Abstract
In this study, we investigated the diversity of fungal endophytes associated with Pinus wallichiana from the Western Himalayas, with emphasis on comparison of endophytic communities harbored by the stem and needle tissues of the host and their antimicrobial potential. A total number of 130 isolates, comprising of 38 different genera, were recovered from 210 fragments of the plant. Among the isolated fungi, only a single isolate, Tritirachium oryzae, belonged to the Phylum Basidiomycota whereas the rest belonged to Ascomycota. Dothideomycetes was the dominant class with the highest isolation frequency of 49.2 %. The most frequent colonizers of the host were Alternaria spp., Pestalotiopsis spp., Preussia spp., and Sclerostagonospora spp. The diversity and species richness were higher in needle tissues than in the stems. Antimicrobial activities were displayed by extracts from a total number of 22 endophytes against one or more pathogens. Endophytes designated as P1N13 (Coniothyrium carteri), P2N8 (Thielavia subthermophila), P4S6b (Truncatella betulae), P7N10 (Cochliobolus australiensis), and P8S4 (Tritirachium oryzae) were highly active against Candida albicans. Broad spectrum antimicrobial activities were obtained with the extracts of P8-S4 (Tritirachium oryzae) and P5-N26 (Coniochaeta gigantospora) that were potentially active against the Gram-positive and Gram-negative bacteria as well as the fungal pathogen, Candida albicans. The most prominent antagonistic activity against fungal pathogens was shown by P8-S4 (Tritirachium oryzae), P5-N31a (Truncatella spadicea), and P5-N20 (Fusarium larvarum). Our findings indicate that Pinus wallichiana harbors a rich endophytic fungal community with potential antimicrobial activities. Further studies are needed to understand the ecology and evolutionary context of the associations between the Himalayan pine and its endophytes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Endophytes are an important component of plant microbiota, residing in the living tissues of almost every plant and do so in a variety of relationships, ranging from symbiosis to balanced antagonism [9, 41]. They are indispensible mutualistic partners of higher plants that provide various benefits to their hosts. Endophytes are known to provide stress tolerance, both biotic and abiotic, to the plants thus helping them to adapt to different environmental conditions [36, 38, 42]. Further, the plant–endophyte interactions influence plant nutrition, growth rate, as well as plant survival and distribution [27, 40, 50]. In addition to their ecological significance, endophytes represent a huge resource for the isolation of novel biomolecules and biocatalysts for applications in medicine, agriculture, and industry [2, 28, 35]. Endophytic microorganisms undergo a constant process of strain development by passing through various stages of plant growth and development and by way of continued efforts to survive in the host tissues [28, 38, 53]. This fact, coupled with their ecological functions, contributes to their immense metabolic proficiency and advocates the ever increasing interest in such microorganisms for bioprospecting of potential natural products. The ubiquity of these cryptic symbionts is clear, but the scale of their diversity, host range, and geographic distributions are unknown. Some endophytes are host-specific while others are able to colonize multiple host species [17, 18, 38]. Owing to the overwhelming diversity of plants, we are still in the early stages of recognizing the functional roles of endophytes in nature and their real potential as a biological resource.
In this study, an effort was made to investigate the biodiversity of fungal endophytes associated with Pinus wallichiana (blue pine/Bhutan pine) and explore their antimicrobial potential. Conifers, including different species of Pinus, harbor diverse microorganisms as endophytes, varying with species as well as with geographical distribution. They seem to be reservoirs of enormous microbial wealth waiting to be characterized and explored for human welfare [5, 22, 26]. The Himalayan blue pine is native to the Himalayas of Nepal, Myanmar, Bhutan, China, India, Pakistan, and Afghanistan, growing at altitudes ranging from 1,500 to 3,400 m above the mean sea level [56]. In the Jammu and Kashmir state, a total of 20,230 km2 is under forest cover with conifers covering an area of 40.87 % of which 9.73 % is occupied by Pinus wallichiana (locally known as kɒ’yʊr) [3]. The wood of this plant is widely used as timber which is valued next to the wood of Cedrus deodara and is regarded as superior to that of Pinus roxburghii. It is one of the most dominant species of the forest ecosystem of the Kashmir Himalayas and has immense commercial, medicinal, and ecological importance [7, 15, 16]. Moreover, this species is a prominent source of oleoresin which is used for the production of turpentine oil, rosin, needle oil, and camphor. The essential oil of this treasured plant, which has been found to possess potential anticancer and antioxidant properties, comprises of β-pinene, α-pinene, myrecene, camphene, limonene, α-phellandrene, trans-caryophyllene, α-humulene, α-cadinol, and α-bisabolol. In addition, it also contains undecane, dodecane, tridecane, abietic acid, and isopimaric acid [14, 16]. Further, the dark-brown resinous substance (known locally as kɪllʊm/kɪllʌm), extracted from this species, is traditionally used by the farmers of Kashmir to protect their skin from insect bites, skin-cracks, and infections while working long hours in the water-logged rice fields.
The study of endophytes of this majestic Himalayan pine may give an insight into their diversity, distribution, and host affinity which, in turn, may guide to a better management of this plant. Such studies are also imperative to explore the bioactive potential of microorganisms associated with this species and achieve their conservation.
Materials and Methods
Sampling
Small cuttings (about 10 cm) of the stems or twigs of ten symptomless plants were collected from a specific location (34° 31′ 13″ N, 74° 22′ 55″ E) in the Western Himalayas of the Lolab Valley. The Lolab Valley is a Himalayan sub-valley and forms the northwestern corner of the Valley of Kashmir. Multiple numbers of samples from individual trees (n = 10) of Pinus wallichiana were collected in the month of May 2012. The plant materials were transported to the lab in an icebox and stored at 4 °C until processed.
Endophytic Fungal Isolation
Endophytic fungi were isolated as described previously by Ezra et al. [20] and Arnold and Lutzoni [6]. Plant materials were thoroughly washed with running tap water, cut under sterile conditions into small pieces (2–3 cm), and surface-sterilized with 1 % sodium hypochlorite for 2–5 min for needles and stems, respectively, followed by 70 % ethanol for 1 min. Before the treatment with alcohol, traces of sodium hypochlorite were removed by washing with sterile distilled water. After drying under the sterile laminar air flow and briefly passing through the flame, the outer tissues of the stem cuttings were removed. The tissues were cut into smaller segments of around 0.5 cm each. A total of 210 segments (from stems and needles) were evenly plated on different microbiological media such as water agar, potato dextrose agar, and Kenknight and Munaier’s medium (dextrose 1 g, KH2PO4 0.1 g, NaNO3 0.1 g, KCl 0.1 g, MgSO4.7H2O 0.1 g, agar 15.0 g, distilled water 1,000 ml, pH 7.2) supplemented with streptomycin (50 mg l−1) to suppress bacterial growth. The effectiveness of the surface sterilization was confirmed by making imprints of disinfected plant fragments on PDA plates from which no fungal growth was observed [45]. The plates were incubated at 25 °C for 3–4 weeks. Hyphal tips of fungi, emerging out of the plant tissues, were picked and grown on potato dextrose agar in pure culture. After the proper incubation of the plates, 15-day-old cultures were preserved by placing pieces of hyphal growth in 30 % glycerol at −70 °C and also by lyophilization. All the media and chemicals were obtained from Hi Media (India). The cultures were also submitted to the CSIR-IIIM Microbial repository.
For morphological analyses, microscopic slides of each endophyte were prepared by staining with lactophenol cotton blue [10, 30] and were examined under light microscope (Olympus, USA). Based on mycelial characteristics, representative of all unique phylotypes were selected for molecular analysis.
Genomic DNA Extraction
Each endophytic fungus was cultured in PD broth at 25 °C with constant shaking for 7 days. The fungal mycelia were freeze-dried, and the genomic DNA was extracted by the cetyltrimethylammonium bromide (CTAB) method [8]. Briefly, 500 mg of fungal mycelia was vigorously crushed in liquid nitrogen to make a fine powder. The cells were lysed in 10 ml of extraction buffer (50 mM Tris–HCl pH 8.0, 50 mM EDTA, 0.7 M NaCl, 2 % cetrimide, 1 % SDS, and 50 μl β-mercaptoethanol), mixed thoroughly, and incubated at 65 °C for 30 min with continuous shaking. The lysate was extracted with an equal volume of chloroform/isoamyl alcohol (24:1) and centrifuged at 10,000×g for 10 min at 4 °C. The aqueous phase was transferred to a sterile tube; the genomic DNA was precipitated in a 2× volume of chilled isopropanol and centrifuged at 4 °C for 10 min at 10,000×g. The resulting pellet was washed twice with 70 % ethanol, air-dried, and dissolved in 20 μl of sterile Millipore water.
PCR Amplification and Sequencing of ITS (ITS1–5.8S–ITS2) Region
Phylogenetic analyses of the endophytes were carried out by the acquisition of the ITS1–5.8S–ITS2 ribosomal gene sequencing. The ITS regions of the fungi were amplified with the universal ITS primers, ITS4 (5′TCCTCCGCTTATTGATATGC3′) and ITS5 (5′GGAAGTAAAAGTCGTAACAA3′) [58], using the polymerase chain reaction (PCR). The 50-μl reaction mixture contained 1× PCR buffer, 200 μM each dNTP, 1.5 mM MgCl2, 10 pmol of each primer, 1–5 ng of DNA, and 2.0 U of Taq DNA polymerase. A negative control, using deionized water instead of template DNA, was included in each batch of the PCR. The PCR conditions used were as follows: initial denaturation at 94 °C for 3 min followed by 30 cycles of 94 °C for 15 s, 55 °C for 30 s, 72 °C for 45 s, and a final extension at 72 °C for 7 min. The amplified products (5 μl) were visualized on 1 % (w/v) agarose gel to confirm the presence of a single amplified band of approximately 600 bp. The amplicons were purified by Amicon Ultra columns (Millipore, USA), and 20–40 ng was used in a 10-μl sequencing reaction using the Big Dye Terminator sequencing kit (v. 3.1). The forward or the reverse primer (2 pmol) was used in the cycle sequencing reactions. Twenty-five cycles of 96 °C for 10 s, 50 °C for 5 s, and 60 °C for 4 min were performed, and the extension products were purified by ethanol precipitation, dissolved in 10 μl of HiDi formamide, incubated at 95 °C for 5 min, and loaded on ABI Prism 377 Genetic Analyzer (Perkin-Elmer) for sequencing. All the reagents for sequencing were from Applied Biosystems, USA. The amplified products were sequenced and aligned with the sequences in the GenBank by BLASTN program [1] to find out the sequence homology with closely related organisms. Sequences from the closely related organisms were downloaded to construct the phylogenetic trees using MEGA software version 4.0 [55]. The neighbor-joining (NJ) method was used to infer the evolutionary history of the fungal isolates, and the bootstrapping was carried out using 1,000 replications. The ITS1–5.8S–ITS2 DNA sequence for each strain was submitted to the GenBank. Accession numbers of the respective strains are provided in Supplementary material Table 1.
Statistical Data Analysis
The colonization frequency (CF) was calculated as the total number of segments colonized by endophytic fungi divided by the total number of incubated segments. The relative species frequency (RF) was calculated as the number of isolates of one species divided by the total number of isolates [59].
The similarity of endophytic fungal assemblages among both tissues was compared using the following similarity indices: Sorensen’s index of similarity (QS), QS = 2a/(2a + b + c), where a is the number of common species in both endophytic populations, while b and c are the number of species specific to stem and needles, respectively [37]. Jaccard’s index of similarity (JS) was calculated using the formula: JS = a/(a + b + c), where a is the number of common species in both mycopopulations, while b and c are the number of species specific to leaf and stem, respectively. Simpson’s diversity was calculated as D = 1 − Ʃpi2, where pi = ni/N, ni being the total no. of isolates of the species i and N being the total no. of isolates of all the species [48]. It gives the probability that two individuals selected at random will belong to the same species. The Shannon–Weaver diversity index (H′) [46] was used to show diversity of the endophytic fungal species and was calculated as: H′ = −Ʃpi (ln pi). Shannon evenness (E) was calculated as H′/H max, where H max = ln(S), with S as the total number of species in the sample. The Shannon evenness index ranges from 0 (when one species is dominant) to 1 (when all species are equally abundant). Species richness was calculated as S/√N, where S = total number of species. The “effective number of species” was calculated as exp (H) where H = Shannon–Weaver diversity index [33].
Antimicrobial Activity of Endophytic Fungal Extracts
All endophytic cultures were cultured at 25 °C for 15 days in 200 ml of PD broth with constant shaking. The fermented broth of each endophyte was homogenized with 10 % methanol, extracted thrice with methylene chloride, concentrated, and dissolved in dimethyl sulfoxide (DMSO) at a concentration of 10 μg/μl. The extracts prepared from the endophytes were evaluated for antimicrobial activity against Escherichia coli ATCC 25922 (Gram negative), Staphylococcus aureus ATCC 29213 (Gram positive), and Candida albicans MTCC 4748 (fungal pathogen). Stock solutions of each culture were prepared in normal saline solution (0.85 % NaCl (w/v)) at a concentration of 108 cells/ml. Two hundred-microliter Mueller–Hinton broth (for Escherichia coli and Staphylococcus aureus) and PD broth (for Candida albicans) were added to each well in the first column, and 100 μl of corresponding media was added to the rest of the wells. Extracts of different endophytes were added to the first row at a concentration of 100 μg/ml and serially diluted to make concentrations of 50, 25, 12.5, and 6.25 μg/ml. Each well was inoculated with 104 cells of the relevant test organism and incubated at 37 °C for 24 h. Each plate had a set of controls: a column with broad-spectrum antibiotics (ciprofloxacin and nystatin) as positive controls, a column without the test extract, and one without the relevant test organism. Absorbance was measured spectrophotometrically at 620 nm, and IC50 of the extracts was calculated from the average percent inhibition of three replicates of each concentration. The percent inhibition by the fungal extract was calculated as [(X − Y)/X] × 100, where X is the change in OD of the control and Y is the change in OD of treated broth. IC50 values were calculated from the graph obtained by plotting the % inhibition against different concentrations of the extract. The cutoff IC50 value was taken as 100 μg/ml [19].
Antimycotic Activity of the Endophytic Fungi
All the endophytes were tested against several plant pathogenic fungi to assess the antifungal potential of their extrolites. Briefly, small plugs (ca. 3 mm diameter) of each test fungi were placed a centimeter away from the edge of a 7-day-old endophytic culture. The plates were wrapped with parafilm and incubated at 25 °C for 24–48 h. Inhibition of the test pathogens was reported as % inhibition as compared to their relevant controls. The diameter of the pathogens was observed after 24 and 48 h to analyze the percentage inhibition of growth as compared to the control plates. The percent growth inhibition (%GI) was calculated as GI (%) = [(KR − R1)/KR] × 100, where KR represents the distance (measured in mm) from the point of inoculation to the colony margin on the control dishes and R1 is the distance of fungal growth from the point of inoculation to the colony margin on the treated dishes in the direction of the antagonist [31].
Results
Diversity and Phylogenetics of Endophytic Fungi
Endophytic fungi were isolated from healthy and symptomless stem and needle segments of Pinus wallichiana to determine the diversity and bioactive potential of these endophytes. A total of 130 endophytic fungal species belonging to 38 different genera were isolated from 210 segments of both needle and stem tissues of the host (Supplementary material Table 1). Three different media were used for the isolation of endophytic fungi, and maximum number of endophytes was obtained from potato dextrose agar (40 %) followed by water agar (30.7 %) and Kenknight and Munaier’s agar (29.2 %), respectively. The molecular identification was supported by the morphological characteristics of the cultures that produced spores under the growth conditioned used in this study. However, many of the endophytes did not sporulate; thus, molecular identification was the only option to ascertain their identity. Among the total number of endophytic fungi (n = 130), isolated from the host, only a single isolate P8S4 (Tritirachium oryzae) belonged to the Phylum Basidiomycota whereas all other isolates belonged to Ascomycota. A total of 49.2 % of isolates belonged to Dothideomycetes followed by Sordariomycetes (33.8 %), Eurotiomycetes (6.9 %), Leotiomycetes (4.6 %), and Pezizomycetes (3.8 %). Thus, the most abundant class of fungi obtained was Dothideomycetes represented by the orders Pleosporales, Capnodiales, and Botryosphaeriales. Sordariomycetes were mostly isolated from needle tissues and represented by the orders Xylariales, Diaporthales, Sordariales, Hypocreales, and Trichosphaeriales. The least abundant classes were Eurotiomycetes represented by the order Eurotiales, Leotiomycetes represented by the orders Rhytismatales and Helotiales, and Pezizomycetes represented by the order Pezizales. The dominant fungi observed were Alternaria spp., Pestalotiopsis spp., Preussia spp., and Sclerostagonospora spp. In the needle tissues, Alternaria spp. (7.6 %) showed the highest isolation frequency followed by Preussia spp. (4.6 %) and Sclerostagonospora spp. (4.6 %) while in the stem tissues, Pestalotiopsis spp. (5.3 %) showed the highest isolation frequency (Table 1). The common species found in both tissue segments were Alternaria spp., Sclerostagonospora spp., Epicoccum spp., Preussia spp., Coniochaeta spp., Trichoderma spp., Pestalotiopsis spp., Phoma spp., Truncatella spp., and Aspergillus spp. Endophytic fungi isolated only from the stem tissues were Cladosporium spp., Rachicladosporium spp., Leptosphaeria spp., Microdiplodia spp., Neurospora spp., Lophiostoma spp., Therrya spp., Lophodermium spp., Glomerella spp., Penicillium spp., Tritirachium spp., and Phomopsis spp. whereas those that were restricted only to the needles were Anthostomella spp., Tricharina spp., Paraconiothyrium spp., Coniothyrium spp., Thielavia spp., Sordaria spp., Lecythophora spp., Rosellinia spp., Pseudoplectania spp., Nigrospora spp., Fusarium spp., Xylaria spp., Sporormiella spp., Cadophora spp., Geopyxis spp., Cochliobolus spp., and Fimetariella spp. (Table 1).
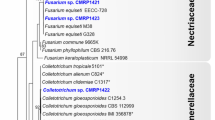
The ITS1–5.8S–ITS2 sequences of some nearest neighbors of the endophytic fungal isolates were retrieved from GenBank (NCBI) to construct the phylogenetic tree. The sequences displayed diverse taxonomic affinities among the isolated endophytes. BLAST and NJ analyses placed the sequences into various groups of Ascomycota belonging to five different classes of Dothideomycetes, Sordariomycetes, Eurotiomycetes, Leotiomycetes, and Pezizomycetes (Fig. 1). Eight isolates P1N13 (Coniothyrium carteri), P4S11 (Therrya fuckelii), P1N6 (Anthostomella conorum), P1S14 (Leptosphaeria sp.), P6N9 (Preussia intermedia), P5N1 (Preussia sp.), P3N33 (Pseudoplectania nigrella), and P6N18 (Cadophora malorum) showed a sequence similarity of less than 95 % with known organisms in the GenBank (Supplementary material Table 1). These endophytes could likely be from novel fungal lineages.
Phylogenetic position of the endophytes of the Himalayan blue pine. The evolutionary history was inferred using the neighbor-joining method [44]. The optimal tree with the sum of branch length = 0.96703271 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) is shown next to the branches [21]. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the maximum composite likelihood method [54] and are in the units of the number of base substitutions per site. All positions containing alignment gaps and missing data were eliminated only in pairwise sequence comparisons (pairwise deletion option). Phylogenetic analyses were conducted in MEGA4 [55]. The basidiomycete isolate and endophytes, P3N33, P6N9, and P6N18, showing <90 % homology with known fungi were excluded from this phylogenetic tree
The biodiversity analysis showed that colonization of endophytic fungi was greater and more diverse in needles (66.9 %) than in stem tissues (53.7 %) (Table 2). The diversity of the endophytic community isolated from both tissues was compared using indices of α-diversity (Shannon–Wiener index and Simpson’s diversity index) and their components, i.e., species richness and evenness (Table 3). The concentration of dominance or Simpson’s dominance of endophytic fungi was almost identical in the foliar (0.041) and stem tissues (0.043). Simpson’s diversity indices of 0.95, in both the tissues, indicate a high diversity of endophytes harbored by the host plant. Shannon–Wiener diversity indices were slightly higher in fungal endophytes of the needle tissues (2.48) than in the stem tissues (2.23). There was a higher species richness of endophytic fungi colonizing the foliar (2.19) than the stem tissues (1.84). Species evenness was uniform in both the tissues (Table 3). Shannon effective number of species was found to be 11.9 in case of the needle tissues and 9.3 in the stem tissues. This is the true diversity of the community in question. These diversity indices reflect the richness of phylotypes in Pinus wallichiana (Table 3). Jaccard’s (Jc) and Sorenson’s similarity indices between the endophytes of both tissues were 0.26 and 0.41 %, respectively (Table 4). This indicates that the biodiversity of fungal endophytes in stem and needle tissues of Pinus wallichiana is quite significant.
Antibacterial and Antifungal Activities of Endophytic Fungal Extracts
Endophytic fungal extracts from Pinus wallichiana were tested for antibacterial and antifungal activities by the micro-titer plate assay against the representatives of Gram-positive and Gram-negative bacteria (Staphylococcus aureus ATCC 29213 and Escherichia coli ATCC 25922) and a fungal pathogen (Candida albicans MTCC 4748). A total of 34 extracts were found to possess significant antibacterial and antifungal activity against one or more pathogens (Table 5). Extracts from fungal isolates, P1N10 (Tricharina hiemalis), P1N13 (Coniothyrium carteri), P1S1b (Epicoccum nigrum), P2N8 (Thielavia subthermophila), P4S6b (Truncatella betulae), P7N10 (Cochliobolus australiensis), and P8S4 (Tritirachium oryzae), exhibited significant activity against the tested pathogens. P1N13 (Coniothyrium carteri), P2N8 (Thielavia subthermophila), P4S6b (Truncatella betulae), P7N10 (Cochliobolus australiensis), and P8S4 (Tritirachium oryzae) were significantly active against Candida albicans with IC50 of 17, 14, 19, 19, and 26 μg/ml, respectively. Several extracts (n = 25) of the endophytic fungi displayed antimicrobial activity against Staphylococcus aureus with IC50 values less than 100 μg/ml. Especially, four of them, P1N10 (Tricharina hiemalis), P1S2 (Alternaria sp.), P1S1b (Epicoccum nigrum), and P8S4 (Tritirachium oryzae), inhibited Staphylococcus aureus strongly, with IC50 values of 10, 5, 4, and 7 μg/ml, respectively. In contrast, only 16 extracts displayed antimicrobial activity against the Gram-negative bacterium, Escherichia coli, with IC50 values less than 100 μg/ml wherein the lowest IC50 values of 20, 19, 31, and 7 μg/ml were obtained with the extracts prepared from P1S1b (Epicoccum nigrum), P3N33 (Pseudoplectania nigrella), P1N10 (Tricharina hiemalis), and P8S4 (Tritirachium oryzae), respectively. Broad spectrum of antimicrobial activities was obtained with the extracts of P8-S4 (Tritirachium oryzae) and P5-N26 (Coniochaeta gigantospora) that were potentially active against the Gram-positive and Gram-negative bacteria as well as the fungal pathogen, Candida albicans.
Antagonistic Activity Against Plant Pathogens
Representative strains were screened for antagonistic activity against eight fungal pathogens. Antimycotic activities of 17 endophytic fungi that showed 50 % or more inhibition against one or more pathogens are presented in Table 6. Three endophytes, P1N13 (Coniothyrium carteri), P5N20 (Fusarium larvarum), and P5-N31a (Truncatella spadicea), displayed broad range of antimycotic activity, inhibiting at least five pathogens by 50 % or more. The most susceptible pathogen was Ceratocystis fimbriata (MTCC 2281) whose growth was inhibited more than 50 % by the endophytic fungi P1-N1 (Alternaria brassicae), P1-S7b (Sclerostagonospora spp.), P1-S14 (Leptosphaeria sp.), P5-N20 (Fusarium larvarum), P5-N31a (Truncatella spadicea), and P8-S4 (Tritirachium oryzae).
Discussion
Endophytes play vital ecological roles, thus influencing plant growth, development, disease resistance and adaptation to unique niches, etc. [38, 40, 50]. Furthermore, the intimate association of endophytism may modulate the biosynthetic pathways of the plant, thus shaping its secondary metabolism [43, 57]. Given the large number of plant species growing in diverse environmental niches on our planet and lack of genuine laboratory models, many aspects of endophytism like host specificity, geographical distribution, phylogenetic affinities, as well as the phenomena mentioned above have been poorly understood; thus, we have limited knowledge about various factors influenced by endophytism and vice versa. Moreover, despite continued efforts to characterize and preserve endophytes, much of this natural treasure is unexplored owing to the amplitude of their diversity and wide distribution.
This study represents the first report of the diversity and bioactivity of endophytes associated with the Himalayan blue pine, particularly from the Kashmir valley. The total number of endophytes isolated (n = 130) from ten trees represented 38 different genera. This implies an average of about four different genera from each plant, thus indicating that the magnitude of diversity harbored by the blue pine is quite significant. The endophytic community was dominated by Dothideomycetes followed by Sordariomycetes, representing 83 % of the isolates, collectively. Similarly, Dothideomycetes was the most prevalent class of endophytes hosted by Pinus halepensis from Spain [11]. Sordariomycetes was the second most abundant class in our study frequently isolated from the foliar tissues (Table 1). It has been previously reported as the dominant class in the foliar endophytes [5].
Most of the fungi isolated in this study from Pinus wallichiana corresponded with those recorded in previous studies of Pinus spp. [5, 11, 12]. However, some species such as Anthostomella conorum, Tricharina hiemalis, Microdiplodia spp., Thielavia subthermophila, Therrya fuckelii, and Tritirachium oryzae were isolated for the first time from Pinus sp. Pestalotiopsis spp., described as endophyte of pines and isolated only from needles in previous studies [24], was isolated from both stems and needle tissues of Pinus wallichiana. Eurotiomycetes and Pezizomycetes constitute the least abundant classes in this study (Table 1) which is in agreement with the previous studies on endophytes of other plants [4].
Furthermore, in contrast to the endophytic community hosted by Pinus monticola from northern Rocky Mountains of the USA, where 95 % of the foliar endophytes belonged to the Rhytismataceae [23], we obtained only four isolates from this family (two belonging to the genus Lophodermium and the other two closely related to Therrya spp.) (Supplementary material Table 1). Lophodermium is regarded among the most common endophytes isolated from Pinus, Abies, and Picea [52] although one species, Lophodermium seditiosum, is regarded as a major pathogen causing needle cast of pines [34, 49]. The striking difference in the endophytic communities between the Pinus spp. may be due to species specificity of the endophytes as well as the conditions provided by the two different ecological niches. Further, a clear differentiation was observed in the community structure of endophytes in the stem and foliar tissues, with the latter supporting a wider diversity of endophytes. Interestingly, several of the endophytes found in the needles did not colonize the stems and vice versa. Thus, tissue specificity was evident among the endophytes of the Himalayan blue pine. These tissues represent two distinct micro-environments consequently shaping their microbiota differently.
A total number of eight isolates displayed less than 95 % sequence (ITS1–5.8S–ITS2) similarity with known organisms. Thus, these may represent novel species. The taxonomic novelty of such organisms may also correspond to chemical novelty of their secondary metabolites [32]. Further, several of these fungi have not been explored for their natural products. Thus, these microbes, along with those that displayed bioactivity, will be given priority for characterization of their secondary metabolites to isolate novel molecules with bioactivity. After further exploration, endophytic cultures showing prominent antimycotic activities may be use to control root rot and other microbial diseases, particularly in the pine seedlings.
Natural products remain a consistent source of drug leads with more than 40 % of new chemical entities (NCEs) reported from 1981 to 2005 having been derived from microorganisms [13, 29, 47]. In comparison to other natural sources like plants, microorganisms are highly diverse but narrowly explored. Studies based on estimation of microbial populations have revealed that only about 1 % of bacteria and 5 % of fungi have been characterized and the rest remain unexplored for their contribution to the human welfare [25, 51]. Further, fungi are proficient producers of bioactive metabolites and drug-like molecules [28]. This is not surprising in the light of their evolution over millions of years in diverse ecological niches and natural habitats.
We have obtained these data on plants growing at a single site, but endophytic communities may differ along the latitudinal gradient [6]. Further, in a previous study, we observed that some conifers from the same sampling site harbored novel bioactive endophytes [32, 39]. Therefore, concerted efforts are needed to study the endophytic diversity of the Himalayan blue pine and other conifers growing at the same site as well as along the latitudinal gradient to understand their distribution, host specificity, and phylogenetic affinity in the northwestern Himalayas. When compared to the endophytes of conifers from other biogeographic regions, the geographic distribution of these endophytes can also be understood. It is also important to tap these endophytes and conserve them ex situ for bioprospecting.
References
Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Aly AH, Debbab A, Proksch P (2011) Fungal endophytes: unique plant inhabitants with great promises. Appl Microbiol Biotechnol 90:1829–1845
Anonymous (2008) A digest of forest statistics. Statistics Division, Department of Forests, Jammu and Kashmir Government, Srinagar, p 55
Arnold AE (2007) Understanding the diversity of foliar endophytic fungi: progress, challenges, and frontiers. Fungal Biol Rev 21:51–66
Arnold AE, Henk DA, Eells RL, Lutzoni F, Vilgalys R (2007) Diversity and phylogenetic affinities of foliar fungal endophytes in loblolly pine inferred by culturing and environmental PCR. Mycologia 99:185–206
Arnold AE, Lutzoni F (2007) Diversity and host range of foliar fungal endophytes: are tropical leaves biodiversity hotspots? Ecology 88:541–549
Aslam M, Reshi Z, Siddiqi TO (2011) Genetic divergence in half-sib progenies of Pinus wallichiana A.B.. Jackson plus trees in the Kashmir Himalaya, India. Trop Ecol 52:201–208
Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (1994) Current protocols in molecular biology. Wiley, New York
Bacon CW, White JF (2000) Microbial endophytes. Dekker, New York, pp 341–388
Barnett HL, Hunter BB (1998) Illustrated genera of imperfect fungi, 4th edn. APS, St. Paul
Botella L, Diez JJ (2011) Phylogenic diversity of fungal endophytes in Spanish stands of Pinus halepensis. Fungal Divers 47:9–18
Chang KK, Ju-Kyeong E, Ahn-Heum E (2013) Diversity and seasonal variation of endophytic fungi isolated from three conifers in Mt. Taehwa, Korea. Mycobiology 41:82–85
Clardy J, Walsh C (2004) Lessons from natural molecules. Nature 432:829–837
Coppen JJW, Robinson JM, Kaushal AN (1988) Composition of xylem resin from Pinus wallichiana and P. roxburghii. Phytochemistry 27:2873–2875
Dar AR, Dar GH (2006) The wealth of Kashmir Himalaya—gymnosperms. Asian J Plant Sci 5:251–259
Dar MY, Shah WA, Mubashir S, Rather MA (2012) Chromatographic analysis, anti-proliferative and radical scavenging activity of Pinus wallichiana essential oil growing in high altitude areas of Kashmir, India. Phytomedicine 19:1228–1233
Doty SL, Oakley B, Xin G, Kang JW, Singleton G, Khan Z, Vajzovic A, Staley JT (2009) Diazotrophic endophytes of native black cottonwood and willow. Symbiosis 47:23–33
Ek-Ramos MJ, Zhou W, Valencia CU, Antwl JB, Kalns LL, Morgan GD, Kerns DL, Sword GA (2013) Spatial and temporal variation in fungal endophyte communities isolated from cultivated cotton (Gossypium hirsutum). PLoS ONE 8:e66049
Ellof JN (1998) A sensitive and quick microplate method to determine the minimal inhibitory concentration of plant extracts for bacteria. Planta Med 64:711–713
Ezra D, Hess WH, Strobel GA (2004) New endophytic isolates of M. albus, a volatile antibiotic-producing fungus. Microbiology 150:4023–4031
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Ganley RJ, Brunsfeld SJ, Newcombe G (2004) A community of unknown, endophytic fungi in western white pine. Proc Natl Acad Sci 101:10107–10112
Ganley RJ, Newcombe G (2006) Fungal endophytes in seeds and needles of Pinus monticola. Mycol Res 110:318–327
Guo LD (2002) Pestalotiopsis besseyi, a new record of endophytic fungi from Pine in China. Mycosystema 21:455–456
Heywood VH (ed) (1995) Global biodiversity assessment. Cambridge University Press, Cambridge
Hoffman MT, Arnold AE (2008) Geographic locality and host identity shape fungal endophyte communities in cupressaceous trees. Mycol Res 112(Pt3):331–344
Iqbal J, Nelson JA, McCulley RL (2013) Fungal endophyte presence and genotype affect plant diversity and soil-to-atmosphere trace gas fluxes. Plant Soil 364:15–27
Keller NP, Turner G, Bennett JW (2005) Fungal secondary metabolism—from biochemistry to genomics. Nat Rev Microbiol 3:937–947
Khosla C (1997) Harnessing the biosynthetic potential of modular polyketide synthases. Chem Rev 97:2577–2590
Kiffer E, Morelet M (2000) Deuteromycetes. Mitosporic fungi. Classification and generic keys. Science, Enfield
Korsten L, Jager ESD, Villiers EED, Lourens A, Kotze JM, Wehner FC (1995) Evaluation of bacterial epiphytes isolated from avocado leaf and fruit surfaces for biocontrol of avocado postharvest diseases. Plant Dis 79:1149–1156
Kumar M, Qadri M, Sharma PR, Kumar A, Andotra SS, Kaur T, Kapoor K, Gupta VK, Kant R, Hamid A, Johri S, Taneja SC, Vishwakarma RA, Riyaz-Ul-Hassan S, Shah BA (2013) Tubulin inhibitors from an endophytic fungus isolated from Cedrus deodara. J Nat Prod 76:194–199
MacArthur R (1965) Patterns of species diversity. Biol Rev 40:510–533
Minter DW, Millar CS (1980) Ecology and biology of three Lophodermium species on secondary needles of Pinus sylvestris. Eur J For Pathol 10:169–181
Mousa WK, Raizada MN (2013) The diversity of anti-microbial secondary metabolites produced by fungal endophytes: an interdisciplinary perspective. Front Microbiol 4:65
Nagabhyru P, Dinkins RD, Wood CL, Bacon CW, Schardl CL (2013) Tall fescue endophyte effects on tolerance to water-deficit stress. BMC Plant Biol 13:127
Osono T, Mori A (2004) Distribution of phyllosphere fungi within the canopy of giant dogwood. Mycoscience 45:161–168
Porras-Alfaro A, Bayman P (2011) Hidden fungi, emergent properties: endophytes and microbiomes. Annu Rev Phytopathol 49:291–315
Qadri M, Johri S, Shah BA, Khajuria A, Sidiq T, Lattoo SK, Abdin MZ, Riyaz-Ul-Hassan S (2013) Identification and bioactive potential of endophytic fungi isolated from selected plants of the Western Himalayas. Springer Plus 2:8
Reinhold-Hurek B, Hurek T (2011) Living in side plants: bacterial endophytes. Curr Opin Plant Biol 14:435–443
Rodriguez R, Redman R (2008) More than 400 million years of evolution and some plants still can’t make it on their own: plant stress tolerance via fungal symbiosis. J Exp Bot 59:1109–1114
Rodriguez RJ, Henson J, Van Volkenburgh E, Hoy M, Wright L, Beckwith F, Kim Y, Redman RS (2008) Stress tolerance in plants via habitat-adapted symbiosis. ISME J 2:404–416
Rudgers JA, Miller TE, Ziegler SM, Craven KD (2012) There are many ways to be a mutualist: endophyte fungus reduces plant survival but increases population growth. Ecology 93:565–574
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Schulz B, Guske S, Dammann U, Boyle C (1998) Endophyte host interactions II. Defining symbiosis of the endophyte host interaction. Symbiosis 25:213–227
Shannon CE, Weaver W (1963) The mathematical theory of communication. University of Illinois Press, Urbana
Sieber SA, Marahiel MA (2005) Molecular mechanisms underlying nonribosomal peptide synthesis: approaches to new antibiotics. Chem Rev 105:715–738
Simpson GG (1964) Species density of North American recent mammals. Syst Zool 13:57–73
Sinclair WS, Lyon HH, Johnson WT (1987) Diseases of trees and shrubs, 4th edn. Cornell University Press, Ithaca, pp 32–33
Singh LP, Gill SG, Tuteja N (2011) Unraveling the role of fungal symbionts in plant abiotic stress tolerance. Plant Signal Behav 6:175–191
Staley JT, Castenholz RW, Colwell RR, Holt JG, Kane MD, Pace NR, Saylers AA, Tiedje JM (1997) The microbial world: foundation of the biosphere. American Academy of Microbiology, Washington, DC, p 32
Stone JK, Bacon CW, White JF Jr (2000) An overview of endophytic microbes: endophytism defined. In: Bacon CW, White JF Jr (eds) Microbial endophytes. Marcel Dekker, New York, pp 3–29
Strobel GA, Daisy B (2003) Bioprospecting for microbial endophytes and their natural products. Microbiol Mol Biol Rev 67:491–502
Tamura K, Nei M, Kumar S (2004) Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci 101:11030–11035
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Bio Evol 24:1596–1599
Troup RS (1921) The silviculture of Indian trees. Vol.III. Clarendon, Oxford
Vandenkoornhuyse P, Baldauf SL, Leyval C, Straczek J, Young JP (2002) Extensive fungal diversity in plant roots. Science 295:2051
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. Academic, San Diego, pp 315–322
Yuan Z, Zhang C, Lin F, Kubicek CP (2010) Identity, diversity, and molecular phylogeny of the endophytic mycobiota in the roots of rare wild rice (Oryza granulate) from a nature reserve in Yunnan, China. Appl Environ Microbiol 76:1642–1652
Acknowledgments
The first author is grateful to the Indian Council of Medical Research, New Delhi for her Senior Research Fellowship. We acknowledge the financial support obtained from CSIR 12th FYP project, “PMSI” (BSC0117) of the Council of Scientific and Industrial Research (CSIR), New Delhi, India. The authors are also thankful to Mr. Fairoz I. Nazki, a resident of Lolab valley, for facilitating the collection of plant samples, and Mr. Jasbir Singh and Rajinder Kumar for preserving the cultures. This article bears the institutional manuscript number IIIM/1626/2013.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Supplementary material Table 1
(DOCX 33 kb)
Rights and permissions
About this article
Cite this article
Qadri, M., Rajput, R., Abdin, M.Z. et al. Diversity, Molecular Phylogeny, and Bioactive Potential of Fungal Endophytes Associated with the Himalayan Blue Pine (Pinus wallichiana). Microb Ecol 67, 877–887 (2014). https://doi.org/10.1007/s00248-014-0379-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-014-0379-4