Abstract
Group-living can promote the evolution of adaptive strategies to prevent and control disease. Fungus-gardening ants must cope with two sets of pathogens, those that afflict the ants themselves and those of their symbiotic fungal gardens. While much research has demonstrated the impact of specialized fungal pathogens that infect ant fungus gardens, most of these studies focused on the so-called higher attine ants, which are thought to coevolve diffusely with two clades of leucocoprinaceous fungi. Relatively few studies have addressed disease ecology of lower Attini, which are thought to occasionally recruit (domesticate) novel leucocoprinaceous fungi from free-living populations; coevolution between lower-attine ants and their fungi is therefore likely weaker (or even absent) than in the higher Attini, which generally have many derived modifications. Toward understanding the disease ecology of lower-attine ants, this study (a) describes the diversity in the microfungal genus Escovopsis that naturally infect fungus gardens of the lower-attine ant Mycocepurus smithii and (b) experimentally determines the relative contributions of Escovopsis strain (a possible garden disease), M. smithii ant genotype, and fungal cultivar lineage to disease susceptibility and colony fitness. In controlled in-vivo infection laboratory experiments, we demonstrate that the susceptibility to Escovopsis infection was an outcome of ant-cultivar-Escovopsis interaction, rather than solely due to ant genotype or fungal cultivar lineage. The role of complex ant-cultivar-Escovopsis interactions suggests that switching M. smithii farmers onto novel fungus types might be a strategy to generate novel ant-fungus combinations resistant to most, but perhaps not all, Escovopsis strains circulating in a local population of this and other lower-attine ants.
Similar content being viewed by others

Avoid common mistakes on your manuscript.
Introduction
The evolution of societies, from simple aggregations to eusocial species, has been shaped by benefits and costs of group-living. Close proximity to other members of the same species, for example, can aid in defense against enemies but can also facilitate the spread of diseases and parasites between group members. In response, strategies may evolve in group-living organisms to prevent and to fight disease threats. Social insects have become popular model systems in recent years to study the epidemiology of infectious diseases [1, 2]. Members of insect societies live in close proximity, sharing food and having close body contact. Additionally, social insect societies most often consist of families, which makes them even more vulnerable to pathogens and parasites because individuals are closely related. Similar to human societies, social insects evolved various strategies to prevent and overcome outbreaks of disease, such as sanitary behavior [3, 4], corpse removal [5], exiling moribund colony members [6], early warning systems in response to pathogen exposure [7], self- and allo-grooming to remove pathogens [8], cleaning of offspring [9,10,11], vaccination-like inoculation [12], and hygienic effects of mutualistic symbionts [13,14,15,16,17,18].
Fungus-growing ants (tribe Attini) are of special interest for disease ecology because they have to cope with dual disease threats to themselves and also to their fungal garden crops. Attine ants evolved fungiculture of fungi of the family Agaricaceae (formerly Lepiotaceae) (Basidiomycota: Agaricales) [19,20,21]. The fungal cultivars are carefully tended by the ants, which are obligate dependent on their fungus as their primary food source. In return, the fungus receives shelter and protection from ants and is dispersed when the ant colonies reproduce (founding queens carry fungal inocula during nuptial flight to seed gardens in new colonies [22,23,24,25]). While most fungal symbionts are cotransmitted vertically within ant lineages, horizontal exchange of fungal symbionts between ant colonies is also possible and differs in frequency between ant lineages [20, 26,27,28]. The mechanisms underlying horizontal exchange are unknown but could include stealing of garden from neighboring colonies after garden loss [20, 29,30,31,32].
While each attine colony appears to grow only one genetic strain of fungal cultivar [33], the ants are essentially managing a consortia of microorganisms because the fungus garden contains also, embedded among the dominant live biomass of cultivated fungus, numerous transitory and resident “auxiliary microbes” [34,35,36,37,38,39,40]. In addition to a diverse bacterial community, ant fungus gardens are host to not only many microfungal species, which includes a number of “weedy” competitor species such as Fusarium, Aspergillus, Mucor, Penicillium, and Trichoderma [41,42,43,44], but also specialized garden parasites in the genus Escovopsis (Ascomycota: anamorphic Hypocreales) [42, 45]. The fungus Escovopsis is thought to consume the garden cultivar [46, 47] and appears to be largely horizontally transmitted between colonies [45, 48, 49]. In some ants in the genus Acromyrmex, the ants are thought to protect their gardens against Escovopsis with the help of antibiotic-secreting bacteria inhabiting the ant integument, which produce antibiotics with general activity against filamentous fungi, such as entomopathogens [40, 50], but the generality of the hypothesized Escovopsis-suppression by integumental bacteria is not known for attine ants at large. While most research has been focused on the so-called higher Attini (the leafcutter ant genera Atta and Acromyrmex [41, 48, 51] and the non-leaf-cutting higher-attine genus Trachymyrmex [42, 49]), relatively few studies have been conducted on the coevolutionary relationships between the so-called lower attine ants and their pathogens [52]. For example, in Cyphomyrmex longiscapus, C. muelleri, and C. costatus, the associated Escovopsis parasites are thought to be cultivar-specific because of clade-to-clade congruency between fungal cultivar and the parasite phylogenies [53, 54]. Likewise, in Apterostigma ants, closely related Escovopsis generally infect closely related fungal hosts, with occasional host-switching by Escovopsis or acquisition of new infections from unknown environmental sources [55]. Moreover, to our knowledge, only one in vivo experiment has been reported on the pathogeny of Escovopsis in one lower attine species (Apterostigma pilosum, [56]).
The lower-attine ant Mycocepurus smithii is a widespread clonal species found throughout Central and South America and many Caribbean islands [23, 57,58,59]. M. smithii has been shown to grow a diverse assemblage of fungal cultivars, including some grown by distantly related Cyphomyrmex species [20, 26]. M. smithii populations in Panamá grow fungal species from two different phylogenetic clades, with occasional horizontal switches between cultivar lineages [26]. The rarity of Actinobacteria found in culture-independent bacterial surveys of M. smithii ants and their gardens [37] suggests that Actinobacteria do not play as prominent a role in disease control as is thought to occur in some Acromyrmex species [60]. Widespread horizontal transmission and frequent de-novo recruitment from environmental sources of fungal and bacterial symbionts, as well as low specificity of symbionts apparent in phylogenetic analyses, should result in weak and diffuse coevolution among ant hosts, fungal symbionts, pathogenic fungi, and any defensive bacterial symbionts in M. smithii [20, 26, 37].
We here examine the disease ecology and coevolutionary relationships found in nature among host ants, fungal cultivar, and Escovopsis parasites associated with M. smithii. The aim of this study was (1) examine infection rates in the field and test for phylogenetic correspondences between M. smithii ants, their fungal cultivars, and associated Escovopsis lineages; (2) determine whether Escovopsis infects gardens grown by M. smithii and if the Escovopsis strains infecting M. smithii are similar to those infecting gardens grown by Cyphomyrmex muelleri, C. longiscapus, and C. costatus ants, which grow fungal cultivars from the same clades as M. smithii [20, 26, 53]; and (3) explore the factors that determine the virulence of Escovopsis infection. These results suggest that horizontal switching onto novel fungus types might be a strategy of the ants to escape the effects of pathogens.
Material and Methods
Collections, Infection Prevalence, Escovopsis Isolation, and Taxonomic Identification
Fungus garden chambers of colonies of M. smithii were collected from 11 locations in the Panama Canal area, Republic of Panamá, by excavating subterranean nests as described in Kellner et al. [26] (see also Table 1). Contents of garden chambers (ants and fungus gardens) from different chambers were collected separately, even if the garden chambers were from the same colony, and transported in a cooler to the laboratory. In total, 36 colonies with ants and gardens from 67 garden chambers were collected. Each colony was housed in the lab in two plastic boxes (7 cm × 7 cm × 2.5 cm), one with sterile plaster bottom as nest chamber and one without plaster bottom as foraging arena [26, 61]. The boxes were connected with a tube. Each of these setups were further enclosed in a covered, plastic shoebox to avoid cross-contamination between different colonies. Colonies were fed a standardized diet of sterile polenta and oats, and the plaster bottoms were regularly moistened with sterile water. Feeding and handling of colonies was performed wearing gloves, which were ethanol-sterilized between handling different colonies. After 1-week habituation to laboratory conditions, each garden was screened for the presence of Escovopsis (Ascomycota) by pulling small fragments (1–3 mm3) of clean cultivar from the fungus gardens with flame sterilized forceps, then placing these on PDA (potato dextrose agar, Difco) Petri plates (five pieces on one plate, one plate for each garden; N = 67 plates). Plates were sealed with parafilm and incubated at room temperature (about 22 °C). Plates were screened daily for the appearance of Escovopsis-like mycelia growing from garden fragments [42]. Possible Escovopsis candidates were subcultured by cutting mycelium from the growth-front and transferring the mycelial isolate onto PDA plates. Thirteen candidates with Escovopsis-like mycelium and spore-production were sequenced for the EF-1α gene (see below) to confirm the visual identification. As in Gerardo et al. [53], Escovopsis isolates were classified into “yellow-spored” and “pink-spored” morphotypes according to spore coloration.
Sequence-Identification of Escovopsis Isolates, Phylogenetic Reconstruction, and Parafit Analyses
DNA was extracted by incubating small pieces of mycelia in 100 μl 10% Chelex resin (Sigma-Aldrich) for 1.5 h at 60 °C, then 10 min at 99 °C. Sequencing targeted a 987-bp region spanning one exon of nuclear elongation factor-1 alpha (EF-1α), using primers EF1-2218R and EF1-983F and a touchdown PCR-protocol developed by Rehner and Buckley [62]. PCR products were purified and cycle-sequenced (ABI BigDye Terminator Kit; ABI PRISM 3100) at the UT Austin core facility using standard procedures (https://icmb.utexas.edu/dna-sequencing-facility). Forward and reverse sequences were assembled in Sequencher 4.6 (GeneCodes, AnnArbor, MI, USA). Sequence information is deposited at NCBI GenBank (accession numbers KX259112–KX259124).
To place the new Escovopsis isolates into a phylogenetic framework, we obtained from NCBI GenBank sequence-information of Escovopsis sp. isolated from other lower-attine ants. Specifically, we downloaded a popset of Escovopsis sequences isolated from Cyphomyrmex colonies (38 sequences; 1529-bp fragment of EF-1 alpha; GenBank accessions AY629361–AY629398 [53], a popset of Escovopsis strains isolated from Apterostigma colonies (54 sequences; 987 bp; GenBank accessions DQ848156–DQ848209 [55]), one sequence of Escovopsis kreiselii isolated from a Mycetophylax morschi colony (KJ808766 [49]), and one Escovopsis strain isolated from a Mycocepurus goeldii colony (KF033128 [63]). The alignments from Gerardo et al. [53, 55] were merged, and the two additional sequences and our new 13 Escovopsis sequences were added using McClade [64], preserving the gaps in Gerardo’s original alignments. The final alignment contained 107 sequences and was 992 bp in length. jModelTest 0.1.1 [65, 66] identified the GTR+I+G model as the most suitable model for phylogenetic analyses. A maximum-likelihood tree was computed using GARLI 0.951 [67]. Ten trees were generated from which the tree with the best likelihood score was chosen (likelihood-score of best tree: − 8884.69, 589 constant characters, 314 parsimony informative characters). Bootstrap support was evaluated in 1000 pseudoreplicate analyses.
Because information on genotypes of ants as well as fungus cultivars was available from a previous study [26], it was possible to test for clade-to-clade correspondences between (i) Escovopsis strains and fungus cultivars of the host colonies and between (ii) Escovopsis strains and ant lineages (asexual ant clones) of the host colonies. We used the Parafit function implemented in the R package “ape” [68] to test for phylogenetic congruence between phylogenies of the M. smithii ants, fungal cultivars, and Escovopsis strains. Parafit performs a test on each single observed link to assess the significance of that particular association. As input files, we used presence/absence matrices of the Escovopsis strain—ant genotypes or fungus genotypes (scored as 1 and 0, respectively), a genetic distance matrix from Kimura2P genetic distances among Escovopsis strains using Mega [69], a genetic distance matrix obtained from allele-sharing distances among ant genotypes, and a genetic distance matrix from Kimura2P genetic distances among fungus cultivars (both of these matrices were available from [26]). p values were obtained through permutations with 9999 pseudoreplicates.
Cultivation of Escovopsis and Preparation of Spore Suspensions
Two Escovopsis strains isolated from two different M. smithii nests were selected for infection experiments (strain Escovopsis sp. HF100409-03-Esco5, a pink-spore type, henceforth called strain Esco5; and Escovopsis sp. KK100413-01 ch3-Esco6, a yellow-spore type, called Esco6) (see Fig. 1). These two strains were chosen because they were distantly related within the Escovopsis diversity known to associate with lower-attine ants, because they differed in spore coloration (pink versus yellow) and because they showed vigorous spore-production on PDA plates. Escovopsis strain Esco5 may be more typically associated with M. smithii, whereas Esco6 may be less typically associated with M. smithii (see “Results” Fig. 1).

The two strains were kept in long-term storage (sterile water) at − 80 °C. Subcultures were started on sterile PDA plates. After approximately 4 weeks of incubating the plates at room temperature (first spores are produced by mycelia after 2 weeks), spores were harvested and suspended in 10 ml sterile 0.005% Tween80. To check spore viability, 5 μl of a 10× dilution of the spore suspension was applied onto a sterile PDA plate. To determine spore concentration in a suspension, 12 μl of a 100× dilution was evaluated in a spore count chamber (type Neubauer improved). The undiluted spore suspensions contained 11.6 × 10^9 spores/ml for strain HF10040903-Esco5 and 10 × 10^9 spores/ml for KK10041301ch3-Esco6. A 10× dilution of the original spore solution was filled in sterile spray bottles (Esco5 and Esco6 one spray bottle each, 11.6 × 10^8 and 10 × 10^8 spores/ml). One puff out of the spray bottle dispensed approximately 100 μl suspension. Fungus gardens in the experimental series below were inoculated by spraying one puff directly onto the garden surface, resulting in infection doses of 11.6 × 10^7 spores in one puff for Esco5 and 10 × 10^7 spores in one puff for Esco6. Both final infection doses have been shown to be in or above lethal range for attine gardens used in prior infection experiments (6000–100,000 spores [41, 51, 56, 70]). To confirm viability of spores in solution, a single puff of each strain was sprayed onto sterile PDA plates, and growth was confirmed in both cases.
Infection Experiments
Infection experiments were performed in vivo using ants and fungus cultivars from colonies with known genetic backgrounds. Ant lineages (A–J) are based on microsatellite genotyping and fungal cultivars (1–7) are based on ITS sequences (details are described in [26]). A cophylogenetic tree illustrates the relationships between ant and fungal lineages (details in [26] and see Fig. 2a).
-
1.
Infection of garden fragments not tended by ants: Fungus-garden fragments of approximately 3 cm3 were placed in Petri dishes with moistened plaster bottoms (UV sterilized) using flame sterilized forceps. Fungus-garden fragments were obtained from different colonies growing different fungus cultivar genotypes (cultivar types 2, 5, 6, and 7, as defined by [26]). Cultivar types 2 and 5 belong to the lower-attine fungus clade 1 (closely related to fungal cultivars grown by C. longiscapus) and cultivar types 6 and 7 to fungus clade 2 (closely related to fungal cultivars grown by C. muelleri) (details in [26], and see Fig. 1). Fragments (n = 87) were randomly assigned to treatment and control groups and sprayed-inoculated with either one puff of Escovopsis strain Esco5, strain Esco6, or control (sterile 0.005% Tween80). Sample sizes consisted of ten replicates of each cultivar type (5, 6, or 7) sprayed with either Esco5 or Esco6 and five replicates of cultivar type 2 sprayed with either Esco5 or Esco6, respectively. Only five replicates were possible for cultivar 2 because less garden material was available in the source nest. Control groups were five replicates each for cultivars 5, 6, 7, and 2 replicates for cultivar 2. All replicates were spray-inoculated on the same day and monitored daily for 10 consecutive days. Escovopsis infection was determined by visual identification of Escovopsis mycelia growing from the sprayed garden fragment (see Fig. 3a and Table 2). All scoring was performed blindly and without knowledge of treatment or fungus genotype [71].
-
2.
Infection of garden fragments tended by different numbers of ants: Fungus-garden fragments from two different colonies: A5 colonies (ants from ant lineage “A” and fungus from fungal cultivar lineage 5) and E6 colonies (lineage E ants growing fungus from fungal cultivar lineage 6) the same size (3 cm3) were placed in Petri dishes with moistened plaster bottoms (UV sterilized) using sterile forceps. Ants were added onto the garden fragments in three experimental groups: ten ants, five ants, and two ants per garden fragment. The three groups were sprayed with one puff spore solution with either Esco5 (five replicates each for cohorts of ten ants; ten replicates each for cohorts of five and two ants), Esco6 (five replicates each for cohorts of ten ants; ten replicates for cohorts of five and two ants), or with one puff sterile 0.005% Tween80 only (control groups) (five replicates each for cohorts of ten, five, and two ants). All replicates were sprayed on the same day and monitored daily for 10 consecutive days. After 10 days, small pieces of each experimental colony were transferred onto sterile PDA plates with sterile forceps and scored for Escovopsis mycelia growing from garden fragments (Fig. 3b). All scoring was performed blindly and without knowledge of treatment [71].
-
3.
Effect of infection on garden mass loss: Subcolonies were obtained from taking fungus garden fragments of approximately the same size (3 cm3) along with five worker ants from five different colonies (composed of the following ant-fungal combinations G7, E6, J5, A5, B5, see [26]) were placed in Petri dishes with moistened plaster bottoms (UV sterilized) using sterile forceps. Subcolonies received either one of three treatments: Spray-inoculation with Esco5 (5 replicates each), with Esco6 (5 replicates each), and control spraying with sterile 0.005% Tween80 (3 replicates each). Thus, there were five ant-fungal combinations, two manipulations (Escovopsis lineage), and a control. All replicates were monitored daily for 10 consecutive days. After 10 days, small pieces of each experimental garden were transferred onto PDA plates with sterile forceps and scored for the appearance of Escovopsis mycelia. Fresh weights of gardens were measured before and after the experiment, and the loss of garden mass was calculated as percentage of the original weight. Data analysis was conducted using two-way ANOVA with ant-fungal combination and Escovopsis lineage each as main effects. Post hoc comparisons were conducted with Tukey’s test. All measurements and scoring were performed blindly and without knowledge of treatment or ant × fungus combination [71]. To meet parametric assumptions, data were arc sine transformed because they were percentages. All statistical analyses were performed in R Version 3.1.3 [72].
Cophylogenetic trees illustrating the symbiotic relationships between a lineages of the clonal fungus-farming ant Mycocepurus smithii and its fungal cultivars; b lineages of M. smithii; and c fungal cultivars and the different isolated Escovopsis strains. Lines connecting the cophylogenies illustrate associations observed in the field; line thickness corresponds to statistical significance of the association as inferred in Parafit analysis. Phylogenetic trees are based on microsatellite genotypes for the ant lineages (A–K), ITS sequences for the fungal cultivar lineages (1–9) (details in [26]), and EF1-α sequences for the Escovopsis lineages (Esco1–EscoA). Ant and fungal cophylogenies have been published previously [26]. The linkages suggest that ants from lineage G, fungus from cultivar lineage 7 and strain Escovopsis 5 have a coevolutionary history
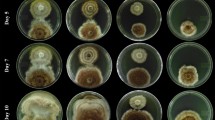
a Escovopsis mycelial growth and spore production of experimentally inoculated garden fragments that are not tended by ants. Left: Escovopsis strain Esco5 pink spore type. Middle: Escovopsis strain Esco6 yellow spore type. Right: Escovopsis strain Esco6 yellow spore type at a younger stage (spores still white and not yet yellow). b Escovopsis infection of garden fragments tended by ants. Top row, left: Diseased garden tended by two ants, overgrown with Escovopsis strain Esco5 mycelia. Top row, right: Fungus garden tended by five ants, Escovopsis strain Esco5 mycelia emerge from refuse pile at top edge of dish, while the garden itself is maintained healthy by the ants. Bottom: Healthy garden tended by ten ants, spray-inoculated with spore solution of Escovopsis strain Esco5, no visible signs of infection
Results
Infection Rate in the Field
A total of 27 isolates were sequenced, and 13 of these were identified by NCBI-BLAST search as Escovopsis sp. and the remaining isolates as Fusarium (n = 6) and Trichoderma (n = 3) or unspecified (n = 5). The 13 Escovopsis sp. had been isolated from 13 different gardens from 13 colonies (i.e., we did not find any double-infection with two different Escovopsis isolates from the same garden) (see Table 1). Because we had sampled 67 gardens, the estimated minimum infection rate of field colonies by Escovopsis is 36% (13 out of 67 gardens). More than one Escovopsis strain was found at several collection locations, with the Gamboa/Apartment 183 location yielding four different strains (see Table 1). Other collection locations in the Panama Canal region (e.g., Beach, Pipeline Road, Achiote, and Gatun, see details in [26]) did not yield any Escovopsis, but it is unclear whether M. smithii gardens at these sites were free of Escovopsis, whether these sites were undersampled, or whether Escovopsis strains prevalent at these sites are more difficult to isolate. For details on all sample locations see [26].
Phylogenetic Affinities of Escovopsis Isolates
A maximum likelihood analysis (Fig. 1) shows that the Escovopsis strains isolated from M. smithii gardens are closely related to strains from C. longiscapus and C. muelleri ants [53], with the Escovopsis strains from the clade 1 and clade 2 fungus groupings corresponding to the respective groupings known for the two Cyphomyrmex species (Fig. 1; see also [20, 26, 27]). Most of the Escovopsis strains (85%, 11 of 13 total) belonged to the pink-spored morphotype, which was also the most frequent morphotype isolated by Gerardo et al. [53] from Cyphomyrmex gardens. Only two of the isolates were visually categorized as yellow-spored morphotype (strains Esco6 and -7), and phylogenetic analyses placed these two into the yellow-spored clade of Escovopsis previously described for Apterostigma gardens ([53, 55]; these Apterostigma grow a phylogenetically very distinct fungal cultivar than M. smithii). As illustrated in Fig. 1, pink-spored Escovopsis strains were isolated from M. smithii gardens of both fungal cultivar clades 1 and 2.
The ParaFitGlobal test, with ant lineages defined as hosts and Escovopsis strains as parasites, was overall significant (ParaFitGlobal = 0.0052, p = 0.016), indicating that observed clade-to-clade correspondences between ant and Escovopsis phylogenies are either due to a coevolutionary history or clade-specific de-novo acquisition and/or persistence of ant-Escovopsis associations. However, only three out of 13 individual association links were significant (Fig. 2b: G ants—Esco5, p = 0.005, G ants—Esco1, p = 0.001, G ants—Esco3, p = 0.021), suggesting that these links represent a coevolutionary history. In contrast, the ParaFitGlobal test for fungus lineages defined as hosts and Escovopsis strains as parasites was overall not significant (ParaFitGlobal = 0.00044, p = 0.0683), indicating no overall coevolutionary history between fungal cultivar lineages and Escovopsis strains. However, four of the 13 host-parasite links gave significant results (Fig. 2c: fungus 6—Esco2, p = 0.035; fungus 7—Esco5, p = 0.006; fungus 7—Esco1, p = 0.003; fungus 7—Esco3, p = 0.030), indicating that these links could be coevolutionary associations in the otherwise overall random association between the fungal cultivar phylogeny and the parasite Escovopsis. Significant association patterns between ants × fungus, ants × Escovopsis, and fungus × Escovopsis (Fig. 2) indicate coevolutionary history between G ants, fungus 7, and Escovopsis strain Esco1, -3, and -5.
Infection Experiments
-
1.
Infection of garden fragment not tended by ants: The earliest signs of infection were observed on day 2 in replicates of the cultivar 2 × Esco5 combination. Signs of infection were scored on day 7 in the cultivar 6 × Esco6 and cultivar 6 × Esco5 combinations in five and seven replicates, respectively. One hundred percent infection rate was reached after 7 days in cultivar 7 × Esco5 and Esco6 combinations and after 8 days in cultivar 5 × Esco5 and Esco6 combinations. The slowest rate of infection was recorded in cultivar 6 × Esco5 combination, where two replicates remained uninfected after 10 days. All control replicates (sprayed with sterile 0.005% Tween80 only) showed no sign of infection (see Fig. 3a and Table 2).
-
2.
Effect of worker number on Escovopsis infection of fungal garden fragments: (i) Cohort of ten ants: On day 1, all replicates except the controls started to accumulate refuse piles. Ant workers were observed grooming the gardens, removing pieces of fungus, and adding them onto refuse piles located at the edges of the Petri dishes (Fig. 3b). While refuse-piles sizes increased and garden sizes decreased, no visible signs of infection or illness were observed during the experiment. Escovopsis growth appeared on PDA plates from all replicates except in the ant × fungus × Escovopsis combination A × 5 × Esco5. (ii) Cohort of five ants: Similar to the ten-ant cohorts, all replicates except the controls started to make refuse depots. While infection of fungus gardens was observed only in a few cases, Escovopsis mycelia was observed to grow on all refuse piles. Escovopsis mycelia growth appeared on PDA plates from all replicates except controls. (iii) Cohort of two ants: All replicates except the controls started to accumulate refuse piles. Infection of fungus gardens was observed in all replicates of all ant × fungus × Escovopsis combinations on days 2 and 3. By day 10, Escovopsis mycelium had taken over each garden fragment and ant workers had abandoned the gardens. Escovopsis mycelium appeared on PDA plates from all replicates except the controls (see Fig. 3b and Table 3).
-
3.
Effect of ant-fungus-Escovopsis combinations on garden mass loss: Similar to the outcome of the five-ant cohort experiment, refuse piles accumulated in all Escovopsis-treated replicates, but none of the Escovopsis-treated and control replicates had any visible signs of Escovopsis mycelia growth or disease outbreak on the fungus gardens or on refuse piles. Escovopsis growth was found on all PDA subculture tests except for the controls plates (experimental gardens were subcultured on day 10), confirming presence of viable spores in the Escovopsis-treated gardens during the experiment. Although no visible signs of infection were found, Escovopsis treatment led to significant losses in garden weights. Escovopsis strain Esco5 appeared to be the more virulent strain because experimental replicates treated with Escovopsis strain Esco5 lost significantly more garden weight than replicates treated with strain Esco6 or the control group (F 2,50 = 9.76, p < 0.0001, Fig. 4). The main effect of ant-fungal combination was marginally significant with J5 colonies experiencing more weight loss than B5 colonies (F 4,50 = 2.74, p = 0.04) (Fig. 5). Additionally, we found evidence of synergistic (interaction) effects: Escovopsis treatments interacted significantly with ant-fungal combination of M. smithii colonies, such that G7 colonies exposed to Escovopsis 5 lost more garden weight (F 8,50 = 2.2, p = 0.04, Fig. 5) than G7 colonies exposed to Escovopsis 6 or the control. G7 colonies exposed to Escovopsis 5 also lost more weight than E6 colonies exposed to Escovopsis 6 and B5 colonies exposed to Escovopsis 5.
Median weight loss of fungus gardens as a function of Escovopsis treatment. Garden weight loss is measured as a percentage of the start weight (the weight at the end of the experiment relative to the original weight). Treatment with Escovopsis strain Esco5 caused significantly greater loss than treatment with strain Esco6 or the control treatment (sterile 0.005% Tween80) (one-way ANOVA, F 2,62 = 7.66, p = 0.001, Tukey HSD test, p < 0.05). Different letters correspond to significant differences (α = 0.05). Garden weight loss is measured as a percentage of the start weight (the weight at the end of the experiment relative to the original weight). Boxplots correspond to first and third quartiles; data beyond the whiskers are outliers and plotted as points
Median weight loss of fungus gardens as a function of synergistic interactions between a Escovopsis lineages and ant fungal combinations (subcolonies composed of naturally occurring combinations of ants and fungi). The significant interaction term appeared to be driven by in that G7 ants exposed to Escovopsis lineage 5 lost more weight than G7 colonies exposed to Escovopsis 6 and E6 colonies growing Escovopsis 6 or B5 colonies growing Escovopsis 5 (F 8,50 = 2.2, Tukey’s HSD test, p = 0.04*). Garden weight loss is measured as the percentage of the weight at the end of the experiment relative to the original weight. Boxplots correspond to first and third quartiles; data beyond the boxplot-whiskers are plotted as points
Discussion
The goals of this study were to (a) describe the Escovopsis lineages that naturally infect M. smithii gardens and (b) experimentally determine the relative contributions of experimental Escovopsis infection, M. smithii ants, and fungal cultivar lineages to disease susceptibility and ant colony fitness. We did not find evidence for universally Escovopsis-resistant or universally Escovopsis-susceptible cultivar lineages or ant genotypes. Rather, colony fitness reduction after Escovopsis infection (i.e., Escovopsis virulence) was dependent on unique ant-cultivar-Escovopsis interactions, such that specific ant-cultivar combinations were susceptible to specific Escovopsis infection.
Within the larger phylogenetic context of Escovopsis strains isolated from other lower-attine ant species, we found no Escovopsis clade specifically associated only with M. smithii colonies (Fig. 1). The Escovopsis types isolated in our survey are known to associate generally with typical lower-attine clade 1 and clade 2 cultivars, such as the fungi grown by mycelium-cultivating Cyphomyrmex ants, among diverse other lower-attine ants (Fig. 1) [20, 26, 42, 53, 55, 73, 74]. For example, two of the Escovopsis strains isolated here were found to be similar to those isolated from Apterostigma ants (Fig. 1, [55]), and another Escovopsis strain is related to an isolate from a garden of Brazilian M. goeldii [63], a species closely related to M. smithii (see Fig. S1 in [58]). The fungus-cultivar and Escovopsis types associated with M. smithii are therefore closely related to those known from other lower-attine ants. This suggests that Escovopsis might be more specialized on certain fungus-cultivars rather than the ant species which tend these gardens. A similar result has been found in Cyphomyrmex ants, where an Escovopsis phylogeny was shown to match with a cultivar phylogeny but not with the corresponding ant-host phylogeny [53].
Notably, the infection rate of Escovopsis in the Panamá Canal area was lower in the field (35%) than the rates reported for other lower-attine ants (up to 60% in Cyphomyrmex [53], up to 67% in Apterostigma [55], and in higher-attine ants: 33–51% across five genera [75]). Although we collected in total 67 colony chambers and isolated Escovopsis from 13 of these, we did not find any gardens that were visually diseased (i.e., overgrown with Escovopsis). Other parasitic fungi, like Trichoderma and Fusarium, were also easily isolated from garden fragments (KK unpublished data), which suggests that Escovopsis is one of many weedy or competitor fungi that can be present as spores or mycelium in gardens, as reported previously for attine gardens [44, 76, 77].
Our Parafit analyses revealed significant association patterns between ant genotype G and three Escovopsis lineages (Fig. 2). While most host-parasite association links seem to be not supported statistically, non-random associations were found among Escovopsis strains 1, 3, and 5 and colonies with ant/fungus combinations G7, which therefore could reflect more specific coevolutionary interactions. Escovopsis strain Esco5 used in our infection experiments was isolated from a colony with ant-fungal combination G7, which is also the most common combination found in our field population (not influenced by sampling efforts, see [26] for rarefaction analysis). Interestingly, Esco5 also appeared to be more virulent toward G7 colonies than Esco6. It has been shown that Escovopsis strains isolated from fungus gardens of higher attines differ in their virulence, with certain strains being more aggressive than others [78, 79].
While other investigations used in vitro bioassays to study the pathology of Escovopsis in lower-attine ants (e.g., testing growth and infection behavior on culture plates [53, 73]), to our knowledge, our study is the first using in vivo whole-colony experiments to investigate pathology of Escovopsis within controlled ant genotype and cultivar genotype backgrounds of lower-attine ants. We found significant interaction effects on garden mass loss among ant-fungal combinations and Escovopsis lineages. The overall pattern suggests that colonies are more susceptible to a parasite with a longer coevolutionary history than a more distantly related (foreign) parasite, which is similar to what Gerardo et al. found in Cyphomyrmex ants and their cultivars [53, 54]. Our experiments illustrated that certain combinations (e.g., G7 colonies × Esco5) cause the severest virulence as measured as the most significant garden weight loss. Additionally, our experiments illustrated that, in the absence of the ants, the fungus garden itself has little defense against viable Escovopsis spore infection. On the other hand, in the presence of ants, the infection can be contained. Interestingly, the ratio between worker number and garden size plays an important role: while two ants were insufficient to protect a garden fragment against Escovopsis in our experiment, ten ants were adequate to prevent a similarly sized garden fragment from becoming overgrown with Escovopsis mycelia (Table 3). Five ants were sufficient to physically remove spores and infected garden parts to prevent outbreak of infection on the garden, even though we observed growth of Escovopsis mycelia in the refuse piles. This suggests that M. smithii ants can remove pathogenic spores and mycelia from the gardens by grooming and weeding and in addition might suppress Escovopsis growth with chemical substances from the ants or ant-associated microbial symbionts. Consequently, stress from a garden size: worker number ratio imbalance may be the main reason why workers sometimes cannot control disease outbreaks, as has been hypothesized previously [80,81,82]. As a result, catastrophic infection by Escovopsis might be rare in natural gardens that are attended by an adequate number of workers. Escovopsis outbreaks may be more likely in nests that experience drastic worker loss, for example, as result of an entomopathogen epidemic or attack by nest-raiding ants or when maintained under stressed conditions in the laboratory.
Occasional horizontal exchange of fungal cultivars reported by Kellner et al. [26], as well as de novo domestication from free-living populations of potential cultivars, may generate novel ant-fungal combinations that are less susceptible to Escovopsis attack. Horizontal switching of ants onto new fungal cultivars (i.e., dependence on a new crop) might therefore be a mechanism to evade diseases through an analog of crop rotation in human agriculture. This symbiont-reassociation mechanism could be very important in a symbiosis that is built on low genetic diversity resulting from clonal ant hosts growing clonal fungal cultivars, perhaps making this symbiosis more vulnerable to diseases than the typical, sexually-reproducing attine host.
References
Kappeler PM, Cremer S, Nunn CL (2015) Sociality and health: impacts of sociality on disease susceptibility and transmission in animal and human societies. Philos T R Soc B 370(1669):20140116. https://doi.org/10.1098/rstb.2014.0116
Stroeymeyt N, Casillas-Pérez B, Cremer S (2014) Organisational immunity in social insects. Current Opinion in Insect Science 5:1–15. https://doi.org/10.1016/j.cois.2014.09.001
Czaczkes TJ, Heinze J, Ruther J (2015) Nest etiquette—where ants go when nature calls. PLoS One 10(2):e0118376. https://doi.org/10.1371/journal.pone.0118376
Richard FJ, Errard C (2009) Hygienic behavior, liquid-foraging, and trophallaxis in the leaf-cutting ants, Acromyrmex subterraneus and Acromyrmex octospinosus. Journal of insect science (Online) 9:1–9. https://doi.org/10.1673/031.009.6301
Sun Q, Zhou X (2013) Corpse management in social insects. Int. J. Biol. Sci. 9(3):313
Heinze J, Walter B (2010) Moribund ants leave their nests to die in social isolation. Curr Biol 20(3):249–252
Rosengaus R, Jordan C, Lefebvre M, Traniello J (1999) Pathogen alarm behavior in a termite: a new form of communication in social insects. Naturwissenschaften 86(11):544–548
Theis FJ, Ugelvig LV, Marr C, Cremer S (2015) Opposing effects of allogrooming on disease transmission in ant societies. Philos T R Soc B 370(1669):20140108. https://doi.org/10.1098/rstb.2014.0108
Tragust S, Mitteregger B, Barone V, Konrad M, Ugelvig Line V, Cremer S (2013) Ants disinfect fungus-exposed brood by oral uptake and spread of their poison. Curr. Biol. 23(1):76–82. https://doi.org/10.1016/j.cub.2012.11.034
Tragust S, Ugelvig LV, Chapuisat M, Heinze J, Cremer S (2013) Pupal cocoons affect sanitary brood care and limit fungal infections in ant colonies. BMC Evol. Biol. 13(1):225
Westhus C, Ugelvig LV, Tourdot E, Heinze J, Doums C, Cremer S (2014) Increased grooming after repeated brood care provides sanitary benefits in a clonal ant. Behav Ecol. Soc. 68(10):1701–1710
Konrad M, Vyleta ML, Theis FJ, Stock M, Tragust S, Klatt M, Drescher V, Marr C, Ugelvig LV, Cremer S (2012) Social transfer of pathogenic fungus promotes active immunisation in ant colonies. PLoS Biol. 10(4):e1001300
Kaltenpoth M (2009) Actinobacteria as mutualists: general healthcare for insects? Trends Microbiol. 17(12):529–535. https://doi.org/10.1016/j.tim.2009.09.006
Kaltenpoth M, Engl T (2013) Defensive microbial symbionts in Hymenoptera. Funct. Ecol. 28(2):315–327
Biani NB, Mueller UG, Wcislo WT (2009) Cleaner mites: sanitary mutualism in the miniature ecosystem of neotropical bee nests. Am. Nat. 173(6):841–847
Evans JD, Armstrong T-N (2006) Antagonistic interactions between honey bee bacterial symbionts and implications for disease. BMC Ecol. 6(1):1
Forsgren E, Olofsson TC, Vásquez A, Fries I (2010) Novel lactic acid bacteria inhibiting Paenibacillus larvae in honey bee larvae. Apidologie 41(1):99–108
Gerardo NM, Parker BJ (2014) Mechanisms of symbiont-conferred protection against natural enemies: an ecological and evolutionary framework. Current Opinion in Insect Science 4:8–14
Chapela IH, Rehner SA, Schultz TR, Mueller UG (1994) Evolutionary history of the symbiosis between fungus-growing ants and their fungi. Science 266:1691–1694
Mueller UG, Rehner SA, Schultz TR (1998) The evolution of agriculture in ants. Science 281:2034–2038
Schultz TR, Sosa-Calvo J, Brady SG, Lopes CT, Mueller UG, Bacci Jr M, Vasconcelos HL (2015) The most relictual fungus-farming ant species cultivates the most recently evolved and highly domesticated fungal symbiont species. Am. Nat. 185(5):693–703
Fernández-Marín H, Zimmermann JK, Wcislo WT (2004) Ecological traits and evolutionary sequence of nest establishment in fungus-growing ants (Hymenoptera, Formicidae, Attini). Biol. J. Linn. Soc. 81:39–48
Fernández-Marín H, Zimmermann JK, Wcislo WT, Rehner SA (2005) Colony foundation, nest architecture and demography of a basal fungus-growing ant, Mycocepurus smithii (Hymenoptera, Formicidae). J. Nat. Hist. 39:1735–1743
Seal JN, Tschinkel WR (2007) Energetics of newly-mated queens and colony founding in the fungus- gardening ants Cyphomyrmex rimosus and Trachymyrmex septentrionalis (Hymenoptera: Formicidae). Physiol. Entomol. 32(1):8–15
Huber J (1905) Über die Koloniegründung bei Atta sexdens. L Biol Zentralbl 25:606–619 624-635
Kellner K, Fernandez-Marin H, Ishak HD, Sen R, Linksvayer TA, Mueller UG (2013) Co-evolutionary patterns and diversification of ant-fungus associations in the asexual fungus-farming ant Mycocepurus smithii in Panama. J. Evol. Biol. 26(6):1353–1362. https://doi.org/10.1111/jeb.12140
Mehdiabadi NJ, Mueller UG, Brady SG, Himler AG, Schultz TR (2012) Symbiont fidelity and the origin of species in fungus-growing ants. Nature Communications 3:840
Mueller UG, Mikheyev AS, Solomon SE, Cooper M (2011) Frontier mutualism: coevolutionary patterns at the northern range limit of the leaf-cutter ant–fungus symbiosis. Proc. R. Soc. B 278:3050–3059
Adams RM, Mueller UG, Schultz TR, Norden B (2000) Agro-predation: usurpation of attine fungus gardens by Megalomyrmex ants. Naturwissenschaften 87(12):549–554
Adams RM, Shah K, Antonov LD, Mueller UG (2012) Fitness consequences of nest infiltration by the mutualist-exploiter Megalomyrmex adamsae. Ecol Entomol 37(6):453–462
Adams RMM, Mueller UG, Holloway AK, Green AM, Narozniak J (2000) Garden sharing and garden stealing in fungus-growing ants. Naturwissenschaften 87:491–493
Green AM, Mueller UG, Adams RMM (2002) Extensive exchange of fungal cultivars between sympatric species of fungus-growing ants. Mol. Ecol. 11:191–195
Mueller UG, Scott JJ, Ishak HD, Cooper M, Rodrigues A (2010) Monoculture of leafcutter ant gardens. PLoS One 5:e12668
Bacci MJ, Anversa MM, Pagnocca FC (1995) Cellulose degradation byLeucocoprinus gongylophorus, the fungus cultured by the leaf-cutting ant Atta sexdens rubropilosa. Anto Leeuw 67(4):385–386
Barke J, Seipke R, Gruschow S, Heavens D, Drou N, Bibb M, Goss R, Yu D, Hutchings M (2010) A mixed community of actinomycetes produce multiple antibiotics for the fungus farming ant Acromyrmex octospinosus. BMC Biol. 8(1):109
Ishak HD, Miller JL, Sen R, Dowd SE, Meyer E, Mueller UG (2011) Microbiomes of ant castes implicate new microbial roles in the fungus-growing ant Trachymyrmex septentrionalis. Sci. Rep. 1:204
Kellner K, Ishak HD, Linksvayer TA, Mueller UG (2015) Bacterial community composition and diversity in an ancestral ant fungus symbiosis. Fems Microbiol Ecol. https://doi.org/10.1093/femsec/fiv073
Pinto-Tomás AA, Anderson MA, Suen G, Stevenson DM, Chu FST, Cleland WW, Weimer PJ, Currie CR (2009) Symbiotic nitrogen fixation in the fungus gardens of leaf-cutter ants. Science 326(5956):1120–1123. https://doi.org/10.1126/science.1173036
Rodrigues A, Mueller UG, Ishak HD, Bacci Jr M, Pagnocca FC (2011) Ecology of microfungal communities in gardens of fungus-growing ants (Hymenoptera: Formicidae): a year-long survey of three species of attine ants in Central Texas. FEMS Microbiol. Ecol. 78(2):244–255. https://doi.org/10.1111/j.1574-6941.2011.01152.x
Sen R, Ishak H, Estrada D, Dowd S, Hong E, Mueller U (2009) Generalized antifungal activity and 454-screening of Pseudonocardia and Amycolatopsis bacteria in nests of fungus-growing ants. Proc. Natl. Acad. Sci. U. S. A. 106:17805–17810
Currie CR, Stuart AE (2001) Weeding and grooming of pathogens in agriculture by ants. P Roy Soc B-Biol Sci 268(1471):1033–1039
Meirelles LA, Solomon SE, Bacci M, Wright AM, Mueller UG, Rodrigues A (2015) Shared Escovopsis parasites between leaf-cutting and non-leaf-cutting ants in the higher attine fungus-growing ant symbiosis. Royal Society Open Science 2(9):150257
Möller AFW (1893) DiePilzgärten einiger südamerikanischer. Ameisen. Gustav Fischer, Jena
Rodrigues A, Bacci M, Mueller U, Ortiz A, Pagnocca F (2008) Microfungal ‘weeds’ in the leafcutter ant symbiosis. Microb. Ecol. 56:604–614
Currie C, Scott J, Summerbell R, Malloch D (1999) Fungus-growing ants use antibiotic-producing bacteria to control garden parasites. Nature 398:701–705
Reynolds HT, Currie CR (2004) Pathogenicity of Escovopsis weberi: the parasite of the attine ant-microbe symbiosis directly consumes the ant-cultivated fungus. Mycologia 96(5):955–959
de Man TJ, Stajich JE, Kubicek CP, Teiling C, Chenthamara K, Atanasova L, Druzhinina IS, Levenkova N, Birnbaum SS, Barribeau SM, Bozick BA, Suen G, Currie CR, Gerardo NM (2016) Small genome of the fungus Escovopsis weberi, a specialized disease agent of ant agriculture. Proc. Natl. Acad. Sci. U. S. A. 113(13):3567–3572. https://doi.org/10.1073/pnas.1518501113
Currie CR, Wong B, Stuart AE, Schultz TR, Rehner SA, Mueller UG, Sung G-H, Spatafora JW, Straus NA (2003) Ancient tripartite coevolution in the attine ant-microbe symbiosis. Science 299(5605):386–388. https://doi.org/10.1126/science.1078155
Meirelles LA, Montoya QV, Solomon SE, Rodrigues A (2015) New light on the systematics of fungi associated with attine ant gardens and the description of Escovopsis kreiselii sp. nov. PLoS One 10(1):e0112067. https://doi.org/10.1371/journal.pone.0112067
Mattoso TC, Moreira DD, Samuels RI (2012) Symbiotic bacteria on the cuticle of the leaf-cutting ant Acromyrmex subterraneus subterraneus protect workers from attack by entomopathogenic fungi. Biol. Lett. 8(3):461–464. https://doi.org/10.1098/rsbl.2011.0963
Little AEF, Murakami T, Mueller UG, Currie CR (2006) Defending against parasites: fungus-growing ants combine specialized behaviours and microbial symbionts to protect their fungus gardens. Biol. Lett. 2:12–16
Schultz TR, Brady SG (2008) Major evolutionary transitions in ant agriculture. Proc. Natl. Acad. Sci. U. S. A. 105:5435–5440
Gerardo N, Mueller U, Price S, Currie C (2004) Exploiting a mutualism: parasite specialization on cultivars within the fungus-growing ant symbiosis. P Roy Soc B-Biol Sci 271:1791–1798
Birnbaum SSL, Gerardo NM (2016) Patterns of specificity of the pathogen Escovopsis across the fungus-growing ant symbiosis. Am. Nat. 188(1):52–65. https://doi.org/10.1086/686911
Gerardo NM, Mueller UG, Currie CR (2006) Complex host-pathogen coevolution in the Apterostigma fungus-growing ant-microbe symbiosis. BMC Evol. Biol. 6(1):88
Little AE, Currie CR (2008) Black yeast symbionts compromise the efficiency of antibiotic defenses in fungus-growing ants. Ecology 89(5):1216–1222
Himler AG, Caldera EJ, Baer B, Fernández-Marín H, Mueller UG (2009) No sex in fungus-farming ants or their crops. P Roy Soc B-Biol Sci 276:2611–2616
Rabeling C, Gonzales O, Schultz TR, Bacci MJ, Garcia MVB, Verhaagh M, Ishak HD, Mueller UG (2011) Cryptic sexual populations account for genetic diversity and ecological success in widely distributed, asexual fungus-growing ant. P Natl Acad Sci USA 108:12366–12371
Rabeling C, Lino-Neto J, Cappellari SC, Dos-Santos IA, Mueller UG, Bacci MJ (2009) Thelytokous parthenogenesis in the fungus-gardening ant Mycocepurus smithii (Hymenoptera: Formicidae). PLoS One 4:e6781
Poulsen M, Bot ANM, Currie CR, Nielsen MG, Boomsma JJ (2003) Within-colony transmission and the cost of a mutualistic bacterium in the leaf-cutting ant Acromyrmex octospinosus. Funct. Ecol. 17(2):260–269. https://doi.org/10.1046/j.1365-2435.2003.00726.x
Sosa-Calvo J, Jesovnik A, Okonski E, Schultz TR (2015) Locating, collecting, and maintaining colonies of fungus-farming ants (Hymenoptera: Myrmicinae: Attini). Sociobiology 62(2):300–320
Rehner SA, Buckley E (2005) A Beauveria phylogeny inferred from nuclear ITS and EF1-alpha sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 97(1):84–98
Masiulionis VE, Cabello MN, Seifert KA, Rodrigues A, Pagnocca FC (2015) Escovopsis trichodermoides sp. nov., isolated from a nest of the lower attine ant Mycocepurus goeldii. Antonie Van Leeuwenhoek 107(3):731–740. https://doi.org/10.1007/s10482-014-0367-1
Maddison DR, Maddison WP (2000) MacClade4: analysis of phylogeny and character evolution, 4.0. edn. Sinauer Associates, Sunderland
Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696–704
Posada D (2008) jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 25:1253–1256
Zwickl DJ (2006) Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. Ph.D. dissertation, The University of Texas at Austin, Austin
Paradis E, Claude J, Strimmer K (2004) APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20:289–290
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739
Currie C, Bot A, Boomsma JJ (2003) Experimental evidence of a tripartite mutualism: bacteria protect ant fungus gardens from specialized parasites. Oikos 101(1):91–102
Kardish MR, Mueller UG, Amador-Vargas S, Dietrich EI, Ma R, Barrett B, Fang C-C (2015) Blind trust in unblinded observation in ecology, evolution, and behavior. Front Ecol Evol 3:51. https://doi.org/10.3389/fevo.2015.00051
Team RC (2015) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
Gerardo NM, Caldera EJ (2007) Labile associations between fungus-growing ant cultivars and their garden pathogens. ISME J 1(5):373–384
Vo TL, Mueller UG, Mikheyev AS (2009) Free-living fungal symbionts (Lepiotaceae) of fungus-growing ants (Attini: Formicidae). Mycologia 101(2):206–210
Currie CR, Mueller UG, Malloch D (1999) The agricultural pathology of ant fungus gardens. Proc. Natl. Acad. Sci. U. S. A. 96(14):7998–8002
Augustin JO, Groenewald JZ, Nascimento RJ, Mizubuti ESG, Barreto RW, Elliot SL, Evans HC (2013) Yet more “weeds” in the garden: fungal novelties from nests of leaf-cutting ants. PLoS One 8(12):e82265. https://doi.org/10.1371/journal.pone.0082265
Montoya QV, Meirelles LA, Chaverri P, Rodrigues A (2016) Unraveling Trichoderma species in the attine ant environment: description of three new taxa. Antonie Van Leeuwenhoek. https://doi.org/10.1007/s10482-016-0666-9
Marfetán JA, Romero AI, Folgarait PJ (2015) Pathogenic interaction between Escovopsis weberi and Leucoagaricus sp.: mechanisms involved and virulence levels. Fungal Ecol. 17:52–61
Wallace DEE, Asensio JGV, Tomás AAP (2014) Correlation between virulence and genetic structure of Escovopsis strains from leaf-cutting ant colonies in Costa Rica. Microbiology 160(8):1727–1736
Pagnocca FC, Masiulionis VE, Rodrigues A (2012) Specialized fungal parasites and opportunistic fungi in gardens of attine ants. Psyche 2012:9. https://doi.org/10.1155/2012/905109
Rodrigues A, Pagnocca F, Bacci M, Hebling M, Bueno O, Pfenning L (2005) Variability of non-mutualistic filamentous fungi associated with Atta sexdens rubropilosa nests. Folia Microbiol 50(5):421–425
Rodrigues A, Pagnocca F, Bueno O, Pfenning L, Bacci Jr M (2005) Assessment of microfungi in fungus gardens free of the leaf-cutting ant Atta sexdens rubropilosa (Hymenoptera: Formicidae). Sociobiol 46(2):329–334
Acknowledgments
We thank the Autoridad Nacional del Ambiente de Panamá, the Smithsonian Tropical Research Institute, and Orelis Arosemena and Annette Aiello for collecting and export permits; Hermogenes Fernández, Ruchira Sen, Nancy Lowe, and Nicole Gerardo for help with field collections; Kate Hertweck for advice on analyses in R; Simon Tragust for sharing his spore-harvesting protocols. Comments from three anonymous reviewers helped improve the manuscript.
Funding
The work was supported by a National Science Foundation award (DEB-0919519) to TAL and UGM and by the W.M. Wheeler Lost Pines Endowment from the University of Texas at Austin. This paper was written and developed while KK was supported by NSF award DEB-1354629 and JNS was supported by IOS-1552822.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kellner, K., Kardish, M.R., Seal, J.N. et al. Symbiont-Mediated Host-Parasite Dynamics in a Fungus-Gardening Ant. Microb Ecol 76, 530–543 (2018). https://doi.org/10.1007/s00248-017-1124-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-017-1124-6