Abstract
Victoria Land permafrost harbours a potentially large pool of cold-affected microorganisms whose metabolic potential still remains underestimated. Three cores (BC-1, BC-2 and BC-3) drilled at different depths in Boulder Clay (Northern Victoria Land) and one sample (DY) collected from a core in the Dry Valleys (Upper Victoria Valley) were analysed to assess the prokaryotic abundance, viability, physiological profiles and potential metabolic rates. The cores drilled at Boulder Clay were a template of different ecological conditions (different temperature regime, ice content, exchanges with atmosphere and with liquid water) in the same small basin while the Dry Valleys site was very similar to BC-2 conditions but with a complete different geological history and ground ice type. Image analysis was adopted to determine cell abundance, size and shape as well as to quantify the potential viable and respiring cells by live/dead and 5-cyano-2,3-ditolyl-tetrazolium chloride staining, respectively. Subpopulation recognition by apparent nucleic acid contents was obtained by flow cytometry. Moreover, the physiological profiles at community level by Biolog-Ecoplate™ as well as the ectoenzymatic potential rates on proteinaceous (leucine-aminopeptidase) and glucidic (ß-glucosidase) organic matter and on organic phosphates (alkaline-phosphatase) by fluorogenic substrates were tested. The adopted methodological approach gave useful information regarding viability and metabolic performances of microbial community in permafrost. The occurrence of a multifaceted prokaryotic community in the Victoria Land permafrost and a large number of potentially viable and respiring cells (in the order of 104–105) were recognised. Subpopulations with a different apparent DNA content within the different samples were observed. The physiological profiles stressed various potential metabolic pathways among the samples and intense utilisation rates of polymeric carbon compounds and carbohydrates, mainly in deep samples. The measured enzymatic activity rates suggested the potential capability of the microbial community to decompose proteins and polysaccharides. The microbial community seems to be appropriate to contribute to biogeochemical cycling in this extreme environment.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Permafrost represents an extreme environment that covers more than 25% of the Earth’s surface. It is extremely sensitive to global climate warming, and speculative research on this ecosystem leads to suppose that climate changes (i.e. increasing temperature and consequent permafrost thaw) will probably result in the release of considerable amounts of carbon and nutrients [6, 48] as well as greenhouse gases [27].
Permafrost hosts a potentially large pool of cold-affected microorganisms, composed by eukaryotes (fungi, mosses, algae) and prokaryotes (bacteria and archaea), which are supposed to be the only life forms known which have retained viability over geological time [17]. Permafrost microorganisms have developed physiological adaptations, which made them able to colonise and survive in extremely cold environments 53]. Thawing renews their physiological activity and exposes ancient life to modern ecosystems [19]. Microbial survival in permafrost raises the question of what constitutes the limit for microbial life. In fact, their adaptation—at species or population level—and physiological flexibility allow them to survive to very low temperatures, cycles of freezing and thawing, high radiation exposure, drying and starvation on geological time scales [47, 52].
Bacterial strains isolated from permafrost are believed to have developed forms of resilience/resistance [47] as a survival strategy. Other studies theorise that their survival is possible thanks to low metabolic rates or stages of anabiosis [41] or dormancy [48], and that their physiological activities are restored during defrosting [19]. A close relationship has been found between the expression of active cellular metabolic activity and long-term survival in 500,000-year-old permafrost samples [28, 30].
Recent studies show that microorganisms isolated from permafrost are psychrotolerant rather than psychrophiles and introduce the eco-physiological concept of eutectophiles, i.e. organisms living at the critical interface inherent to the phase change of water to ice [9, 40]. Metabolic biomarkers, used to understand the ecology and evolution of both permafrost and overlying active layer, showed microbial enzymatic and respiratory activities in permafrost samples [3, 40] stimulating new questions about the nature of the source and sink of CO2 in such environments. Extracellular enzymes of psychrophilic bacteria, which are involved in the decomposition of dissolved and particulate organic carbon, seem to have a higher specific activity at lower temperatures [36]. However, the mechanisms regulating the degradation of the dissolved organic matter, including permafrost-derived organic carbon, remain still unclear [34].
Direct counts by epifluorescence microscopy of prokaryotic cells in Antarctic permafrost are in the order of 105–106 cells g−1 [18], pointing out cell abundances two orders lower than in Arctic permafrost [53].
Although significant advances have been made in the last years, gaps exist to date in the knowledge of ecological and physiological properties of microorganisms in the permafrost of the Victoria Land, region that has poorly been surveyed [1, 35].
Avoiding the use of bacterial isolates whom reactivation/cultivation raises some bias [37], our study aims at providing novel insights on the distribution and metabolic potential of the prokaryotic community in the permafrost of the Northern Victoria Land, a large ice-free area in Continental Antarctica. In particular, Boulder Clay, where the soils are glacic haplorthels and not sufficiently wet for the development of thick organic layers at the surface, was studied. In comparison, another permafrost sample, collected from the Dry Valleys—so called because of their extremely low humidity, where the soils are typic haploturbels and lack of snow or ice cover in the Upper Victoria Valley—was studied.
This paper focused on the following research questions: (1) how much prokaryotic cells inhabited the studied permafrost, (2) how much were alive and respiring, (3) which metabolic activities they could potentially carry out and (4) whether the different nature of soil could influence the microbial colonisation and behaviour and if there were differences among the sites.
Materials and Methods
Site Description and Characteristics of Permafrost
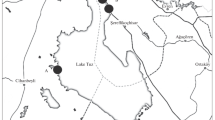
Permafrost samples were collected from one site in the Northern Victoria Land (i.e. Boulder Clay), close to the Italian Antarctic research station “Mario Zucchelli” (MZS), and one site in the Upper Victoria Valley within the McMurdo Dry Valleys (Fig. 1).
a Location of the sampling sites: Boulder Clay (BC), Upper Victoria Valley (DY); b main core characteristics: A Lake ice, B frozen sediments that in BC-3 have some intrusive ice lenses while in the others contain only interstitial or segregated ice, C glacier ice rich in debris, D pure glacier ice, E hypersaline brine and F, sample depth
The two selected sites were chosen because they have similar climatic and permafrost conditions (e.g. ice-cemented permafrost in a very cold and dry environment), but they differ according to the nature and amount of ice along the cores.
Boulder Clay Site
The Boulder Clay site is a debris-covered glacier with several perennially frozen lakes on its surface [18, 19]. The soils here are mainly glacic haplorthels where no evidence of cryoturbation has been observed and scattered mosses and epilithic lichens constitute the vegetation types according to Cannone et al. [5]. The mean annual air temperature (MAAT) is around −14 °C. Permafrost is ice cemented, and the mean annual permafrost temperature (MAPT; at the permafrost table around 30 cm of depth) is around −16.5 °C, although in continuous warming in the last decade [21, 25]. Active layer, monitored in the adjacent circumarctic active layer monitoring (CALM) grid, ranges between 0 and around 90 cm and is continuously thickening [25].
Upper Victoria Valley
The borehole 11 m deep was drilled close to the border of a large polygon within the Packard unit not far from the eastern shore of the Upper Victoria Valley. The soils here are mainly typic haploturbels or typic haplorthels according to Bockheim and McLeod [4]. MAAT is around −18 °C while in the closest borehole monitored to this site, in Wright Valley, MAPT is −18.8 °C [24]. Permafrost in the site of the analysed borehole is ice cemented with an active layer of ca. 30 cm.
Collection and Preliminary Treatment of Samples
With the aim of studying the microbial distribution with depth, in November 2003, samples were obtained from three permafrost cores (coordinates, 74° 4404500 S–164° 0101700 E) drilled at the Boulder Clay (BC) site at different depths, i.e. BC-Lake (sample BC-1, 60–74 cm), BC-2 (sample BC-2, 265–275 cm) and BC-Frost Mound (sample BC-3, 533–543 cm bottom). The three different cores (diameter, 100 mm) were drilled in three different positions (not far from 20 m from each other) in the same catchment basin in order to understand possible connections between the frozen lake, the surrounding permafrost terrain and the frost mound in the middle of the lake. BC-1 was formed by frozen lacustrine silt sediments on the bottom of the lake. Here, gas exchanges with atmosphere were still possible, as well as infiltration of liquid water. BC-2 was drilled immediately outside of the frozen lake, and this core revealed an upper part (94 cm thick) characterised by glacial frozen sediments overlying 296 cm of massive buried glacier ice quite rich in debris, followed by a massive pure glacier ice unit. It is a sample of the debris-rich glacier ice and virtually had no gas exchanges. BC-3 core was drilled on the side of a frost mound [1, 22, 23], located in the same frozen lake of the BC-1 core. The BC-3 core had a more complex permafrost stratigraphy, underlying below 42 cm of lake ice. Below that lake ice, 158 cm of permafrost constituted mainly by silty-sand with intrusive ice lenses occurred followed by 125 cm of the same unit of buried glacier ice rich in debris of the BC-2 sample. Below there is a talik with some hypersaline brines - thick 50 cm - overlaid almost 2 m of pure glacier ice. The sample from Upper Victoria Valley (DY; coordinates, 77° 2000300 S–161° 3702300 E) was collected from an 11-m-deep permafrost core achieved in November 2003. The core was characterised by quite homogeneous frozen sediments mainly composed by sand and silty-sand sediments with little fresh-water ice and water [24, 57].
The main characteristics of the permafrost samples are reported in Table 1. Except for BC-1 that is clearly more recent, the surfaces where these cores were carried out possibly had the same age, although the ages available were not totally conformable (especially for DY) according to Zucconi et al. [57] and Bockheim and McLeod [4].
At lab, the frozen core samples were fractured in a hood with sterile knife and only internal fragments were taken by sterile forceps and placed in sterile containers to be processed according to standard techniques developed for sediments. They were thawed and homogenised and then, to extract the associated microflora, two different protocols were adopted accordingly with the best result performances and replicability. In protocol A, 0.5 g of each permafrost sample were aseptically treated in triplicate with the addition of 5 μl of Tween 80 (10%) and then of tetrasodium pyrophosphate (10 mM), following sonication with a Branson sonifier (at 75% of ultrasounds frequency = 58 W, three times for 1 min, with a break of 30 s every minute), and finally, centrifugation at 800 rpm for 1 min [8, 12]. In protocol B, 1 g of permafrost sample (in triplicate) was diluted in 9 ml of phosphate buffer saline (PBS) and homogenised by vortexing for 3 min. The best performances for the prokaryotic abundances by image analysis (IA) and flow cytometry (FC), the morphmetrical analyses of cells (VOL) and the counts of live/dead cells (L/D) were obtained using protocol A, while for the respiring cell abundance (CTC+), the assessment of the physiological profiles by Biolog-Ecoplate™ and of the potential rates of ectoenzymatic activities (EEA), protocol B was chosen.
Prokaryotic Cell Abundances
Estimation of Total Prokaryotic Cell Abundance, Cell Volume and Morphotypes by Image Analysis
Prokaryotic cell abundance, size and shape were quantified by IA. The three replicated samples, after treatment with the above-reported protocol A, were immediately fixed with filter-sterilised formaldehyde (0.22 μm porosity; final concentration 2%) and stored at 4 °C in the dark. Thereafter, they were filtered through polycarbonate black membranes (porosity 0.22 μm; GE Water & Process Technologies) and stained for 10–20 min with 4′,6-diamidino-2-phenylindole (DAPI; Sigma, final concentration 10 μg ml−1) according to Porter and Feig [42]. A Zeiss AXIOPLAN 2 Imaging (magnification: Plan-Neofluar ×100 objective and ×10 ocular; HBO 100 W lamp; filter sets: G365 excitation filter, FT395 chromatic beam splitter, LP420 barrier filter) equipped with the digital camera AXIOCAMHR (Zeiss) and AXIOVISION 3.1 software for the subsequent morphometric analysis was used. Details of the technical features and calibration were reported in La Ferla et al. [32] and references herein. The volume of individual cells was derived from two manually obtained linear dimensions (width (W) and length (L)). Curved objects were drawn by curve spline. VOL of a single cell was calculated according to the geometrical formula adopted by La Ferla et al. [31] and references herein, i.e. VOL (μm3) = (π/4) × W2 × (L − W/3) (for coccal forms, W = L), assuming that the cells are cylindrical straight rods with hemispherical or, in the case of coccoid forms, spherical caps. The mean cell VOL was calculated by averaging all the cell VOL and the total biovolume by multiplying the mean cell VOL to IA abundances of each sample. The cell shape was operationally defined as cocci if their length and width differed by less than 0.10 μm, coccobacilli if their length and width differed by more than 0.10 μm, and rods if their length was at least double their width; V-shaped, C-shaped and S-shaped cells were defined as vibrios, curved rods and spirillae, respectively.
Estimation of Prokaryotic Cell Abundance and Subpopulation by Flow Cytometry
The three replicates, treated according to protocol A, were fixed with sterile paraformaldehyde at 2% (final concentration), incubated for 30 min at 4 °C and then frozen at −20 °C. After thawing, they were centrifuged and each supernatant was diluted 10× and then stained with Sybr Green I (at a final concentration of 5 × 10−4 of the commercial stock solution; Molecular Probes). The samples were then analysed using a FACSCalibur flow cytometer (BD Biociences) equipped with 488 nm argon laser [2, 16].
Quantification of Viable and Respiring Cells
Live/Dead
The viability of prokaryotic cells—in terms of membrane integrity—was analysed using the Molecular Probes Live/Dead Bac Light Bacterial Viability Kit™ that utilises two separate solutions of SYTO® 9 (green-fluorescent nucleic acid stain) and propidium iodide (red-fluorescent nucleic acid stain). Stock solutions were prepared for each dye using 5 ml of the sample subjected to protocol A, and the final concentration of each dye was 6 μM SYTO 9 stain and 30 μM propidium iodide. The two solutions were mixed thoroughly and incubated at 4 °C in the dark for 1 h. After filtration of 1 ml through polycarbonate black membranes, the above-referred procedure for DAPI staining was performed on the replicates per each sample. Cell microscope counts were done with the specific sets for fluoresceine (BP450-490; FT510; LP520) and rhodamine (BP546/12; FT580; LP590). All the measurements were done under aerobic conditions.
CTC+
The Bac Light Redox Sensor CTC Vitality Kit™ (Molecular Probes) was adopted to quantify the prokaryotic respiring cells. In this case, protocol B was adopted and 1 ml of the pretreated sample was added to 0.1 ml of 5-cyano-2,3-ditolyl-tetrazolium chloride (CTC; final concentration 5 mM) in three replicates per each sample. The solution was incubated at 4 °C in the dark for 5 h—that resulted to be the optimum incubation time chosen among trials of 5, 10 and 18 h. Healthy cells respiring via the electron transport chain absorbed and reduced CTC into a water-insoluble, red-fluorescent formazan product. Incubation was stopped with filter-sterilised (0.22 μm porosity) formalin (final concentration 2%). For each sample, a killed control was prepared by injecting formalin before CTC addition. Slides were prepared in two replicates according to the DAPI procedure earlier described using the rhodamine specific filter set (BP546/12; FT580; LP590). All the measurements were done under aerobic conditions.
Metabolic Profiles and Activities
Physiological Profiles
Following the standard procedure of Garland [14] and Garland and Mills [15], Biolog-Ecoplate™ microplates were used to determine the functional diversity of permafrost samples and the physiological profiles of microbial assemblage by differences in the metabolic potential. Ninety-six-well microtiter plates containing 31 carbon sources and a control in triplicate together with the redox dye tetrazolium violet were inoculated with 150 μl of sample pretreated by protocol B. Each plate was incubated at 4 °C in the dark under aerobic conditions. The oxidation of the carbon sources produced formazan whose absorbance was quantified at 590 nm using a microplate-reader spectrophotometer (MICROTITER ELX-808, Bio Whittaker, Inc.) equipped with an Automatic Microplate Reader and the specific software (WIN KQCL) for data processing. Optical density (OD) was measured immediately after inoculation (at time T0), after 1 day (T1) and then once a week (T7, T14, T21) until OD values showed a saturation of the utilisation rate in all samples.
The colour development for each plate was expressed as averaged substrate colour development (ASCD) according to Sala et al. [43], i.e. ASCD = Σ ((R − C)/31) where R is the averaged absorbance of the three wells with substrate and C is the averaged absorbance of the control wells (without substrate). The absorbance percentages of each substrate were determined according to Sala et al. [44], and 2% absorbance of the total absorbance per plate was used as threshold for substrate utilisation.
Potential Rates of Ectoenzymatic Activities
Microbial ectoenzymatic activities on proteinaceous (leucine-aminopeptidase (LAP)) and glucidic (ß-glucosidase (ß-GLU)) organic matter and on organic phosphates (alkaline-phosphatase (AP)) were estimated on samples subjected to protocol B, in order to quantify the whole (both particulate and dissolved) enzymatic contents. Enzyme assays were performed using specific fluorogenic substrates, which are methylcoumarine (MCA) or methylumbelliferone (MUF) derivates and are analogues to natural compounds, according to the Hoppe’s [26] method. Particularly, l-leucine-4-methylcoumarinylamide hydrochloride, 4-methylumbelliferyl B-d-glucopyranoside and 4-methylumbelliferyl phosphate (Sigma) were used for LAP, ß-GLU and AP measurements. According to a multi-concentration protocol used to reach enzyme saturation, increasing amounts of each substrate were added to triplicate 5-ml subvolumes of each sample and the fluorescence released by substrate hydrolysis was measured with a Turner TD-700 model fluorimeter, at 0 time (initial time) and 3 h after incubation at 4 °C. Calibration curves with the standards 7-amino-4-methylcoumarin (MCA) or 4-methylumbelliferone (MUF) were performed for LAP or for ß-GLU and AP, respectively. Data were reported in terms of maximum reaction velocity (Vmax) of hydrolysis of the substrates, namely as nanomoles of leucine, glucoside and PO4 potentially released per gramme and per hour, by LAP, ß-GLU and AP, respectively, taking into account the initial dilution of each sample.
Statistical Analyses
Statistical analyses of datasets were performed using the Sigma-Stat software version 3.1 (SYSTAT Software, Inc.). Pearson correlation analysis was used to explore the correlations among the assayed microbial variables, and particularly, whether the prokaryotic abundance (IA and FC counts), cell size (VOL) as well as microbial activity data (LAP, ß-GLU, AP), were significantly related to cell physiological properties (live, dead, CTC cells) or cell morphology (vibrios, spirillae, rods, curved rods, cocci, coccobacilli).
Prior to the analysis, the normal distribution of the data set was assessed by normality test; data that did not meet this assumption was logarithmically transformed, in order to satisfy the criteria of normality and homogeneity of variances.
Results
Cell Abundances, Volumes and Morphotypes
The prokaryotic cell abundances detected with IA, FC, CTC+ and L/D are reported in Table 2. IA total counts were in the order of 105 cells g−1 (min and max in DY and BC-1, respectively). FC analyses estimated abundances one order higher than IA; cytograms showed four subpopulations with different apparent DNA content in BC-1, two in BC-2 and BC-3, and only one in DY (Fig. 2).
In the BC samples, cell lengths increased with increasing depth, while cell widths, mean cell VOL and total biovolume decreased. In DY, mean cell VOL was similar to that of BC-2 (Table 3) but total biovolume was smaller. As concerns the size frequency, the size 0.110–0.190 μm3 was prevalent in BC-1; 0.020–0.049 μm3 in BC-2 and BC-3; and 0.05–0.079 μm3 in DY. The cell length was mostly represented by the size classes 0.4–0.8 μm in BC-1, BC-2 and DY (cell percentages 29, 28 and 26% of the total cells, respectively) and 0.8–1.16 μm in BC-3 (63% of the total cells; data not shown). Various cell morphologies were overall observed. Rods, cocci and curved rods showed abundances higher than those of vibrios, coccobacilli and spirillae in the BC samples. Differently, vibrios prevailed over curved rods in DY (data not shown). Spirillae, coccobacilli and curved rods accounted for the greatest VOL (Table 4) followed by rods and vibrios, while the smallest sizes featured cocci.
Quantification of potentially Viable and Respiring Cells
The results of L/D analysis are reported in Table 2. The sum of both alive and dead cells in all samples fell in the order of 105 cells g−1, and dead cells were always lower than potentially live cells. These cells contributed for the 70, 78 and 86% of the total L/D counts in BC-1, BC-2 and BC-3, respectively. In DY, the lowest values of both alive and dead cells were found and potential living cells accounted for the 75% of the total L/D count, similarly to BC-2. Comparing potentially live cells to IA total counts, the proportion of recorded viable cells was again the highest in the BC-3 (94% of total IA counts) followed by DY (84%). The BC-2 and BC-1 samples were characterised by lower amounts of living cells (48 and 32% of the total IA).
Respiring cells (CTC+) in the BC samples were in the order of 105 cells g−1(Table 2) with the highest count again in BC-3 (79% of the total IA count). In BC-1 and BC-2, CTC+ cells accounted for the 40 and 66% of the total IA counts, respectively. In DY, CTC+ resulted in the order of 104 cells g−1, accounting for only the 11% of the total IA count.
Physiological Profiles of Community
The kinetic profiles of the ASCD showed increases over time, from the day of inoculation (T0) to the 21st day of reading (Fig. 3). The overall plate response stressed different patterns in each sample, with high substrate utilisation rates in DY and BC-2, intermediate in BC-3 and low in BC-1.
In Table 5, the utilisation of sole carbon sources by the prokaryotic assemblage of each sample in terms of percentage absorbance of the total absorbance of the plate at the 21st day of reading are reported. The number of utilised substrates (with absorbance >2%) was 9, 13, 21 and 23 in BC-2, DY, BC-3 and BC-1, respectively. High mean absorbance (>10% of the total absorbance of the plate) was detected in all samples, and the number of utilised substrates was 1, 2, 3 and 5 in BC-1, BC-3, DY and BC-2, respectively. Sharing the percent utilisation of the carbon sources into the main source categories, the 69% of the total complex carbon compounds was utilised, particularly Tween 80, Tween 40, and α-cyclodextrin. The 57% of total carbohydrates was used but only d-cellobiose in BC-2 and DY, and d-mannitol in BC-1 were intensely used. A similar percentage of carboxylic acids was largely (55%) but not intensely used, with the exception of the 4-hydroxy benzoic acid, mainly in BC-2 and DY. The 54% of amino acids were used but quite weakly like as happens for the 25% of both phosphate-carbon sources and amines. Finally, six carbon sources (α-d-lactose, β-methyl-glucoside, d,l-α-glicerol phosphate, d-galactonic acid γ-lactone, d-malic acid and l-arginine) were not utilised at all. Differences among samples were recorded: BC-2 was characterised by high utilisation of polymers, amino acids, phenolic compounds and low of carboxylic acids, unlike BC-1 and BC-3, which were also similar in carbohydrates utilisation. DY showed a trend similar to BC-2 with respect to amino acids and phenolic compounds.
Potential Rates of Ectoenzymatic Activities
Enzymatic activities were on average in the order ß-GLU > AP > LAP (Fig. 4), although in the examined samples a different relative importance of the three activities was observed. Higher LAP activity was measured in samples BC-1 and BC-2, and the vertical distribution of enzymatic profiles depicted a decreasing pattern with depth within the BC samples. ß-GLU activity was expressed mostly in the samples BC-1 and DY, while AP showed peaks of activity in the samples BC-1 and BC-3. Within sample DY, ß-GLU activity was twice that of LAP, while AP showed activity rates about one order of magnitude lower than the other enzymes.
No clear relationship among the enzymatic rates and the fraction of both living and actively respiring cells was found. However, in samples BC-1 and DY, the highest enzymatic activity was associated with a lower abundance of live and respiring cells (Table 2).
Statistical Analyses
Pearson correlation analysis yielded the outputs shown in Table 6; only the significant Pearson correlation coefficients (r) are reported, together with their level of significance (P). IA counts were significantly correlated to FC and CTC ones. Significant relationships were observed also between FC counts and VOL, as well as between IA and dead cell counts. All the microbial activities (LAP, ß-GLU, AP) correlated significantly to IA counts and to the fraction of dead cells; in addition, there were significant relationships between LAP and FC abundances as well as between LAP and cell VOL.
Among the organic substrates assayed by Biolog metabolic profiles, complex carbon sources such as Tween 40 and Tween 80 were correlated to live and CTC cell abundances. Significant correlations were found also between complex carbon sources (glycogen), carbohydrates (d-mannitol), carboxylic acids (d-glucosamic acid), aminoacids (phenylalanine), amines (putresceine) and both IA and dead cell counts.
With respect to cell morphological types, a significant relationship linked FC abundances to the vibrios morphotype, which correlated also to different organic polymers. The coccobacilli fraction correlated significantly to live and CTC cells, whereas the dead fraction was correlated to the rods component. VOL, LAP and GLU activity rates were significantly related to the vibrios, cocci and curved rods fraction, AP to rods.
The utilisation profiles of carbohydrates (xylose and mannitol), carboxylic acids (d-glucosamic and d-galactonic acids) and amino acids (l-serine and l-threonine) were significantly associated to the spirillae and rod morphotypes, putrescine to cocci.
Discussion
The present study aims at establishing the amount of prokaryotic cells inhabiting the studied permafrost, and the percentage of those cells that were alive and respiring. To date, the prokaryotic communities inhabiting permafrost in the Victoria Valley have mainly been investigated by the isolation and characterisation of the microbiota [1, 6, 38]. Differently, this study refers for the first time on the living and metabolic properties of the microbial community associated to permafrost samples from Victoria Land and Victoria Valley. The characterisation of the studied samples pointed out differences or similarities among them. The sample BC-1 is clearly a more recent permafrost than the others, while the samples BC-2, BC-3 and DY are presumed to be of the same age. Nevertheless, BC-1 and BC-3 undergo to external exchanges since BC-1 is interconnected with the active layer and periodically can suffer infiltration of liquid water from the surface; BC-3 instead suffers the flow of saline brine from the underground talik. Conversely, BC-2 and DY permafrost have in common the lack of exchange with external input of liquid water because the samples are more than 2 m deeper than the maximum active layer depth and the ice content is much lower(<35% ) and are further characterised by a substantial absence of exchanges with atmosphere. Moreover, the yearly temperature variations result to be similar in the BC-2 and DY samples, high in BC-1 and much smaller in BC-3. Essentially, BC-2 and DY are more stable than BC-1 and BC-3. It is very interesting that BC-2 and DY are very similar, but they have a very different geological history because DY was above the grounding line during the Last Glacial Maximum and therefore the sediments occurred there did not have any contamination with the sea that, on the contrary, occurred at Boulder Clay site. Therefore, the DY sample is very important to understand if the sea contamination during the Last Glacial Maximum and, probably, also in the Late Pleistocene at Boulder Clay influenced the microbial communities. This different geological history is also reflected by the occurrence of relict glacier ice partially of marine origin at Boulder Clay and its absence at DY where only segregated ice occurs.
Comparing our data on prokaryotic abundance with existing direct counts is difficult due to the different analytical methods often used. The prokaryotic abundance, determined by IA direct counts and ranging in the order of 105 cells g−1 is similar to that found by epifluorescence microscopy with DAPI stain in the Antarctic Dry Valleys permafrost [18]. Other indirect estimates by adenosine tri phosphate (ATP) measurements [7] in the Antarctic Dry Valleys soils as well as by phospholipid fatty acids (PLFA) and DNA-based biomass by PCR in the McMurdo Dry Valleys [50] gave similar results. In our study, IA highlights similar counts among the BC samples and a low value in DY. In any case, the Antarctic permafrost always gave a lower prokaryotic abundance than the Arctic permafrost [51–53].
The quantitative discrepancy between IA and FC counts in a similar sample has often been reported in aquatic environments (lake, estuaries and seawater); this has been assumed to depend on technical constrains of each method [16]. However, the absolute comparison between different counting methods is beyond the scope of this research. Although the side scatter (SSC; x-axis; Fig. 2) of the entire population is similar, the bidirectional plot of scatter × green fluorescence allows the separation of different bacterial subpopulations. Instead, the finding by FC of different microbial subpopulations inhabiting the different samples suggests a range from a homogenous sample with only one population (DY) to a heterogenous one with up to four different groups (BC-1).
In all the samples, mean cell VOL results in very small sizes, i.e. <0.1 μm3, with the only exception of BC-1. The BC samples show decreasing cell sizes with increasing depths; in DY, cell VOL is comparable with that of BC-2.The occurrence of various cell morphotypes—including cell attached to detritus or in chain or clustered or free—has been observed. Spirillae are only lacking in both BC-3 and DY. Observing each morphotype (Table 4), the small size of cells is particularly noticeable not only in cocci but also in vibrios and rods, raising the question whether they are ultramicrobacteria (UMB) and name species which maintain their size and volume regardless of their growth condition, if they proliferate and possess small genomes [11], or if they ultramicrocells, i.e. cells which reduce their size and volume as a consequence of life cycle changes or of extreme environmental factors and increase in size and volume when cultivated [30]. Temporary size reduction has been observed in viable but non-culturable state cells [51, 56] and in stressed, starved, dormant or under osmotic pressure cells as strategies to maintain cell integrity and function in extremely cold and salty environments [33, 41]. A study of permafrost permanently remained between −10 and −12 °C showed long-term survival of native UMC cells that accounted for 40–46% of the microbial population [10]. To gain further insights in the species affiliation of ultramicrobacteria, both molecular systematics and microscopic analysis would be advocated.
As regards prokaryotic viability, viable and respiring cells inhabit all the analysed permafrost samples. In the BC samples, the potentially alive cells—determined by L/D procedure—show an increasing trend with increasing depth and also the DY sample highlights high amount of viable cells. In all BC samples, CTC+ staining confirms the increasing trend of respiring cells with increasing depth while in DY the percentage of respiring cells on the total IA cell counts is very low. This discrepancy could be due to the different information on microbial metabolism provided by L/D to CTC+ estimates. Since CTC+ quantifies only very actively growing cells of natural assemblages [45], the findings of this study suggest that actively growing (CTC+) cells are more abundant in the BC samples than in DY, where a high proportion of alive—but probably in a stationary phase—cells are present. Therefore, unlike Steven et al. [47] considerations, in our study, case-reduced life at depth has not been observed. The intrinsic sample characteristics, i.e. ice content, lithology and external exchanges, are presumed to be the main factors determining the observed trend. For instance, the sample BC-3, located below saline brines with which exchanges are possible, shows the highest percentages of both viable and respiring cells.
The patterns of community metabolism reveal different metabolic potentials in the examined samples. Differences in the quality of substrates used in the samples collected from different depths have been observed, highlighting similar ASCD profile development in the BC-2 and DY samples with respect to BC-1 and BC-3 ones. Considering the percentages of used substrates as an index of potential functional diversity, the best exploited carbon sources—in terms of absorbance—are the polymeric substances and carbohydrates as already observed in Siberian permafrost-affected soils [53]. Conversely, in our study, case deep samples (from 265 to 543 cm) show the highest utilisation. Low molecular weight compatible solutes such as polyols are a common means of maintaining positive turgor pressure and metabolic function in continuously changing environment in terms of temperature, salinity and water potential [13]. The efficient utilisation of Tween 40 and Tween 80, easily degradable antifreeze substances that generally occur in high amounts within the Antarctic soil nutrient pool, suggests that bacteria intensively use them [29]. Among carbohydrates, antifreeze agents too, the disaccharides of glucose d-cellobiose and d-mannitol are also highly used by the microbial community. The former, produced by hydrolysis of cellulose, suggests again the occurrence of organic carbon of eukaryotic origin in the BC-2 and DY samples. In BC-2, algae might be present as remnants of sea ice while d-mannitol in DY is likely to be of fungal origin, as yeasts have been previously isolated [57]. The highest utilisation of d-mannitol in the BC-1 sample, at the expense of the other substrates, might be dependent on the multiple roles that this compound has such as carbohydrate storage, reservoir of reducing power, stress tolerance and spore dislodgement and/or dispersal in filamentous fungi [46]. Amino acids appear to be not intensely used by microbial communities, with the only exception of l-asparagine in both BC-2 and DY, confirming the findings in deep Siberian permafrost horizons [53]. Among the carboxylic acids, 4-hydroxy benzoic acid, an allelopathic substance used by plants in their survival strategy, is intensely used in BC-2 and DY. Phenolic compounds are widely spread in nature and represent secondary metabolites of vascular plant tissue and lignine being therefore allochthonous for the study sites. However, they can be synthesised by mosses as antioxidants or allelopathic agents [29]. Grannas et al. [20] suggested the deposition of atmospheric particulate organic matter containing vascular tissue as a source of phenolic compounds in the snow phase of organic matter in the Arctic.
As regards the potential enzyme activity measurements, the examined permafrost samples host a prokaryotic assemblage equipped with a range of enzymes able to hydrolyse organic polymers, particularly proteins and polysaccharides, and therefore showing an active metabolism. Being LAP and β-GLU both involved in carbon cycle, this result gives an indication of the amount of permafrost organic carbon potentially available for microbial metabolism, after its thawing [54], which is still poorly known. As evidences suggest that extensive permafrost thaw could result in a shift toward more readily available organic carbon for metabolism and, consequently, increase mineralisation of these substrates, the microbial processing of permafrost-derived organic polymers could be actively involved in permafrost carbon and climate feedbacks with respect to global climate change [34]. In this context, the study of the metabolism of indigenous psychrophilic microbial communities can give insights on the physiological activities that microorganisms still retain in cold environments [55]. With respect to the distribution of metabolic activity rates with depth, the decreasing vertical pattern of LAP activity inside the BC stations reflects a progressive decomposition of labile organic matter. Decreased enzymes with increased depth and consequently increased burial time were also found by Takano et al. [49], who indicated AP activity as a possible new biomarker of microbial activity in core samples of terrestrial sediments over the past 10,000 years collected from Hokkaido, Japan.
In the DY sample, both ectoenzymatic measurements and physiological profiles highlight the occurrence of enhanced ß-GLU activity. This activity is involved in the degradation of more refractory substrates, such as cellulose. So, the increase of this enzyme especially in the BC-1 sample may be interpreted as an adaptation of the microbial community to exploit organic compounds of different quality, although no clear relationship between the morphological types and the enzymatic activity has been observed. This discrepancy, however, is a feature quite frequently observed in the microbial community.
The uncoupling observed between enzymatic activity levels and viable bacterial counts (live and CTC+ cells) led us to suppose that in all the examined permafrost samples, free enzymes, probably released by dead cells, account for the measured enzymatic activities. This finding has also been supported by the outputs of the Pearson correlation analysis. Free/dissolved enzymes released from dead cells are likely to contribute significantly to the whole enzymatic activity measured in the permafrost samples.
Conclusions
The overall upshot of this study reveals that the permafrost of Victoria Land, located in the depth levels less than 550 cm, harbours a community that is rich in viable cells possessing large metabolic capabilities as well as intense levels of functionality. The observed differences among the examined samples suggest that the different nature of soils influences the microbial colonisation and activity in the different sites. High viability and activity levels appear to be linked with organic matter of eukaryotic origin, presumably algae or yeasts, and with intrinsic sample characteristics among which the external exchanges and ice content. The detected microbial communities seem to be efficiently appropriate to contribute to biogeochemical cycling or ecological processes under the extreme conditions they live, sharing new light on the research of limits of life as well as of cold-adapted enzymes.
References
Abramovich RS, Pomati F, Jungblut AD, et al (2012) T-RFLP fingerprinting analysis of bacterial communities in debris cones, Northern Victoria Land, Antarctica. Permafrost Periglac Proc 23(3):244–248
Andrade L, Gonzalez AM, Araujo FV, et al (2003) Flow cytometry assessment of bacterioplankton in tropical marine environments. J Microbiol Meth 55:841–850
Blanco Y, Prieto-Ballesteros O, Gómez MJ, et al (2012) Prokaryotic communities and operating metabolisms in the surface and the permafrost of Deception Island (Antarctica). Environ. Microbiol. 14(9):2495–2510
Bockheim JG, McLeod M (2013) Glacial geomorphology of the Victoria Valley System, Ross Sea Region, Antarctica. Geomorphology 193:14–24
Cannone N, Wagner D, Hubberten HW, et al (2008) Biotic and abiotic factors influencing soil properties across a latitudinal gradient in Victoria Land, Antarctica. Geoderma 144(1–2):50–65
Cary S, McDonald I, Barret JE, et al (2010) On the rocks: the microbiology of Antarctic Dry Valley soils. Nat Rev Microbiol 8:129–138
Cowan D, Russell N, Mamais A, et al (2002) Antarctic Dry Valley mineral soils contain unexpectedly high levels of microbial biomass. Extremophiles 6:431–436
Danovaro R, Dell’Anno A, Trucco A, et al (2001) Determination of virus abundance in marine sediments. Appl. Environ. Microbiol. 67:1384–1387
Deming JW (2002) Psychrophiles and polar regions. Curr. Opin. Microbiol. 5:301–309
Dmitriev VV, Suzina NE, Rusakova TG, Gilichinskii DA, Duda VI (2001) Ultrastructural characteristics of natural forms of microorganisms isolated from permafrost grounds of Eastern Siberia by the method of low-temperature fractionation. Doklady Biol. Sci. 378:304–307
Duda VI, Suzina NE, Polivtseva VN, Boronin AM (2012) Ultramicrobacteria: formation of the concept and contribution of ultramicrobacteria to biology. Microbiology 81(4):379–390
Duhamel S, Jacquet S (2006) Flow cytometric analysis of bacteria- and virus-like particles in lake sediments. J Microbiol Meth 64:316–332
Fillinger S, Ruijter G, Tamás MJ, et al (2001) Molecular and physiological characterization of the NAD-dependent glycerol 3-phosphate dehydrogenase in the filamentous fungus Aspergillus nidulans. Mol. Microbiol. 39:145–157
Garland JL (1996) Analytical approaches to the characterization of samples of microbial communities using patterns of potential C source utilization. Soil Biol. Biochem. 28(2):213–221
Garland JL, Mills AL (1991) Classification and characterization of heterotrophic microbial communities on the basis of patterns of community-level sole-carbon-source utilization. Appl. Environ. Microbiol. 57(8):2351–2359
Gasol JM, Del Giorgio PA (2000) Using flow cytometry for counting natural planktonic bacteria and understanding the structure of planktonic bacterial communities. Sci. Mar. 64:197–224
Gilichinsky D, Rivkina E, Bakermans C, et al (2005) Biodiversity of cryopegs in permafrost. FEMS Microbiol. Ecol. 53(1):117–128
Gilichinsky DA, Wilson GS, Friedmann EI, et al (2007) Microbial populations in Antarctic permafrost: biodiversity, state, age, and implication for astrobiology. Astrobiology 7(2):275–311
Gilichinsky D, Vishnivetskaya T, Petrova M (2008) Bacteria in permafrost. In: Margesin R, Schinner F, Marx JC, Gerday C (eds) Psychrophiles: from biodiversity to biotechnology. Springer-Verlag, Berlin Heidelberg, pp. 83–102
Grannas AM, Shepson PB, Filley TR (2004) Photochemistry and nature of organic matter in Arctic and Antarctic snow. Global Biochem Cycles 18:1–10
Guglielmin M, Cannone N (2012) A permafrost warming in a cooling Antarctica? Clim. Chang. 111:177–195
Guglielmin M, Biasini A, Smiraglia C (1997) The contribution of geoelectrical investigations in the analysis of periglacial and glacial landforms in ice free areas of the Northern Foothills (Northern Victoria Land, Antarctica). Geographiska Annaler: Series A, Physical Geography 79:17–24
Guglielmin M, Lewkowicz A, French HM, et al (2009) Lake-ice blisters, Terra Nova Bay area, Northern Victoria Land, Antarctica. Geogr. Ann. 91A(2):99–111
Guglielmin M, Balks MB, Adlam L, et al (2011) Permafrost thermal regime from two 30-m deep boreholes in Southern Victoria Land, Antarctica. Permafrost Periglac Proc 22:129–139
Guglielmin M, Dalle Fratte M, Cannone N (2014) Permafrost warming and vegetation changes in continental Antarctica. Environ. Res. Lett. doi:10.1088/1748-9326/9/4/045001
Hoppe HG (1993) Use of fluorogenic model substrates for extracellular enzyme activity (EEA) measurement of bacteria. In: Kemp PF, Sherr BF, Sherr EB, Cole JJ (eds) Handbook of methods in aquatic microbial ecology. FL Lewis Publisher, Boca Raton, pp. 423–432
Jansson J, Taş N (2014) The microbial ecology of permafrost. Nat Rev Microbiol 12:414–425
Johnson SS, Hebsgaard MB, Christensen TR, et al (2007) Ancient bacteria show evidence of DNA repair. PNAS 104(36):14401–14405
Kenarova A, Encheva M, Chipeva V, et al (2013) Physiological diversity of bacterial communities from different soil locations on Livingston Island, South Shetland archipelago, Antarctica. Pol Biol 36:223–233
Kuhn E, Ichimura AS, Peng V, et al (2014) Brine assemblages of ultrasmall microbial cells within the ice cover of Lake Vida, Antarctica. Appl. Environ. Microbiol. 80(12):3687–3698
La Ferla R, Maimone G, Azzaro M, et al (2012) Vertical distribution of the prokaryotic cell size in the Mediterranean Sea. Helgol. Mar. Res. 66(4):635–650
La Ferla R, Maimone G, Lo Giudice A, et al (2015) Cell size and other phenotypic traits of prokaryotic cells in pelagic areas of the Ross Sea (Antarctica). Hydrobiologia 761:181–194
Litchfield CD (1998) Survival strategies for microorganisms in hypersaline environments and their relevance to life on early Mars. Meteorit. Planet. Sci. 33:813–819
Mann PJ, Sobczak W, Larue MM, et al (2014) Evidence for key enzymatic controls on metabolism of Arctic river organic matter. Glob. Chang. Biol. 20:1089–1100
Michaud L, Caruso C, Mangano S, et al (2012) Predominance of Flavobacterium, Pseudomonas, and Polaromonas within the prokaryotic community of freshwater shallow lakes in the Northern Victoria Land, East Antarctica. FEMS Microbiol. Ecol. 82:391–404
Mock T, Thomas DN (2005) Recent advances in sea-ice microbiology. Environ. Microbiol. 7(5):605–619
Mulyukin AL, Demkina EV, Manucharova NA, Akimov VN, Andersen D, McKay C, Gal’chenko VF (2014) The prokaryotic community of subglacial bottom sediments of Antarctic Lake Untersee: detection by cultural and direct microscopic techniques. Mikrobiologiya 83(2):215–224
Niederberger TD, Sohm JA, Gunderson TE, et al (2015) Microbial community composition of transiently wetted Antarctic Dry Valley soils. Front. Microbiol. 6:1–12
Orombelli G, Baroni C, Denton GH (1991) Late Cenozoic glacial history of the Terra Nova Bay region, Northern Victoria Land, Antarctica. Geogr. Fis. Din. Quat. 13(2):139–163
Panikov NS, Sizova MV (2007) Growth kinetics of microorganisms isolated from Alaskan soil and permafrost in solid media frozen down to −35 °C. FEMS Microbiol. Ecol. 59(2):500–512
Ponder MA, Thomashow MF, Tiedje JM (2008) Metabolic activity of Siberian permafrost isolates, Psychrobacter arcticus and Exiguobacterium sibiricum, at low water activities. Extremophiles 12(4):481–490
Porter KG, Feig YS (1980) The use of DAPI for identifying and counting aquatic microflora. Limnol. Oceanogr. 25:943–948
Sala MM, Arin L, Balagué V, et al (2005) Functional diversity of bacterioplankton assemblages in Western Antarctic seawaters during late spring. Mar. Ecol. Prog. Ser. 292:13–21
Sala MM, Estrada M, Gasol JM (2006) Seasonal changes in the functional diversity of bacterioplankton in contrasting coastal environments of the NW Mediterranean. Aquat. Microb. Ecol. 44:1–9
Sherr BF, del Giorgio P, Sherr EB (1999) Estimating abundance and single-cell characteristics of respiring bacteria via the redox dye CTC. Aquat. Microb. Ecol. 18:117–131
Solomon PS, Waters ODC, Oliver RP (2007) Decoding the mannitol enigma in filamentous fungi. Trends Microbiol. 15:257–262
Steven B, Léveillé R, Pollard WH, et al (2006) Microbial ecology and biodiversity in permafrost. Extremophiles 10:259–267
Steven B, Briggs G, McKay CP, et al (2007) Characterization of the microbial diversity in a permafrost sample from the Canadian high Arctic using culture-dependent and culture-independent methods. FEMS Microbiol. Ecol. 59:513–523
Takano Y, Mori H, Kaneko T, et al (2006) Phosphatase and microbial activity with biochemical indicators in semi-permafrost active layer sediments over the past 10.000 years. Appl. Geochem. 21:48–57
Tamppari LK, Anderson RM, Archer Jr PD (2012) Effects of extreme cold and aridity on soils and habitability: McMurdo Dry Valleys as an analogue for the Mars Phoenix landing site. Ant Sci 24(3):211–228
Vorobyova E, Minkovsky N, Mamukelashvili A, et al (2001) Micro-organisms and biomarkers in permafrost. In: Paepe R, Melnikov VP (eds) Permafrost response on economic development, environmental security and natural resources. Kluwer Academic Publishers, Dordrecht, pp. 527–541
Wagner D (2008) Microbial communities and processes in Arctic permafrost environments. In: Dion P, Shekhar Nautiyal C (eds) Soil biology microbiology of extreme soils. Springer-Verlag, Berlin Heidelberg, pp. 133–154
Wagner D, Kobabe S, Liebner S (2009) Bacterial community structure and carbon turnover in permafrost-affected soils of the Lena Delta, Northeastern Siberia. Can. J. Microbiol. 55(1):73–83
Waldrop MP, Wickland KP, White III R, et al (2010) Molecular investigations into a globally important carbon pool: permafrost-protected carbon in Alaskan soils. Glob. Chang. Biol. 16:2543–2554
Wilkins D, Yau S, Williams TJ, et al (2013) Key microbial drivers in Antarctic aquatic environments. FEMS Microbiol. Rev. 37:303–335
Zhang DC, Brouchkov A, Griva G, et al (2013) Isolation and characterization of bacteria from ancient Siberian permafrost sediment. Biology 2(1):85–106
Zucconi L, Selbmann L, Buzzini P, et al (2012) Searching for eukaryotic life preserved in Antarctic permafrost. Polar Biol. 35(5):749–757
Acknowledgements
This work was supported by grants from the National Antarctic Research Program (PNRA), Italian Ministry of Education and Research (Research Project PNRA 2013/AZ1.05) and from the National Council of Research (CNR) in the frame of the Short Term Mobility 2015 (STM, AMMCNT-CNR N. 0058167). Our thanks to all the staff at “Mario Zucchelli” Station, for the logistic help and support, which made possible the expedition. We want to thank Dr. Fabio Baio and Dr. Andrea Strini for their role in the drilling and sampling procedures, Dr. Giulia Maricchiolo and Mr. Michele Furnari (CNR-IAMC) for laboratory availability and for technical instrument helps, respectively, as well as Dr. Daniela Zampino (CNR-IPCB) and Dr. Fabrizio Ioppolo for their friendly technical support.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
La Ferla, R., Azzaro, M., Michaud, L. et al. Prokaryotic Abundance and Activity in Permafrost of the Northern Victoria Land and Upper Victoria Valley (Antarctica). Microb Ecol 74, 402–415 (2017). https://doi.org/10.1007/s00248-017-0955-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-017-0955-5