytes (Pigozzi and Solari 2005; Torgasheva et al. 2019; Malinovskaya et al. 2020). However, a recent study in zebra finch showed that the GRC is not always co-inherited with maternally inherited mitochondria, suggesting the possibility of occasional paternal inheritance (Pei et al. 2022). The same study also demonstrated that a small proportion of spermatozoa carry GRC-specific sequences in their nucleus, suggesting that the elimination of the GRC during spermatogenesis may not always be successful and that the whole or partial GRC may sometimes remain in the sperm nucleus (Pei et al. 2022). Interestingly, the proportion of sperm carrying the GRC varied among different zebra finch matrilines, which suggests that certain GRC haplotypes are more likely to be inherited through sperm than others. Such haplotypes might carry genes hindering the GRC elimination during spermatogenesis, facilitating their spread in the population in a selfish manner (Pei et al. 2022). However, an evidence for such a hypothesis is lacking to date. Our analysis of F1 hybrids between Lonchura species with distinct GRC size represents one of the first direct genetic tests of the inheritance mode of the GRC and adds to the evidence that GRC is typically maternally inherited. However, given the low sample size, we cannot rule out occasional paternal inheritance in our system. Analysis of more F1 hybrids will thus be needed to test and quantify this possibility.
The GRC is typically present in a single copy in male germ cells and two copies in female germ cells, which presumably arise by duplication of a single copy inherited from the mother (Pigozzi and Solari 1998, 2005; Itoh et al. 2009; del Priore and Pigozzi 2014; Torgasheva et al. 2019). However, in some species, variability in GRC copy number also exists within the same sex (Pigozzi and Solari 2005; Malinovskaya et al. 2020) and sometimes even among different germ cells of a single individual (Malinovskaya et al. 2020; Torgasheva et al. 2021). Our results add to this evidence by demonstrating mosaicism in GRC copy number in one Lonchura species. We also show, for the first time, mosaicism in GRC size. In one individual of L. atricapilla, we found primary spermatocytes with a single macro-GRC, a single micro-GRC, one micro- and one macro-GRC, or two micro-GRCs. Analysis of expelled GRC micronuclei suggests that some germ cells in this individual might also carry two macro GRCs. For now, we can only speculate how polymorphism between individuals and mosaicism within individuals in GRC copy number and size could arise. There are, however, several mechanisms which might theoretically contribute to the origin of these phenomena.
First, occasional paternal GRC inheritance and/or inheritance of two GRC copies from the mother (as a possible consequence of female meiotic drive) could result in more than one GRC copy in the zygote and create polymorphism in GRC copy number among individuals (Malinovskaya et al. 2020; Pei et al. 2022). If unstable mitotic inheritance occurs during germline cell divisions, as has been observed for B chromosomes (Johnson Pokorná and Reifová 2021), mosaicism in GRC copy number may arise within a single individual. This may happen for example through non-disjunction of GRC sister chromatids or through chromosome lagging during mitosis, leading to the loss or gain of GRC copies in the daughter cells. Such losses and amplifications of GRC copies during the mitotic propagation of germ cells were proposed to explained mosaicism in GRC copy number in pale martin (Malinovskaya et al. 2020) and great tit (Torgasheva et al. 2021) and could also explain mosaicism in GRC copy number in L. atricapilla.
The origin of polymorphism and mosaicism in GRC size requires yet another explanation. We suggest that shorter copies of the GRC could arise in a population through fragmentation of the GRC during its elimination from spermatocytes (Schoenmakers et al. 2010) and the subsequent paternal inheritance of GRC fragments which remained in sperm cells. If fragments of the GRC contain the centromere, they could be regularly transmitted to daughter cells during mitosis as other chromosomes. Alternatively, the shorter GRC might be the result of GRC fragmentation and loss of its parts during germline mitotic divisions. Such large deletions would normally be deleterious on standard chromosomes, but given the enormous variability in GRC size, even among closely related species, it is possible that large parts of this chromosome are in fact non-essential and thus their loss might not have large effects on the fitness of the carrier. Such mutations shortening the GRC together with continuous additions of new sequences copied from standard chromosomes might be the source of the GRC size polymorphism within species which can then be translated into GRC size differences between species. Polymorphism in GRC size in L. atricapilla could also originate from interspecific introgression of differently sized GRCs between the Lonchura species as reproductive isolation between many species is still incomplete (McCarthy 2006). Evidence for interspecific introgression of another type of germline-restricted chromosome has been recently obtained in Sciarid flies (Hodson et al. 2022). Once polymorphism in GRC size exists in a species, mosaicism in GRC size within individuals could simply arise if the zygote inherits two GRCs of different sizes, which are then unstably inherited during germline mitotic divisions.
Conclusion
Our results together with previous studies indicate that the GRC is the fastest evolving avian chromosome in terms of changes in its size and genetic content. The GRC shows unexpectedly large variation in size, ranging from a tiny micro-chromosome to a very large macro-chromosome, even among very closely related species and sometimes also within the same species or even individual. This suggests that although the GRC seems to be indispensable for passerines, large parts of this chromosome might in fact be non-essential and species-specific, potentially harboring genes facilitating the spread of GRC haplotypes in a selfish manner. Given that the GRC carries many testes and ovary expressed genes, changes in the genetic content of this chromosome might have important implications for the origin of postzygotic as well as postmating prezygotic isolation in passerine birds.
Data availability
All data was submitted.
References
Albrecht T, Opletalová K, Reif J, Janoušek V, Piálek L, Cramer ERA, Johnsen A, Reifová R (2019) Sperm divergence in a passerine contact zone: indication of reinforcement at the gametic level. Evolution 73:202–213. https://doi.org/10.1111/evo.13677
Biederman MK, Nelson MM, Asalone KC, Pedersen AL, Saldanha CJ, Bracht JR (2018) Discovery of the first germline-restricted gene by subtractive transcriptomic analysis in the zebra finch, Taeniopygia guttata. Curr Biol 28:1620–1627. https://doi.org/10.1016/j.cub.2018.03.067
Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu C, Xie D, Suchard MA, Rambaut A, Drummond AJ (2014) BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput Biol 10:1–6. https://doi.org/10.1371/journal.pcbi.1003537
Camacho JPM, Sharbel TF, Beukeboom LW (2000) B-chromosome evolution. R Soc Lond B Biol Sci 355:163–178. https://doi.org/10.1098/rstb.2000.0556
Cramer ER, Ålund M, McFarlane SE, Johnsen A, Qvarnström A (2016) Females discriminate against heterospecific sperm in a natural hybrid zone. Evolution 70:1844–1855. https://doi.org/10.1111/evo.12986
Darriba D, Taboada GL, Doallo R, Posada D (2012) jModelTest 2: more models, new heuristics and high-performance computing. Nat Methods 9(8):772
Dedukh D, Krasikova A (2022) Delete and survive: strategies of programmed genetic material elimination in eukaryotes. Biol Rev Camb Philos Soc 97(1):195–216. https://doi.org/10.1111/brv.12796
Del Priore L, Pigozzi MI (2014) Histone modifications related to chromosome silencing and elimination during male meiosis in Bengalese finch. Chromosoma 123:293–302
Drummond AJ, Rambaut A (2007) BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7:214
Drummond AJ, Suchard MA, Xie D, Rambaut A (2012) Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 29:1969–1973
Ellegren H (2010) Evolutionary stasis: the stable chromosomes of birds. Trends Ecol Evol 25:283–291. https://doi.org/10.1016/j.tree.2009.12.004
Fjeldså J, Christidis L, Ericson GP (2020) The largest avian radiation the evolution of perching birds, or the order Passeriformes. Lynx, Barcelona
Goday C, Pigozzi MI (2010) Heterochromatin and histone modifications in the germline-restricted chromosome of the zebra finch undergoing elimination during spermatogenesis. Chromosoma 119:325–336
Griffin DK, Robertson LBW, Tempest HG, Skinner BM (2007) The evolution of the avian genome as revealed by comparative molecular cytogenetics. Cytogenet Genome Res 117:64–77
Hodson CN, Jaron KS, Gerbi S, Ross L (2022) Evolution of gene-rich germline restricted chromosomes in black-winged fungus gnats through introgression (Diptera: Sciaridae). PLoS Biol 20(2):e3001559. https://doi.org/10.1371/journal.pbio.3001559
Itoh Y, Kampf K, Pigozzi MI, Arnold AP (2009) Molecular cloning and characterization of the germline-restricted chromosome sequence in the zebra finch. Chromosoma 118:527–536
Johnson Pokorná M, Reifová R (2021) Evolution of B chromosomes: from dispensable parasitic chromosomes to essential genomic players. Front Genet 12:727570. https://doi.org/10.3389/fgene.2021.727570
Kinsella CM, Ruiz-Ruano FJ, Dion-Côté AM, Charles AJ, Gossmann TT, Cabrero D et al (2019) Programmed DNA elimination of germline development genes in songbirds. Nat Commun 10:1–27
Malinovskaya LP, Zadesenets KS, Karamysheva TV, Akberdina EA, Kizilova EA, Romanenko MV et al (2020) Germline-restricted chromosome (GRC) in the sand martin and the pale martin (Hirundinidae, Aves): synapsis, recombination and copy number variation. Sci Rep 10:1–10
McCarthy EM (2006) Handbook of avian hybrids of the world. Oxford University Press, New York
Nanda I, Benisch P, Fetting D, Haaf T, Schmid M (2011) Synteny conservation of chicken macrochromosomes 1–10 in different avian lineages revealed by cross-species chromosome painting. Cytogenet Genome Res 132:165–181
O’Connor RE, Kiazim L, Skinner B, Fonseka G, Joseph S, Jennings R et al (2019) Patterns of microchromosome organization remain highly conserved throughout avian evolution. Chromosoma 128:21–29
Pei Y, Forstmeier W, Ruiz-Ruano FJ, Mueller JC, Cabrero J, Camacho JPM et al (2022) Occasional paternal inheritance of the germline-restricted chromosome in songbirds. Proc Natl Acad Sci 119(4):e2103960119. https://doi.org/10.1073/pnas.2103960119
Peona V, Palacios-Gimenez OM, Blommaert J, Liu J, Haryoko T, Jønsson KA, Irestedt M et al (2021) The avian W chromosome is a refugium for endogenous retroviruses with likely effects on female-biased mutational load and genetic incompatibilities. Philos Trans R Soc B 376:20200186. https://doi.org/10.1098/rstb.2020.0186
Peters AHFM, Plug AW, Van Vugt MJ, De Boer P (1997) A drying-down technique for the spreading of mammalian meiocytes from the male and female germline. Chromosom Res 5:66–68
Pigozzi MI, Solari AJ (1998) Germ cell restriction and regular transmission of an accessory chromosome that mimics a sex body in the zebra finch, Taeniopygia guttata. Chromosom Res 6:105–113
Pigozzi MI, Solari AJ (2005) The germ-line-restricted chromosome in the zebra finch: recombination in females and elimination in males. Chromosoma 114:403–409
Poignet M, Johnson M, Altmanová M, Majtanova Z, Dedukh D, Albrecht T et al (2021) Comparison of karyotypes in two hybridizing passerine species: conserved chromosomal structure but divergence in centromeric repeats. Front Genet 12:768987. https://doi.org/10.3389/fgene.2021.768987
Price TD, Bouvier MM (2002) The evolution of F1 postzygotic incompatibilities in birds. Evolution 56:2083–2089
Rambaut A (2012) Figtree 1.4.0. http://tree.bio.ed.ac.uk/software/figtree/
Schneider C, Rasband W, Eliceiri K (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675
Schoenmakers S, Wassenaar E, Laven JSE, Grootegoed JA, Baarends WM (2010) Meiotic silencing and fragmentation of the male germline restricted chromosome in zebra finch. Chromosoma 119:311–324
Smith JJ, Timoshevskiy VA, Saraceno C (2021) Programmed DNA elimination in vertebrates. Annu Rev Anim Biosci 9:173–201
Stryjewski KF, Sorenson MD (2017) Mosaic genome evolution in a recent and rapid avian radiation. Nat Ecol Evol 1:1912–1922
Suh A, Dion-Côté AM (2021) New perspectives on the evolution of within-individual genome variation and germline/soma distinction. Genome Biol Evol 13:6–11. https://doi.org/10.1093/gbe/evab095
Torgasheva AA, Malinovskaya LP, Zadesenets KS, Karamysheva TV, Kizilova EA, Akberdina EA et al (2019) Germline-restricted chromosome (GRC) is widespread among songbirds. Proc Natl Acad Sci 116:11845–11850. https://doi.org/10.1073/pnas.1817373116
Torgasheva A, Malinovskaya LP, Zadesenets K, Shnaider E, Rubtsov N, Borodin P (2021) Germline-restricted chromosome (GRC) in female and male meiosis of the great tit (Parus major, Linnaeus, 1758). Front Genet 12:1–7. https://doi.org/10.3389/fgene.2021.768056
Wang J, Davis RE (2014) Programmed DNA elimination in multicellular organisms. Curr Opin Genet Dev 27:26–34
Zhang G, Li C, Li Q, Li B, Larkin DM, Lee C et al (2014) Comparative genomics reveals insights into avian genome evolution and adaptation. Science 346:1311–1320
Acknowledgements
We thank Alexander Suh, Anna Torgasheva, Wolfgang Forstmeier, and other members of their laboratories for very inspiring discussions about the GRC.
Funding
This research was funded by the Grant Agency of Charles University (grant 1264120 to MS-M), the Czech Science Foundation (grant 20-23794S to RR and TA), and the Charles University grant PRIMUS/19/SCI/008 to RR. The participation of DD and KJ was funded by Institutional Research Concept RVO67985904.
Author information
Authors and Affiliations
Contributions
RR, TA, and MS-M designed the experiment. TA and OK performed breeding of the Lonchura species and dissected the birds. MS-M, MP, and DD carried out the cytogenetic experiments. MS-M did the microscopic work and analyzed the photos. MS-M and KJ performed the phylogenetic analysis. MS-M, RR, MP and SS interpreted the data. MS-M and RR wrote the manuscript with feedback from all the other authors.
Corresponding authors
Ethics declarations
Ethics approval
The present work with birds followed the Czech law on protection of animals (law no. 246/1992 Sb.) and was approved by the ethical committee of the Charles University (permission no: UKPRF/28830/2021).
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sotelo-Muñoz, M., Poignet, M., Albrecht, T. et al. Germline-restricted chromosome shows remarkable variation in size among closely related passerine species. Chromosoma 131, 77–86 (2022). https://doi.org/10.1007/s00412-022-00771-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00412-022-00771-6