Abstract
Muscodor spp. are proficient producers of bioactive volatile organic compounds (VOCs) with many potential applications. However, all members of this genus produce varying amounts and types of VOCs which suggests the involvement of epigenetics as a possible explanation. The members of this genus are poorly explored for the production of soluble compounds (extrolites). In this study, the polyketide synthase (PKS) and non-ribosomal peptide synthetase (NRPS) genes from an endophyte, Muscodor yucatanensis Ni30, were cloned and sequenced. The PKS genes belonged to reduced, partially reduced, non-reduced, and highly reduced subtypes. Strains over-expressing PKS genes were developed through the use of small-molecule epigenetic modifiers (suberoylanilide hydroxamic acid (SAHA) and 5-azacytidine). The putative epigenetic variants of this organism differed considerably from the wild type in morphological features and cultural characteristics as well as metabolites that were produced. Each variant produced a different set of VOCs distinct from the wild type, and several VOCs including methyl 3-(3,5-di-tert-butyl-4-hydroxyphenyl)hexane-2,4-diol and 2-carboxymethyl-3-n-hexylmaleic appeared in the variant strains, the production of which could be attributed to the activity of otherwise silent PKS genes. The bioactive extrolite brefeldin A was isolated and characterized from the wild type. However, this metabolite was not detected in EV-1, but instead, two other products were isolated and characterized as ergosterol and xylaguaianol C. Hence, M. yucatanensis has the genetic potential to produce several previously undetectable VOCs and organic solvent soluble products. It is also the case that small-molecule epigenetic modifiers can be used to produce stable variant strains of fungi with the potential to produce new molecules. Finally, this work hints to the prospect that the epigenetics of an endophytic microorganism can be influenced by any number of environmental and chemical factors associated with its host plant which may help to explain the enormous chemical diversity of secondary metabolic products found in Muscodor spp.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Endophytic fungi have gained extensive attention for the production of novel bioactive metabolites with a wide range of pharmaceutical applications. These secondary metabolites are structurally and functionally diverse, mostly because of the various ecological challenges imposed by their natural habitats [1]. Regulation of secondary metabolism in fungi involves various mechanisms such that energy and precursors are used only when their production is advantageous to their environment [2]. In standard fermentation conditions, some of these chemical or environmental signals for inducing secondary metabolite genes are missing as a result of which encoded secondary metabolites are not produced or have low production rates and thus escape detection [3]. This fact has been revealed by modern genome sequencing of several fungi showing that most of the biosynthetic gene clusters coding for secondary metabolites are not expressed and thus remain cryptic [4–6]. For example, genome sequencing of eight Aspergillus species revealed a large number of gene clusters coding for polyketide synthases and non-ribosomal peptide synthetases. However, this number is much higher than the number of secondary metabolites identified from this group of fungi [7]. Thus, despite the incredible record of new taxonomic discoveries [8, 9], the chemistry and utility of fungal products are largely unexplored.
Several efforts have been made to develop strategies to induce these suppressed biosynthetic pathways for the one strain-many compounds (OSMAC) approach through variation of fermentation parameters, genetic engineering, and epigenetic manipulation for the production of novel metabolites [10–12]. Another important strategy involves simulation of natural environment by co-cultivation of microorganisms from the same ecosystem [6]. All these approaches attempt to achieve the same goal, that is, change the chromatin condensation level in such a way that new genes are expressed resulting in the production of novel compounds. The complete analyses of fungal genomes in recent times indicate that many putative biosynthetic gene clusters are located in the distal regions of the chromosomes and exist in a heterochromatin state with the constitutive genes often transcriptionally controlled by epigenetic regulation such as histone deacetylation and DNA methylation [13]. Epigenetic regulation of gene transcription is a relatively simple method and can be applied to a wide range of fungi without prior knowledge of the genome sequence. In fungi, both class I and class II histone deacetylases (HDACs) and lysine- and arginine-specific MTs have been identified [14]. Both the modification of histones, acetylation, and methylation have been demonstrated to have important effects on the production of fungal secondary metabolites [13, 14], sometimes inducing heritable epigenetic changes [15, 16]. Histone modifications and DNA methylation operate to modify chromatin, thereby regulating gene expression or silencing in fungi and other organisms [17]. The addition of small-molecule epigenetic modulators has been applied in several fungi, thus inducing the expression of gene clusters responsible for the production of novel secondary metabolites [17, 18]. In some cases, this approach increases the yield of secondary metabolites already produced by the fungal species under classical conditions [16, 19].
As Muscodor spp. are known to be proficient producers of volatile organic compounds (VOCs) and no natural product (extrolite) has been isolated and characterized from Muscodor yucatanensis, we undertook a study to characterize the extrolites of this fungus and to find out if the use of epigenetic modifiers can induce the expression of secondary metabolism-related genes (PKS genes) and consequently modify the production of extrolites and VOCs. In this case, in the PKS-over-expressing variant, new compounds were found whereas some compounds present in the wild type were no longer detectable.
Materials and Methods
Isolation and Molecular Characterization of the Endophyte
Leaf cuttings of Elleanthus sp. were procured from the Rio San Juan area of Central America (11°02′602″ N, 83°50′355″ W), and the endophyte Ni30 was isolated using the protocol previously described [20, 21]. The endophyte was cultured in potato dextrose (PD) broth at 25 °C with constant shaking for 7 days. The fungal mycelia were freeze-dried, and the genomic DNA was extracted by the cetyl trimethylammonium bromide (CTAB) method [22]. The ITS1–5.8S–ITS2 ribosomal gene of the fungi was amplified with the universal internal transcribed spacer (ITS) primers [23], as described previously for other endophytes [24]. The sequence obtained was aligned with the sequences in GenBank by the BLASTN program [25] to find out the similarity with closely related organisms. Sequences from the closely related organisms were downloaded to construct the phylogenetic tree using the MEGA software version 4.0 [26]. The ITS1–5.8S–ITS2 ribosomal gene sequence of the strain was submitted to GenBank with accession no. KR905059. The identification was supported by cultural and microscopic characteristics of the fungus [27]. The fungal culture was also submitted to the National Fungal Culture Collection of India (NFCCI) under the accession no. NFCCI 2858.
Development of Epigenetic Variants of the Endophyte Ni30
The wild-type fungus Ni30, growing on potato dextrose agar, was hyphal tipped in order to obtain a genetically pure strain of the fungus. The endophyte was treated with epigenetic modulators, 5-azacytidine (AZA), a DNA methyl transferase (DNMT) inhibitor, and suberoylanilide hydroxamic acid (SAHA), an HDAC inhibitor [5]. The fungus was grown on potato dextrose agar (PDA) plates supplemented with 50 μM of AZA and SAHA for 15 days. Hyphal tips of the fungus growing on PDA containing the modulators were picked up and also grown on plain un-supplemented PDA to study the effect without further exposure to the modulators. Thus, four different variants were created as given below:
-
1.
EV-1: A strain selected after a 15-day exposure of Ni30 to 50 μM AZA, hyphal tipped, and transferred to PDA and studied without any subsequent exposure to AZA
-
2.
EV-2: The wild-type strain exposed to 50 μM SAHA and always maintained on PDA supplemented with the same concentration of SAHA
-
3.
EV-3: A strain selected after a 15-day exposure of Ni30 to 50 μM SAHA, hyphal tipped, and transferred to PDA and studied without any subsequent exposure to SAHA
-
4.
EV-4: The wild-type strain exposed to 50 μM AZA and always maintained on PDA supplemented with the same concentration of SAHA
PCR Amplification of Polyketide Synthase Domains of the Endophyte Ni30
The endophyte Ni30 was explored by PCR for the presence of polyketide synthase (PKS) and non-ribosomal peptide synthetase (NRPS) genes. Different pairs of previously designed primers, KA [28], LC [29], CmeT1/T2 [30], and A3(1)/A5 [31], were used for the amplification of ketosynthase (KS) and carboxy-methyl transferase (CMT) domains of PKS genes and adenylation (A) domain of NRPS. The amplicons were purified from agarose gels using the QIAquick PCR Purification Kit (Qiagen, Germany), cloned in pGEM®-T Easy vector (Promega, USA), and sequenced.
Identification of PKS and NRPS Domains and Their Phylogenetic Analyses
The forward and reverse PKS sequences obtained were aligned using DNASTAR’s MegAlign software (Lasergene 8, USA) to obtain the complete sequence. The vector sequences were removed using VecScreen [25]. The partial PKS and NRPS genes were then detected using the GENSCAN software [32]. These peptide sequences were aligned with the sequences in GenBank using BLASTP program [25] to find out the sequence homology with closely related PKS and NRPS sequences. Sequences from the closely related PKS and NRPS were downloaded from the NCBI protein data bank to construct the phylogenetic trees using MEGA software version 4.0 [26]. The neighbor-joining (NJ) method was used to infer the evolutionary history, and the bootstrapping was carried out using 500 replications. The partial PKS and NRPS protein sequences were submitted to GenBank with accession nos. KR905060, KR905061, KR905062, KR905063, KR905064, and KR905065.
Structure Analyses of Putative Polyketide Synthases and Non-Ribosomal Peptide Synthetase Domains
Multiple sequence alignment of the translated KS and CMT domains of polyketide synthases and the adenylation (A) domain of NRPS sequences was carried out with related polypeptide sequences from different origins to assess the degree of homology using the ClustalW2 tool [33], and secondary structures were predicted using SOPMA [34]. The sequences were submitted for structure homology analyses using the Phyre server [35]. Three-dimensional structure modeling of the polypeptides was done using the automated mode of the SWISS-MODEL tool on the ExPASy website [36]. Structure predictions for these KSs were made using the following templates: 2hg4.1.A (PDB code for 6-deoxyerythronolide B synthase) for Ni30-PKS1 and Ni30-PKS2, 2qo3.1.A (PDB code for EryAII erythromycin polyketide synthase modules 3 and 4) for Ni30-PKS-5, and 4na1.1.A (PDB code for polyketide synthase PKSJ) for Ni30-PKS6. For CMT, structure prediction was made using template 4pip.3.A (PDB code for histidine-specific methyltransferase EgtD) for Ni30-PKS-7 and 3vnq.1.A (PDB code for NRPS adenylation protein CytC1) was used as template for Ni30-NRPS.
Comparative Expression of Selected Genes Through Quantitative Reverse-Transcription Polymerase Chain Reaction
Cultures were grown in Erlenmeyer flasks on 250 ml PD broth. PD broth was supplemented with 50 μM of SAHA and AZA for EV-2 and EV-4, respectively. All the cultures were incubated in a shaker incubator at 25 °C with 200 rpm for 15 days. Mycelia were filtered and used for isolation of RNA using the TRIzol® (Invitrogen, USA) method. Prior to complementary DNA (cDNA) synthesis, RNA was treated with DNase I (Qiagen, Germany), to remove trace amounts of DNA. The RNA yield was determined by measuring the absorbance of the RNA solution at 260 nm using a NanoDrop Spectrophotometer 2000c (Thermo Scientific, USA). The purity of the RNA was ascertained by measuring the ratio of absorbance of the RNA solution at 260 and 280 nm. The integrity of the RNA was ascertained by electrophoresis using 1.2% agarose gel containing formaldehyde. cDNA synthesis was achieved by using the First Strand cDNA Synthesis Kit (Fermentas, Germany).
Various primers were designed from the cloned sequences of PKS (PKS 1a, PKS 6a, PKS 7al, PKS 7a) representing reduced (R-PKS), partially reduced (PR-PKS), and highly reduced PKS (HR-PKS) of the endophyte Ni30 (Table 1). The 28S ribosomal gene was chosen as the internal control. Real-time PCR reactions were performed in triplicate using SYBR® Green JumpStart™ Taq ReadyMix™ (Sigma, India) using Stratagene Mx3000p QPCR System (Agilent Technologies, USA). The 20 μl reaction assay contained 1× SYBR® Green JumpStart™ Taq ReadyMix™ (Sigma, India), 1–8 ng of cDNA, and 0.5 μM of each primer. The final volume was made to 20 μl using nuclease-free water. The PCR conditions used were as follows: initial denaturation at 95 °C for 10 min followed by 45 cycles of 95 °C for 15 s, 55 °C for 30 s, and 72 °C for 30 s. The specificity of each primer pair was validated by a dissociation curve (a single peak with specific Tm was observed for each primer pair). The real-time PCR amplification data were exported into Microsoft Excel, and gene expression levels were calculated based on the comparative Ct (2∆∆CT) method [37].
Qualitative GC/MS Analysis of Fungal VOCs
The analysis of VOCs of the wild-type and epigenetic variants was performed using a Varian 3800 GC, equipped with a CP-Sil 8 capillary column (30 m × 0.32 mm × 0.25 μm film thickness), and an API 4000 mass spectrometer [38]. The carrier gas was helium, at a flow rate of 1 ml/min. The column split ratio was 1:150, and the column oven temperature was 60 °C for 5 min, with 250 at 3 per minute and a hold time of 7 min. For GC/MS detection, an electron ionization system was used with ionization energy of 70 eV. The extracts were prepared as 10 mg/ml in ethyl acetate, and 2 μl of the sample was injected automatically in split mode. The injector and detector temperatures were set at 280 °C. Initial identification of the compounds produced by the wild-type and epigenetic variants was made via library comparison using the National Institute of Standards and Technology (NIST) database, and all the chemical compounds described in this report use the NIST database chemical terminology. Only those compounds showing a match of 75% or more were included.
Isolation of Secondary Metabolites From Wild-Type Ni30 and the Epigenetic Variant EV-1
For the isolation of metabolites, Ni30 and EV-1 were initially grown on PDB at 25 °C in an incubator shaker (New Brunswick, USA) for 15 days. A 10% seed was used to inoculate 5 l PDB in a fermentor (New Brunswick, USA) which was run for 15 days with 100 rpm and 1 vvm aeration. The culture broth was extracted with equal volume of ethyl acetate twice and concentrated under reduced pressure to get 300 mg crude extract. The extract was redissolved in ethyl acetate and subjected to preparative TLC (30% ethyl acetate in hexane) to obtain pure compounds.
Results
Isolation and Phylogenetic Position of the Endophyte Ni30
Ni30 was isolated as an endophyte from a leaf of Elleanthus sp. The fungus was isolated on a water agar medium as a cottony white colony. The isolate developed whitish mycelia on PDA. The culture displayed slow growth at 25 °C with a colony size of 4–4.5 cm in 20 days of incubation and did not sporulate on PDA. The ITS1–5.8S–ITS2 ribosomal gene sequence of Ni30 showed highest similarity of 99% with M. yucatanensis (Fig. 1). Morphological and microscopic characteristics were in confirmation to those reported previously for the species [27].
The phylogenetic position of the endophyte Ni30 inferred using the neighbor-joining method [26]. The endophyte is closely related to Muscodor yucatanensis. Numbers on branches represent the bootstrap values
Phenotypic Variations in Epigenetic Variants of the Endophyte Ni30
The putative epigenetic variants, EV-1 and EV-4, produced the most striking changes in the growth rate, morphology, pigmentation, and bioactivity (Table 2). On transferring these variants to normal PDA plates, they maintained the modified morphological characteristics, which were different from the wild-type and epigenetic variants growing on media supplemented with epigenetic modifiers. The variants EV-1 and EV-4 displayed decreased growth rates, producing colonies with diameters of 2.9 and 2.5 cm compared to 3.5 cm in the wild type after 20 days of growth on PDA at 25 °C. EV-1 and EV-4 also produced a brownish pigment on PDA plates and in PD which was not observed in the wild type. Slight variations in morphology and bioactivity were also observed in epigenetic variants EV-2 and EV-3, while a variation in pigmentation was not observed in these variants (Supplementary Material Fig. S1).
Characterization of Polyketide Synthase and Non-Ribosomal Peptide Synthetase Domains of Ni30
A total of five putative PKS genes and one NRPS gene were amplified by PCR: two genes were detected by the KA series of primers, two genes by LC series, one by CmeT1/T2, and one by NRPS pair of primers (Table 3). For amplification using KAF1 (HR-specific or lovastatin type) in KA series as a forward primer, primer pairs KAF1/KAR1 and KAF1/KAR2 detected two PCR products (one by each pair) in this endophytic fungus. The translated sequences of both fragments showed strong sequence similarity of 60 and 61%, respectively, with known PKS gene sequences (Table 3). In contrast, no products were obtained using KAF2/KAR2, KS3/4, and XKS1/2 primer pairs. Four PKS gene fragments were amplified using primers LC1/2, LC3/5, CmeT1/T2, and A3(1)/A5. Each primer pair detected one single band in Ni30. The translated sequences of these genes displayed similarities of 83, 65, 65, and 84%, respectively, to other known PKS genes (Table 3). Thus, from the fungus, we obtained partial sequences of five putative PKS genes and one NRPS gene. Four products shared sequence similarity with known β-ketoacyl synthase domains; one sequence displayed similarity to a known fungal C-methyltransferase domain and one sequence with the adenylation domain of NRPS.
Phylogenetic Analysis of the Putative PKS and NRPS Domains
The phylogenetic analysis of the putative KS and CMT domains of PKSs showed that these sequences could be grouped into the established clades [39] representing fungal-reduced PKS (R-PKS), non-reduced PKS (NR-PKS), partially reduced PKS (PR-PKS), and highly reduced PKS (HR-PKS) (Fig. 2).
The evolutionary relationship between the putative polyketide synthases of Ni30 with their close relatives from other fungi [26]. The translated sequences grouped into reduced PKS (R-PKS), non-reduced PKS (NR-PKS), partially reduced PKS (PR-PKS), and highly reduced PKS (HR-PKS)
Two sequences could be grouped within the established clade, representing KS domains from R-PKSs. These sequences were obtained with primers KAF1/KAR1 and KAF1/KAR2. The sequence Ni30-PKS-1 shared 60% sequence similarity with a polyketide synthase from Thielavia terrestris NRRL 8126. The sequence Ni30-PKS-2 shared 61% sequence similarity with polyketide synthase from Eutypa lata UCREL1 (Table 2). One sequence, Ni30-PKS-5, could be grouped within the established clade representing NR-PKSs (Fig. 2). This sequence shared 83% sequence similarity with a non-reducing polyketide synthase from Phomopsis sp. CBMAI 1019 (Table 2). The sequence Ni30-PKS-6 amplified with the primer pair LC3/5 grouped in a separate clade representing partially reduced PKS showing 65% sequence similarity with a reducing polyketide synthase from Diaporthe sp. CBMAI 1020. The sequence Ni30-PKS-7 shared 45% homology with the C-methyl transferase domain from Penicillium janthinellum and was grouped in a separate clade representing highly reduced PKS. Non-ribosomal peptide synthetase sequence Ni30-NRPS-1 was grouped in a separate clade and displayed 84% homology with the putative lovastatin nonaketide synthase protein from E. lata UCREL1 and 74% homology with a hybrid NRPS/PKS from Aspergillus clavatus NRRL 1.
Structure Analyses of Deduced Amino Acid Sequences of Polyketide Synthase and Non-Ribosomal Peptide Synthetase Domains
Secondary and tertiary protein structure predictions were performed in order to confirm the putative functions of putative genes. A multiple sequence alignment of the translated sequences of ketosynthase (KS) and carboxy-methyl transferase (CMT) domains of polyketide synthases and the adenylation (A) domain of non-ribosomal peptide synthetase displayed alignment with other known polyketide synthase and non-ribosomal peptide synthetases displaying significant similarity (Supplementary Material Figs. S2, S3, and S4). Secondary structure analyses revealed that Ni30-PKS-1 and Ni30-PKS-2 were similar although they were obtained with different pairs of primers and consisted of 25.36% alpha helix, 19.40% beta sheet, 7.46% beta turn, and 47.76% random coil. Similarly, Ni30-PKS-5 consisted of 36.08% alpha helix, 20.25% beta sheet, 11.39% beta turn, and 32.28% random coil; Ni30-PKS-6 consisted of 37.40% alpha helix, 18.70% beta sheet, 14.63% beta turn, and 29.27% random coil; Ni30-PKS-7 consisted of 35.78% alpha helix, 19.27% beta sheet, 16.51% beta turn, and 28.44% random coil; and Ni30-NRPS consisted of 26.23% alpha helix, 33.61% beta sheet, 9.02% beta turn, and 31.15% random coil. SWISS-MODEL software was used for construction of 3D models of the KS and CMT domains of polyketide synthases and the A domain of NRPS identified from Ni30 (Fig. 3). All the predicted models overlapped with their templates showing alpha helices and beta sheets and gave a conformational pattern similar to that of the known PKSs and NRPS. In addition, ligand ATP was found in contact with chains A, G1, S2, T3, K7, G116, E117, and T118 in the 3D model of Ni30-NRPS using template 3vnq.1.A (PDB code for NRPS adenylation protein CytC1) which is important for the catalytic activity of non-ribosomal peptide synthetases.
SWISS-MODEL; 3D comparative models of KS, CMT, and A domain sequence fragments using templates 2hg4.1.A (PDB code for 6-deoxyerythronolide B synthase) for Ni30-PKS1 and Ni30-PKS2, 2qo3.1.A (PDB code for EryAII erythromycin polyketide synthase modules 3 and 4) for Ni30-PKS-5, 4na1.1.A (PDB code for polyketide synthase PKSJ) for Ni30-PKS6, 4pip.3.A (PDB code for histidine-specific methyltransferase EgtD) for Ni30-PKS-7, and 3vnq.1.A (PDB code for NRPS adenylation protein CytC1; ligand ATP in contact with chains A, G1, S2, T3, K7, G116, E117, T118) for Ni30-NRPS
Comparative Expression of Polyketide Synthase Genes
The epigenetic variant, EV-1, displayed maximum expression in all the selected genes with a 145-fold increase in the expression of NR-PKS (PKS-3) followed by PR-PKS (60-fold), C-methyl transferase domain of HR-PKS (45-fold), and R-PKS (6.5-fold) (Fig. 4). However, the epigenetic variant, EV-4, displayed decreased expression in all genes except PR-PKS, where the relative expression was 3-fold that of the wild-type Ni30. SAHA-treated fungus, when grown with and without the treatment of SAHA (EV-2 and EV-3, respectively), displayed decreased expression in all the PKS genes. Thus, the effect of AZA treatment was more profound than SAHA on the over-expression of PKS genes. Our efforts to quantify the change in expression of the NRPS gene did not yield any results as no amplification was obtained from the cDNA of either the wild-type or the variant strains.
VOCs of Ni30 and Its Variant Strains
Ni30 produced an array of bioactive volatile organic compounds. However, several VOCs were not detected in the variants, namely, trans-3-dodecene, tau-gurjunene, benzenepropanoic acid, 3,5-bis(1,1-dimet), 1-chloroheptacosane, and 2-hexyl-1-octanol. The unique VOCs that appeared in EV-1 were tetradecane, 2-carboxymethyl-3-n-hexylmaleic acid, 1,2-dipentylcyclopropene, methyl 3-(3,5-di-tert-butyl-4-hydroxyphenyl) hexane-2,4-diol, tritetracontane, and 17-pentatriacontene. Likewise, EV-2, EV-3, and EV4 also produced some volatiles which were absent in the control and other variants (Table 4). Also, several of the VOCs produced by the Ni30 wild type were retained by one or more variant strains.
Characterization of Extrolites Isolated From the Wild-Type Ni30 and EV-1
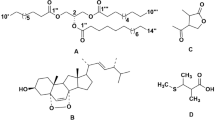
Several extrolites and polyketides were isolated and characterized from these fungi as follows: (1) (12 mg, Rf 0.55) from Ni30 as the only major extrolite, (2) (30 mg, Rf 0.7), and (3) (5 mg, Rf 0.4) from EV-1 as major secondary metabolites. Purified compounds were identified and characterized as (1) brefeldin A, (2) ergosterol, and (3) (1R,4S,5S,7R,10R,11R)-guaiane-10,11,12-triol (xylaguaianol C) by IR, UV, MS, and NMR data as presented below:
-
Compound 1 (brefeldin A): chemical formula, C16H24O4; molecular weight, 280.1675; 1H NMR (400 MHz, CDCl3) δ 7.36 (qt, J = 2 Hz, 1H), 5.91 (qt, J = 9.9 Hz, 1H), 5.69 (m, 1H), 5.29 (m, 1H), 4.86 (m, 1H), 4.34 (m, 1H), 4.10 (m, 1H), 2.36 (m, 1H), 2.20 (m, 1H), 2.08 (m, 1H), 2.02 (m, 1H), 1.95 (m, 1H), 1.87 (m, 1H), 1.84 (m, 1H), 1.81 (m, 1H), 1.74 (m, 1H), 1.53 (m, 1H), 1.51(m, 1H), 1.26 (d, J = 5.2 Hz, 3H), 0.95 (m, 1H)
-
Compound 2 (ergosterol): chemical formula, C28H44O; molecular weight, 396.33; 1H NMR (400 MHz, CDCl3) δ 5.50 (d, J = 3.7 Hz, 1H), 5.32 (d, J = 2.1 Hz, 1H), 5.13 (t, J = 7.8 Hz, 2H), 3.56 (dd, J = 13.1, 9.1 Hz, 1H), 2.40 (d, J = 12.5 Hz, 1H), 2.21 (t, J = 12.8 Hz, 1H), 2.05–1.86 (m, 3H), 1.85–1.75 (m, 4H), 1.74–1.47 (m, 6H), 1.47–1.36 (m, 2H), 1.36–1.10 (m, 6H), 0.97 (d, J = 6.5 Hz, 3H), 0.92–0.82 (m, 6H), 0.76 (t, J = 6.3 Hz, 6H), 0.56 (s, 3H)
-
Compound 2 (xylaguaianol C): chemical formula, C15H28O3; molecular weight, 256.2038; 1H NMR (400 MHz, CDCl3) δ 3.94 (d, J = 10 Hz, 1H) 1H), 3.85 (d, J = 10 Hz, 1H) 1H), 2.55 (m, 1H), 2.34 (m, 1H), 2.29 (m, 1H), 2.24 (m, 1H), 2.21 (m, 1H), 2.08 (m, 1H), 1.64 (m, 1H), 1.70 (m, 1H), 1.62 (m, 1H), 1.29 (m, 1H), 1.99 (m, 1H), 1.16 (m, 1H), 1.45 (m, 1H), 1.69 (m, 1H), 1.36 (s, 3H), 1.34 (s, 3H), 0.98 (d, J = 6.8 Hz, 3H)
-
Assignments were confirmed by comparison with the values reported in literature [40–42].
Discussion
Filamentous fungi are viewed as an important reservoir of novel bioactive natural products, many of which are discovered by the traditional natural product screening approach [43]. However, the advent of genome sequencing has revealed that fungi have the genetic capacity to produce far more molecules than have ever been detected [4, 6]. Among the several strategies to stimulate these cryptic biosynthetic gene clusters, one of the most promising methods is epigenetic modulation [17].
Endophytic fungi belonging to the genus Muscodor are known for the production of volatile organic compounds (VOCs) with bioactive and fuel potential [44]. However, non-volatile secondary metabolites (extrolites) have rarely been reported from this group of fungi. We attempted to isolate non-volatile secondary metabolites from the endophyte Ni30 isolated from Elleanthus sp. and were able to isolate one compound (compound 1) from the strain which was characterized as brefeldin A that was found as the major compound in the culture broth. This compound has been previously reported from Eupenicellium brefeldianum, Paecilomyces sp., and A. clavatus [45, 46]. Brefeldin A, a polyketide, has antibacterial, antiviral, antinematode, and antifungal activities [46, 47]. It is also widely used in biological research to study protein transport as it blocks protein secretion by causing disassembly of Golgi apparatus [48].
Due to the absence of any genomic information about the biosynthetic pathways in Muscodor, it was not possible to study the genome-wide expression of secondary metabolism-related genes. Thus, we cloned and sequenced the PKS and NRPS genes to study the effect of small-molecule epigenetic modifiers on their expression for proof of concept. We were able to amplify five putative polyketide synthase genes and one non-ribosomal peptide synthetase gene from the endophyte Ni30. The sequences were grouped into major groups, comprising reducing PKSs (R-PKS), highly reducing (HR-PKS), and non-reducing (NR-PKS) PKSs, and they were classified by comparison to a diverse set of fungal KSs, CMT, and A domains derived from PKS gene clusters having well-described biosynthetic functions [39]. This indicates that Ni30 possesses remarkable genetic information for the production of polyketides and non-ribosomal peptides. However, the chemical investigation of Ni30 revealed that these gene clusters remained cryptic as no related products were obtained from the fungal culture.
Thus, an attempt was made to modulate the expression of these cryptic biosynthetic genes of Ni30 using HDAC (SAHA) and DNA methyltransferase (AZA) inhibitors for the production of new compounds. Significant variations in morphological and biological properties of the fungus were observed. Two of the newly developed variants did not revert to the wild-type fungus when grown without the modulators even after a series of sub-culturing, indicating that stable variants were created with morphological characteristics different from that of the wild-type.
The PKS genes belonging to different classes were upregulated in the epigenetic variant EV-1, and nearly all of the PKS genes were downregulated in other epigenetic variants EV-2 and EV-3. However, in the epigenetic variant EV-4, there was a 3-fold increase in the expression of partially reduced PKS gene while a decreased expression was observed in the rest of the PKS genes studied. Previous studies on Aspergillus niger have also shown that addition of epigenetic modifiers to cultures resulted in increased transcriptional rates among most of its PKS, NRPS, and hybrid PKS-NRPS (HPN) biosynthetic gene clusters, whereas less than 30% of these gene clusters were transcribed when the organism was grown in the absence of the modifiers [49]. Interestingly, the variants EV-1 and EV-4 demonstrated remarkable brownish pigmentation which suggests that genes involved in pigment synthesis were induced. As many of the pigments produced by fungi are polyketides [50], the pigmentation in these variants may be as a result of the increased expression of PKS-2 (Fig. 4). The appearance of new VOCs in EV-1, such as 2-carboxymethyl-3-n-hexylmaleic acid and methyl 3-(3,5-di-tert-butyl-4-hydroxyphenyl) hexane-2,4-diol, may also be attributed to the over-expression of PKS genes [51]. Each variant produced a different set of VOCs distinct from the wild type and the other strains developed. EV-1 produced some long-chain hydrocarbons that were absent in the wild type, such as tetradecane (C14H30), 1,2-dipentylcyclopropene (C13H24), tritetracontane (C43H88), and 17-pentatriacontene (C 35H72). The mechanism syntheses of alkanes and alkenes have not been elucidated in fungi [52]. Thus, in addition to PKS, it seems that other secondary metabolism-related genes are modulated in the variants resulting in the subsequent production of new VOCs and loss of others (Fig. 5).
Due to the increased expression of all the PKS genes in the epigenetic variant EV-1, this strain was further selected for isolation of secondary metabolites. The TLC profile of EV-1 displayed maximum number of metabolites, not previously observed in the wild-type strain. Consequently, we were able to isolate two major compounds, ergosterol and xylaguaianol C, which were not detected in the parent strain. Xylaguaianol C has been previously isolated from the sister genus Xylaria [42], indicating that fungi from the Xylariaceae family may have a common biosynthetic pathway to produce this compound. The study indicates that in addition to PKS, genes from other biosynthetic pathways also get activated in EV-1 which resulted in the production of VOCs and soluble compounds.
Conclusion
Modulation of cryptic biosynthetic genes through small-molecule epigenetic modulators in M. yucatanensis Ni30 led to the expression of several biosynthetic pathways, in addition to the PKS genes, thus producing new secondary metabolites. Hence, small-molecule epigenetic modifiers can be used to produce stable variant strains with the potential to produce new molecules. Further, efforts are needed to perform an in-depth chemical investigation of EV-1 and other epigenetic strains to get an insight into the potential of Muscodor spp. for the production of known and novel secondary metabolites. It may be also interesting to learn if the changes in the secondary metabolism are due to epigenetic modifications, gene mutations, or a cumulative effect of both.
References
Sun X, Guo L-D, Hyde KD (2011) Community composition of endophytic fungi in Acer truncatum and their role in decomposition. Fungal Divers. 47:85–95
Hoffmeister D, Keller NP (2007) Natural products of filamentous fungi: enzymes, genes, and their regulation. Nat. Prod. Rep. 24:393–416
Chiang YM, Lee KH, Sanchez JF, Keller NP, Wang CC (2009) Unlocking fungal cryptic natural products. Nat. Prod. Commun. 11:1505–1510
Scherlach K, Hertweck C (2009) Triggering cryptic natural product biosynthesis in microorganisms. Org Biomol Chem 7:1753–1760
Cichewicz RH (2010) Epigenome manipulation as a pathway to new natural product scaffolds and their congeners. Nat. Prod. Rep. 27:11–22
Brakhage AA, Schroeckh V (2011) Fungal secondary metabolites—strategies to activate silent gene clusters. Fungal Genet. Biol. 48:15–22
Sanchez JF, Chiang YM, Szewczyk E, Davidson AD, Ahuja M, Oakley CE, Bok JW, Keller NP, Oakley BR, Wang CC (2010) Molecular genetic analysis of the orsellinic acid/F9775 gene cluster of Aspergillus nidulans. Mol. BioSyst. 6:587–593
Heywood VH (1995) Global biodiversity assessment. Cambridge University Press, Cambridge
Hawksworth DL (2001) The magnitude of fungal diversity: the 1.5 million species estimate revisited. Mycol. Res. 105:1422–1432
Bode HB, Bethe B, Hofs R, Zeeck A (2002) Big effects from small changes: possible ways to explore nature’s chemical diversity. Chem Bio Chem 3:619–627
Bills GF, Platas G, Fillola A, Jimenez MR, Collado J, Vicente F, Martın J, Gonzalez A, Bur-Zimmermann J, Tormo JR, Pelaez F (2008) Enhancement of antibiotic and secondary metabolite detection from filamentous fungi by growth on nutritional arrays. J. Appl. Microbiol. 104:1644–1658
Chiang Y, Szewczyk E, Nayak T, Davidson AD, Sanchez JF, Lo H, Ho W, Simityan H, Kuo E, Praseuth A, Watanabe K, Oakley BR, Wang CCC (2008) Molecular genetic mining of the Aspergillus secondary metabolome: discovery of the emericellamide biosynthetic pathway. Chem Biol 15:527–532
Shwab EK, Bok JW, Tribus M, Galehr J, Graessle S, Keller NP (2007) Histone deacetylase activity regulates chemical diversity in Aspergillus. Eukaryot Cell 6:1656–1664
Bok JW, Chiang Y-M, Szewczyk E, Reyes-Domingez Y, Davidson AD, Sanchez JF, Lo H-C, Watanabe K, Strauss J, Oakley BR, Wang CC, Keller NP (2009) Chromatin-level regulation of biosynthetic gene clusters. Nat. Chem. Biol. 5:462–464
Birch PRJ, Sims PFG, Broda PJ (1998) A reporter system for analysis of regulatable promoter functions in the basidiomycete fungus Phanerochaete chrysosporium. Appl. Microbiol. 85:417–424
Riyaz-Ul-Hassan S, Strobel GA, Booth E, Knighton B, Floerchinger C, Sears J (2012) Modulation of volatile organic compound formation in the Mycodiesel producing endophyte Hypoxylon sp. C1-4. Microbiology 158:464–473
Asai T, Chung YM, Sakurai H, Ozeki T, Chang FR, Yamashita K, Oshima Y (2012) Tenuipyrone, a novel skeletal polyketide from the entomopathogenic fungus, Isaria tenuipes, cultivated in the presence of epigenetic modifiers. Org. Lett. 14:513–515
Wang Q, Yuan F, Pan Q, Li M, Wang G, Zhao J, Tang K (2010) Isolation and functional analysis of the Catharanthus roseus deacetylvindoline-4-O-acetyltransferase gene promoter. Plant Cell Report 29:185–192
Williams RB, Henrikson JC, Hoover AR, Lee AE, Cichewicz RH (2008) Epigenetic remodeling of the fungal secondary metabolome. Org. Biomol. Chem. 6:1895–1897
Ezra D, Hess WM, Strobel GA (2004) New endophytic isolates of Muscodor albus, a volatile-antibiotic-producing fungus. Microbiology 150:4023–4031
Riyaz-Ul-Hassan S, Strobel G, Geary B, Sears J (2013) An endophytic Nodulisporium sp. from Central America producing volatile organic compounds with both biological and fuel potential. J. Microbiol. Biotechnol. 23:29–35
Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (1994) Current protocols in molecular biology. Wiley, New York City
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. Academic, San Diego, pp. 315–322
Qadri M, Rajput R, Abdin MZ, Vishwakarma RA, Riyaz-Ul-Hassan S (2014) Diversity, molecular phylogeny and bioactive potential of fungal endophytes associated with the Himalayan blue pine (Pinus wallichiana). Microb. Ecol. 67(4):877–887
Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25(17):3389–3402
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599
González MC, Anaya AL, Glenn AE, Macías-Rubalcava ML, Hernández-Bautista BE, Hanlin RT (2009) Muscodor yucatanensis, a new endophytic ascomycete from Mexican chakah, Bursera simaruba. Mycotaxon 110:363–372
Amnuaykanjanasin A, Punya J, Paungmoung P, Rungrod A, Tachaleat A, Pongpattanakitshote S, Cheevadhanarak S, Tanticharoen M (2005) Diversity of type I polyketide synthase genes in the wood-decay fungus Xylaria sp. BCC 1067. FEMS Microbiol. Lett. 251:125–136
Bingle LE, Simpson TJ, Lazarus CM (1999) Ketosynthase domain probes identify two subclasses of fungal polyketide synthase genes. Fungal Genet. Biol. 26:209–223
Nicholson TP, Rudd BA, Dawson M, Lazarus CM, Simpson TJ, Cox RJ (2001) Design and utility of oligonucleotide gene probes for fungal polyketide synthases. Chem Biol 8:157–178
Paungmoung P, Punya J, Pongpattanakitshote S, Jeamton W, Vichisoonthonkul T, Bhumiratana S, Tanticharoen M, Linne U, Marahiel MA, Cheevadhanarak S (2007) Detection of nonribosomal peptide synthetase genes in Xylaria sp. BCC1067 and cloning of XyNRPSA. FEMS Microbiol. Lett. 274:260–268
Burge C, Karlin S (1997) Prediction of complete gene structures in human genomic DNA. J Mol Bio 268:78–94
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948
Geourjon C, Deleage G (1995) SOPMA: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Comput. Appl. Biosci. 11:681–684
Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10:845–858
Arnold K, Bordoli L, Kopp J, Schwede T (2006) The SWISS-MODEL Workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22:195–201
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-∆∆CT method. Methods 25:402–408
Qadri M, Deshidi R, Shah BA, Bindu K, Vishwakarma RA, Riyaz-Ul-Hassan S (2015) An endophyte of Picrorhiza kurroa Royle ex. Benth, producing menthol, phenylethyl alcohol and 3-hydroxypropionic acid, and other volatile organic compounds. World J. Microbiol. Biotechnol. 31:1647–1654
Kroken S, Glass NL, Taylor JW, Yoder OC, Turgeon BG (2003) Phylogenomic analysis of type I polyketide synthase genes in pathogenic and saprobic ascomycetes. Proc. Natl. Acad. Sci. U. S. A. 100:15670–15675
Glaser R, Shiftan D, Froimowitz M (2000) NMR structure determination of brefeldin-A, a 13-membered ring fungal metabolite. Magn. Reson. Chem. 38:274–280
Tao R, Wang CZ, Kong ZW (2013) Antibacterial/antifungal activity and synergistic interactions between polyprenols and other lipids isolated from Ginkgo biloba L. leaves. Molecules 18:2166–2182
Wu SH, He J, Li XN, Huang R, Song F, Chen YW, Miao CP (2014) Guaiane sesquiterpenes and isopimarane diterpenes from an endophytic fungus Xylaria sp. Phytochemistry 105:197–204
Newman DJ, Cragg GM (2012) Natural products as sources of new drugs over the 30 years from 1981 to 2010. J. Nat. Prod. 75:311–335
Strobel GA, Dirkse E, Sears J, Markworth C (2001) Volatile antimicrobials from Muscodor albus, a novel endophytic fungus. Microbiology 147:2943–2950
Harri E, Loeffler W, Sigg HP, Staehelin S, Tamm C (1963) Uber die isolierung der stoffwechselprodukte aus Penicellium brefeldianum Dodge. Helv Chim Acta 46:1235–1243
Wang F, Jiao R, Cheng A, Tan S, Song Y (2007) Antimicrobial potentials of endophytic fungi residing in Quercus variabilis and brefeldin A obtained from Cladosporium sp. World J. Microbiol. Biotechnol. 23:79–83
Betina V (1992) Biological effects of the antibiotic brefeldin A (decumbin, cyanein, ascotoxin, synergisidin): a retrospective. Folia Microbiol (Praha) 37:3–11
Misumi I, Misumi Y, Miki K, Takntsuki A, Tamura G, lkehara Y (1986) Novel blockade by brefeldin A of intracellular transport of secretory proteins in cultured rat hepatocytes. J. Biol. Chem. 261:11398–11403
Fisch KM, Gillaspy AF, Gipson M, Henrikson JC, Hoover AR, Jackson L, Najar FZ, Wagele H, Cichewicz RH (2009) Chemical induction of silent pathway transcription in Aspergillus niger. J. Ind. Microbiol. Biotechnol. 36:1199–1213
Atanasova L, Knox BP, Kubicek CP, Druzhinina IS, Baker SE (2013) The polyketide synthase gene pks4 of Trichoderma reesei provides pigmentation and stress resistance. Eukaryot Cell 12:1499–1508
Koch L, Lodin A, Herold I, Ilan M, Carmeli S, Yarden O (2014) Sensitivity of Neurospora crassa to a marine-derived Aspergillus tubingensis anhydride exhibiting antifungal activity that is mediated by the MAS1 protein. Mar Drugs 12:4713–4731
Spakowicz DJ, Strobel SA (2015) Biosynthesis of hydrocarbons and volatile organic compounds by fungi: bioengineering potential. Appl. Microbiol. Biotechnol. 99:4943–4951
Acknowledgements
This work was supported by the CSIR 12th FYP project (Grant Number PMSI-BSC0117) of the Council of Scientific and Industrial Research (CSIR), New Delhi, India, and the Major Lab Project MLP1008 of the institute. MQ is grateful to ICMR, India, for a Senior Research Fellowship. We are thankful to Miss Palak Arora for maintaining the growth of the cultures. We are also thankful to Mr. Chand Raina for running the fermentors and to Mrs. Kushal Bindu for recording the GC/MS. The article bears the institutional publication number IIIM/1955/2016.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
ESM 1
(DOCX 398 kb.)
Rights and permissions
About this article
Cite this article
Qadri, M., Nalli, Y., Jain, S.K. et al. An Insight into the Secondary Metabolism of Muscodor yucatanensis: Small-Molecule Epigenetic Modifiers Induce Expression of Secondary Metabolism-Related Genes and Production of New Metabolites in the Endophyte. Microb Ecol 73, 954–965 (2017). https://doi.org/10.1007/s00248-016-0901-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-016-0901-y