Abstract
Marine macroalgae cannot develop normal morphology under axenic conditions although normal morphogenesis can be sustained when certain bacteria are present. In this study, bacteria that induced normal morphogenesis in the red alga Pyropia yezoensis (Nori) were identified. The bacteria were isolated from algal media, thalli, tissue debris, and purified protoplasts during protoplast isolation from P. yezoensis laboratory cultures. 16S rRNA gene sequence analysis showed these bacterial isolates belonged to α-Proteobacteria (12 groups), γ-Proteobacteria (3 groups), and Flavobacteria (2 groups). Axenic protoplasts of P. yezoensis generated by removing epiphytic bacteria were co-cultured along with the bacterial isolates. Most axenic protoplasts showed irregular morphogenetic and anaplastic cells; cells with normal morphology were scarce. However, inoculation with 11 strains of Hyphomonas (α-Proteobacteria) led to significantly higher normal morphogenetic rates (4.5–7.3 %, P < 0.01 or 0.05) compared to axenic protoplasts (0.06 %). These Hyphomonas strains were recovered from all experiments; thus, certain Hyphomonas strains can induce normal morphogenesis in P. yezoensis protoplasts. Direct inoculation of the Hyphomonas strain exhibited higher morphogenetic activity than inoculation of its extracellular and intracellular products. This is the first study demonstrating the influence of specific bacteria on protoplast morphology in marine macroalgae.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Marine green macroalgae such as Ulva and Monostroma cannot develop normal morphology when they are grown in the absence of bacteria. For example, axenic thalli of Ulva lactuca lose typical morphology resulting in small colonies of uniseriate branching filaments [1]. Similar observations have been made of axenic calli of Ulva linza [2] and callus-like masses of Ulva pertusa [3]. Monostroma oxyspermum cells also do not develop typical morphology under aseptic conditions, growing solely as single cells or aggregates of single cells [4]. However, the typical morphology of Ulva and Monostroma was restored by reinfecting them with appropriate bacteria or adding the filtrates of bacterial cultures [1–4]. These previous studies therefore suggest that the presence of certain bacteria is essential for normal morphological growth of marine macroalgae, and that specific bacteria play important roles in algal morphology and growth.
The production of the marine red algae Pyropia and Porphyra (Bangiales, Rhodophyta), known as “Nori” in Japan, is one of the largest aquaculture industries in East Asia [5]. Pyropia and Porphyra are important edible species of seaweeds in Japan and East Asia and are often used in Japanese cuisine, e.g., sushi [6]. In particular, Pyropia yezoensis (former name “Porphyra yezoensis”) is a highly valued seaweed crop. The life cycle of P. yezoensis is heteromorphic with two alternating generations, a leaf gametophyte and a filamentous sporophyte called conchocelis phase [7]. Furthermore, the gametophyte has a secondary life cycle characterized by the production of haploid monospores and the development into new, leafy gametophytes [7]. The complexity of the natural life cycle is additionally compounded through the required release and germination of conchospores and carpospores.
The artificial isolation of protoplasts from thalli of Pyropia and Porphyra using enzyme treatment was established in the 1980s [8, 9]. Pyropia and Porphyra protoplasts have since been employed in cell biological, physiological, and genetic investigations [10, 11]. Studies have demonstrated that protoplast cultures developed into various morphologies and grew into leafy thalli [9, 12, 13]. Recently, axenic protoplast cultures of P. yezoensis were generated by removing epiphytic bacteria through a washing process and enzymatic preparation [14]. These cultures were then used as experimental materials to determine the draft genome sequence of P. yezoensis as whole genome sequencing had proven difficult due to contamination from bacterial DNA.
While numerous studies of bacteria associated with macroalgal morphogenesis have focused on green macroalgae such as Ulva and Monostroma [1–4], little research has examined this relationship with economically import red algae. Previously, it was demonstrated that P. yezoensis conchospores released from axenic filamentous thalli lost their typical morphology and germinated into callus-like masses [15]. Subsequently, Yamazaki et al. [16] isolated bacterial strains, classified as α-Proteobacteria, showing morphogenetic activity against the conchospores. However, there was no quantitative approach to assess the bacterial effects on morphogenesis and growth of the alga compared to the axenic control, and the recovery of incubated bacteria and presence of bacterial contaminants were not examined. Due to constraints of the method employed by Yamazaki et al. [16], namely the 2-week conchospore preparation with multiple antibiotics and the low yield of 30–60 cells, this approach was not suitable for testing a large number of bacterial strains. The use of protoplasts in place of conchospores, however, has two advantages: (i) large numbers of protoplasts (106 cells/mL) are produced from a few thalli within 6 h and (ii) the process used for removing bacteria attached on the cell surface results in the generation of axenic protoplasts. This is essential for the examination of the effects of many bacterial strains on morphological changes in protoplasts. As such, this study utilized protoplasts of P. yezoensis to select bacteria capable of inducing morphogenesis of this alga and represents the first report on bacteria that induce normal morphogenesis in the protoplasts of marine plants.
In this study, we hypothesize that the bacterial community inherent to the laboratory cultures of P. yezoensis may be associated with the growth of the host. The purpose of our investigation is to determine bacterial isolates associated with normal morphogenesis in P. yezoensis protoplasts. As a first step, we isolated various bacterial strains from laboratory cultures of P. yezoensis by adding different antibiotics to the algal culture in order to exploit the “hidden” bacteria. A total of 259 isolates from P. yezoensis were evaluated in co-culture experiments with axenic protoplasts. In addition, we also recovered and identified bacteria from experiments where bacteria were introduced and successfully demonstrated that the cultures have been free of cross contamination.
Material and Methods
Cultivation of P. yezoensis
Monospores of P. yezoensis U-51 were cultured in sterile modified half-strength SWM-III medium for approximately 5 weeks. The algal culture was incubated at 17 °C under illumination of white fluorescent lamps (50 μmol m−2 s−1, 10:14 h light:dark cycle), and the algal medium was replaced every week.
Protoplast Isolation from P. yezoensis
For isolation of protoplasts, a sample of the thalli weighing about 50–100 mg was used. The isolation procedure was done with sterile instruments under a clean bench, according to the modified method of Araki et al. [17]. In brief, the thalli were immersed in 0.5 % citric acid (pH 2.0–2.3) for 90 s, and rinsed twice in sterile 90 % natural seawater (NSW). The cleaned thalli were cut with a microtome blade, and shaken with 2 % papain solution for 30 min at room temperature (20–25 °C). The supernatant was removed, and the precipitated cells were washed thrice in 90 % NSW containing 0.7 M mannitol. Enzyme solutions of agarase, β-1,4-mannanase, and β-1,3-xylanase (1 unit/8 mL, Yakult pharmaceutical industry, Tokyo, Japan) were added to the washed cells, and then the cell suspension was shaken to lyse the cell walls for 60–90 min at room temperature. The suspension was filtered with 20-μm mesh filter to remove the undigested tissue debris (tissue debris sample). The filtrate fraction containing the protoplast cells was centrifuged, and then the protoplast pellet was washed thrice in 90 % NSW containing 0.7 M mannitol (protoplast sample).
Sample Preparation of P. yezoensis and Bacterial Isolation
After thalli were incubated in sterile modified half-strength SWM-III medium for approximately 4 weeks, the thalli were cultured in three different algal media for 1 week; one without antibiotics as a control, one with ampicillin (10 mg/L w/v), and one with neomycin (10 mg/L w/v). Since the presence of the antibiotics ampicillin and neomycin in the algal medium did not individually influence the growth of P. yezoensis, these antibiotics were used to counter select against sensitive species and enrich low-abundance insensitive bacteria species, which may subsequently be isolated through plating. After 1 week incubation in each algal medium, the samples of algal media and thalli (13–19 mg) were collected directly from culture flasks, and then tissue debris and protoplasts were isolated from remained thalli (53–61 mg) according to the method described above. The samples of thalli and tissue debris were mixed with 8 and 20 ml of sterile 75 % NSW, respectively, and vigorously shaken by hand. Each sample was serially diluted 10-fold and a 0.1 ml subsample of optimum dilution was spread on marine agar 2216 (BD, Franklin Lakes, NJ) and 10-fold diluted marine agar 2216, and then incubated at 20 °C for 14 days. In addition, 1 and 0.1 ml of each dilution of the tissue debris and protoplast samples were cultivated in three replicate tubes containing of marine broth 2216 by the most probable number (MPN) technique to increase the detection limit. After incubation, viable bacterial counts by plating on marine agar 2216 and 10-fold diluted marine agar 2216, and MPN values were measured. The MPN counts of tissue debris and protoplast samples were enumerated from the weight of thalli used in the experiment. Colonies from each sample were randomly selected and purified.
DNA Extraction and PCR Amplification
For DNA extraction from bacterial isolates, a single colony was suspended in TE buffer (10 mM Tris, 1 mM EDTA, pH 8.0) containing 1 % Triton X-100 (v/v) and boiled at 100 °C for 5 min. An equal volume of chloroform-isoamylalcohol (24:1 v/v) was added to the suspension, and then the protein was removed by centrifugation at 15,000 rpm for 15 min. The suspension was used as template DNA. PCR amplifications of 16S rRNA gene sequence were performed using the universal primer set 27F and 1492R [18]. PCR mixtures consisted of 2 μL of template DNA, 1× Ex Taq Buffer, dNTP mixture containing 0.2 mM of each nucleotide, 0.5 units of Takara Ex Taq (Takara Bio, Shiga, Japan), 0.5 μM of each primer in a total volume of 20 μL. PCR was performed by initial denaturation at 95 °C for 3 min followed by 30 cycles of denaturation at 95 °C for 30 s, annealing at 55 °C for 30 s, extension at 72 °C for 1 min, and a final extension at 72 °C for 7 min, in a C1000 thermal cycler (Bio-Rad Laboratories, Hercules, CA). Approximately 700 bp of the 5′ end of the 16S rDNA sequence was determined by automated DNA sequencing (Model 3100; Applied Biosystems, Foster City, CA) using the sequencing primer 27F.
Sequence and Phylogenetic Analysis
The partial 16S rRNA gene sequences obtained from each sample were aligned using Clustal X programs [19]. The phylogenetic distances among these sequences were calculated using MEGA version 4 [20]. Isolates showing >98 % identity were assigned to the same phylogenetic group. The representative isolates were selected from each phylogenetic group, and the near full-length sequences of 16S rRNA gene were determined using a combination of sequence primers 27F, 519F (positions 519–537 of E. coli CFT073 16S rRNA gene), 800F (positions 768–787 of E. coli), 1190F (positions 1,169–1,188 of E. coli), 530R (positions 517–536 of E. coli), 760R (positions 716–735 of E. coli), 920R (positions 908–926 of E. coli), and 1492R [18, 21]. These sequences were compared against the GenBank database constructed in the local BLAST database with BLASTN search [22]. A type strain in the database showing the highest homolog with the representative sequence has been described in the results below. When the representative sequence was having >95 % similarity to the type strain, the representative isolate was assumed to belong to the same genus [23].
Nucleotide Sequence Accession Number
The 16S rRNA gene sequences of the representative strains were deposited in GenBank/EMBL/DDBJ nucleotide sequence database under accession numbers AB583774 to AB583776, AB689190, AB689191, AB695286 to AB695288, and AB758562 to AB758592.
Co-cultivation of Protoplasts and Bacteria
Two hundred fifty-nine strains isolated from P. yezoensis were used in the co-culture experiment with axenic protoplasts. Each isolate was cultured in marine broth 2216, and the culture was diluted 10-fold with sterile 75 % NSW. In co-cultured systems, sterile 90 % NSW supplemented with NaNO3 (100 mg/L w/v) and Na2HPO4·12H2O (20 mg/L w/v) was used as the culture medium (NP medium). Dilute bacterial culture (10 μL; 105–106 CFU/well) was inoculated in a 24-well culture plate containing 1 mL of NP medium pre-inoculated with 0.1 mL of the protoplast sample (102 cells/well). In the control experiment, 10 μL of 10-fold diluted marine broth 2216 with 75 % NSW was added to the mixture. The incubation was performed at 17 °C for 6 weeks in the same conditions as described above. For selected strains, the same experiments were replicated four times independently.
Morphological Observation of Protoplasts and Recovery of Bacteria
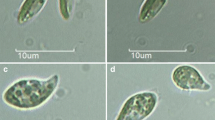
After 6 weeks of incubation, the morphology and number of the protoplasts were observed under 40× magnification using an inverted light microscope (ECLIPSE Ti-S; Nikon Instech, Tokyo, Japan). Surviving protoplasts were categorized into four morphological groups: normal morphogenetic, irregular morphogenetic, callus-like, and anaplastic cells (Fig. 1). A normal morphogenetic cell was defined as a cell possessing one rhizoid and normal cell division (Fig. 1d–f). An irregular morphogenetic cell was defined as a cell showing irregular cell division (Fig. 1b, e). The survival and normal morphogenetic rates were calculated by dividing the each cell number with the inoculated cell number. Data were subjected to one-way ANOVA with Dunnett’s multiple comparison post hoc test. P < 0.05 was considered statistically significant. In order to recover the inoculated bacteria, one loop filled with the co-culture medium incubated in each well was cultured on marine agar 2216 at 20 °C for 2 weeks, and then three representative colonies were selected and further purified. In experiments inoculated with Hyphomonas sp. strain LNM10-16, which showed higher morphogenetic activity, and Algimonas sp. type II strain 14A-2-1, which showed low normal morphogenetic activity, changes of these viable counts were investigated at initial inoculum and 6 weeks after incubation.
Representative photographs showing the morphologies of 6-week-old protoplasts isolated from P. yezoensis. a Anaplastic cell without added bacteria. b Irregular morphogenetic cell without added bacteria. c Callus-like cell without added bacteria. d Normal morphogenetic cell inoculated with Hyphomonas sp. strain LNM-9. e Normal (arrow) and irregular morphogenetic cells inoculated with Hyphomonas sp. strain LNM10-16. f Normal (arrow) and callus-like cells inoculated with Hyphomonas sp. strain LNM10-16. Bars 40 μm
Development of Hyphomonas-Specific PCR Method
Hyphomonas-specific PCR detection method was developed to identify isolates recovered from the culture wells in which Hyphomonas strains were inoculated. Near complete sequences of Hyphomonas strains were compared with those of representative strains in other phylogenetic groups, and then a Hyphomonas-specific primer set of HyphoF1 (5′-TTTCACTACGGAATAGCTCTT-3′; positions 137–157 of E. coli) and HyphoR1 (5′-CTCCTGGTCTCTAGACTTCC-3′; positions 645–664 of E. coli) was designed. The specificity of this primer set was evaluated using genomic DNAs of the representative strains in the 17 phylogenetic groups isolated from P. yezoensis. The PCR condition consisted of an initial denaturation at 95 °C for 3 min followed by 30 cycles of denaturation at 95 °C for 30 s, annealing at 55 °C for 30 s, extension at 72 °C for 1 min, and a final extension at 72 °C for 5 min. The PCR product was analyzed by electrophoresis on a 2.0 % agarose gel. After the specificity was confirmed by the representative genomic DNAs, further isolations were identified according to the description above. Accuracy of Hyphomonas identification was confirmed by 16S rDNA sequencing of bacterial isolations showing PCR-positive, using the sequencing primer 27F.
Survival and Morphogenetic Activities of a Hyphomonas Strain
Effects of viable cells and extracellular and intracellular products of Hyphomonas sp. on the survival and normal morphogenetic activities of P. yezoensis protoplasts were examined. Hyphomonas sp. strain LNM10-16 that show the highest normal morphogenetic activity was used for the experiments. A pre-culture (2.5 mL) was inoculated into 600 mL of marine broth 2216 and cultivated at 20 °C for 4 days with shaking at 120 rpm. The bacterial culture was centrifuged (10,000 rpm, 10 min), and the supernatant was filtered through a sterile 0.2-μm-pore-size filter (Cellulose acetate; Toyo Roshi Kaisha, Tokyo, Japan) and used as culture supernatant. The cell pellets were washed thrice in sterilized 75 % artificial seawater and then resuspended in 10 mL of sterilized distilled water. The resuspended cells were sonicated at 20 kHz and 150 W for 45 min using a sonicator (VP-30S; Taitec, Saitama, Japan), and the undisrupted cells were removed by centrifugation (10,000 rpm, 30 min, 4 °C). The filtered sample was used as intracellular extract. A sample of viable cells was prepared as 10-fold dilution of the bacterial culture. In order to adjust to the concentration of viable cells, samples of the culture supernatants and intracellular extracts were diluted 10-fold and 600-fold, respectively. Then, 10 μL of each sample was added to 1 mL of NP medium containing 102 cells of protoplasts. Three independent experiments were performed as described above. The statistical difference was determined by Student’s t test. Difference with P < 0.05 was considered significant.
Results
Enumeration of Viable Bacteria in P. yezoensis Samples
Details of the samples used for the bacterial isolation and viable bacterial counts are shown in Table 1. When each antibiotic was added to the algal medium, viable counts in the samples with ampicillin and neomycin were not reduced compared with those from algal medium without antibiotics. The bacterial colony counts in the algal media ranged from 7.3 × 105 to 3.6 × 106 CFU/mL on marine agar 2216, and from 5.2 × 105 to 9.6 × 105 CFU/mL on 10-fold diluted marine agar 2216. Viable counts on the thallus surface ranged from 2.3 × 106 to 3.3 × 106 CFU/g on marine agar 2216, and from 2.7 × 105 to 1.1 × 106 CFU/g on 10-fold diluted marine agar 2216. Bacterial abundance of the tissue debris without antibiotics was less than detectable levels, but MPN counts of the tissue debris with ampicillin and neomycin were 3.0 × 103 and 3.4 × 102 MPN/g, respectively. MPN counts of the protoplasts without antibiotics and with ampicillin were not detected, but MPN counts of the protoplasts with neomycin were 7.5 × 102 MPN/g.
Bacterial Community Composition in P. yezoensis Samples
Taxonomic compositions of bacterial isolates from algal media, thalli, tissue debris, and protoplasts of P. yezoensis are shown in Fig. 2. The predominant bacteria in the algal media without antibiotics, with ampicillin, and with neomycin, were Maritalea sp. type I, Alteromonas sp., and Marinobacter sp. type II on marine agar 2216, respectively, and Alteromonas sp., Roseovarius sp., and Hyphomonas sp. on 10-fold diluted marine agar 2216, respectively. On the surface of the thalli cultured without antibiotics, with ampicillin, and with neomycin, the dominant bacteria were represented by Maritalea sp. type I, Alteromonas sp., and Maritalea sp. type I on marine agar 2216, respectively, and Alteromonas sp., Alteromonas sp., and Marinobacter sp. type II on 10-fold diluted marine agar 2216, respectively. Bacteria were not isolated from the tissue debris and protoplasts without antibiotics, while Sphingopyxis sp. was isolated from the tissue debris with ampicillin, and the tissue debris and protoplasts with neomycin. Based on the similarity in the partial sequences of the 16S rRNA gene, 326 isolates from P. yezoensis were divided into 17 groups within the class α-Proteobacteria, γ-Proteobacteria, and Flavobacteria (Table 2). The most frequently represented class α-Proteobacteria (12 groups) consisted of the genus Maritalea (types I and II) in the family Hyphomicrobiaceae, the genera Hyphomonas and Algimonas (types I and II) in the family Hyphomonadaceae, the genera Ahrensia, Antarctobacter, Roseovarius, Sulfitobacter (types I and II), and Tropicibacter in the family Rhodobacteraceae, and the genus Sphingopyxis in the family Sphingomonadaceae. The class γ-Proteobacteria (three groups) was represented by the genera Alteromonas and Marinobacter (types I and II) in the family Alteromonadaceae, and the class Flavobacteria (two groups) was restricted to the genera Kordia and Polaribacter in the family Flavobacteriales.
Effects of Bacteria on Survival and Morphogenesis of P. yezoensis Protoplasts
Two hundred fifty-nine bacterial isolates were selected from 326 isolates based on the number of isolates in each phylogenetic group. Each of the 259 bacterial isolates was separately inoculated with axenic protoplasts in co-culture, and the average values of the survival and normal morphogenetic rates in 17 phylogenetic groups are shown in Fig. 3. In controls without bacterial incubation, the survival and normal morphogenetic rates of the protoplasts were 5.4 and 0 %, respectively. The mean survival rates of protoplasts were higher in Hyphomonas sp. (n = 24, 17.7 %), Algimonas sp. type I (n = 6, 18.2 %), and Algimonas sp. type II (n = 12, 21.2 %) than those of other phylogenetic groups (Fig. 3a). The inoculation with Hyphomonas sp. showed the highest mean rates of normal morphogenesis (n = 24, 3.7 %) (Fig. 3b).
a Average survival rates and b normal morphogenetic rates of P. yezoensis protoplasts at 6 weeks after incubation with 259 each strain divided into 17 phylogenetic groups. Control experiments were axenic protoplast cultures without added bacteria. The numbers on the top of each graph show the strain numbers used in co-cultured experiments. Values are means plus standard error from each inoculated experiment
Among the 259 isolates, 18 isolates that showed normal morphogenetic rates of more than 3 % in the genera Hyphomonas and Algimonas were used to analyze the role of these strains in increased morphological changes (Fig. 4). When the protoplasts were not inoculated with bacteria, the survival and normal morphogenetic rates were 3.6 and 0.06 %, respectively. Among the 18 strains, the incubation of Hyphomonas sp. strains SCM-2, LNM-9, LNM-21, and LNM10-16, Algimonas sp. type I strain LNM-3, and Algimonas sp. type II strain 14A-2-1 significantly increased the survival rates (20.0–24.3 %; P < 0.01 or 0.05) compared to that of the control without bacteria (Fig. 4a). Eleven strains of Hyphomonas sp. (strains SCM-2, SNM-14, LNM-9, LNM-21, LNM-24, SNM10-6, SNM10-12, SNM10-13, SNM10-16, SNM10-19, and LNM10-16) showed significantly higher normal morphogenetic rates (4.5–7.3 %; P < 0.01 or 0.05) as compared with that of the control (Fig. 4b).
a Survival rates and b normal morphogenetic rates of P. yezoensis protoplasts at 6 weeks after incubation with 18 each strain. Control experiments were axenic protoplast cultures without added bacteria. Values are means plus standard error from four independent experiments. Asterisks indicate that the values are significantly different from the control values (*P < 0.05; **P < 0.01)
Identification of Hyphomonas Strains Recovered from Co-cultivation and Changes of Bacterial Abundance
The HyphoF1-HyphoR1 primer set was designed to completely match the sequences of 11 strains of Hyphomonas sp. The primer set of HyphoF1-HyphoR1 was able to specifically amplify DNA of Hyphomonas strains among the 17 phylogenetic groups isolated from P. yezoensis. When the co-culture medium with 11 strains of Hyphomonas sp. (strains SCM-2, SNM-14, LNM-9, LNM-21, LNM-24, SNM10-6, SNM10-12, SNM10-13, SNM10-16, SNM10-19, and LNM10-16) was cultured on marine agar 2216, colonies with same morphology were observed in each experiment. When a total of 132 isolates were identified using the specific primer set, all of these isolates were confirmed as Hyphomonas sp. Furthermore, 16S rRNA sequences of 36 isolates from the co-culture experiments with strains LNM10-16, SCM-2, and LNM-9 showed almost 100 % similarities with those of the inoculated strain, supporting that HyphoF1 and HyphoR1 primers were able to accurately detect Hyphomonas strains.
Changes of viable counts of Hyphomonas sp. LNM10-16 (a high normal morphogenetic strain) and Algimonas sp. 14A-2-1 (a low normal morphogenetic strain) were investigated in co-culture experiments. The initial viable count 2.2 × 106 CFU/well of Hyphomonas sp. strain LNM10-16 changed to 1.7 × 106 CFU/well at incubation of 6 weeks. The initial abundance 1.2 × 106 CFU/well of Algimonas sp. strain 14A-2-1 changed to 9.2 × 105 CFU/well at incubation of 6 weeks.
Survival and Morphogenetic Activities of a Hyphomonas Strain
Survival and normal morphogenetic activities were higher with viable cells than with culture supernatants and intercellular extracts. The survival and normal morphogenetic activities of culture supernatants relative to those of viable cells were significantly reduced by 10.5 % (P < 0.01) and 9.3 % (P < 0.01), respectively. The survival activity of intercellular extracts was significantly reduced by 30.0 % (P < 0.01). The normal morphogenetic activity of intercellular extracts was not significantly different in that of viable cells (Fig. 5).
Comparison of survival and normal morphogenetic activities of Hyphomonas sp. strain LNM10-16 against P. yezoensis protoplasts among viable cells, culture supernatants, and intercellular extracts. The data represent means and standard error of measurements performed in three independent experiments. Asterisks indicate significant decreases compared with activities of the viable cells (**P < 0.01)
Discussion
In this study, we identified the bacteria that induce morphological changes in the protoplasts of the red alga P. yezoensis. Incubation of Hyphomonas strains with axenic protoplasts showed a significant increase in normal morphogenesis (Fig. 4b). These strains themselves were isolated from the healthy red alga and recovered in all co-culture experiments. In the present study, the Hyphomonas strains fulfilled Koch’s postulates; therefore, these strains are considered to be associated with the development of normal morphology in protoplasts of P. yezoensis.
We isolated bacterial cultures from various samples in laboratory cultures of P. yezoensis. The addition of antibiotics to algal cultures drastically changed bacterial floras and enabled us to isolate Maritalea sp. type II, Algimonas sp. type II, Roseobacter sp., and Tropicibacter sp., which were, in general, less frequent and “hidden” in cultures without antibiotics (Fig. 2). Within 17 phylogenetic groups associated with P. yezoensis, Maritalea porphyrae (Maritalea sp. type I), Algimonas porphyrae (Algimonas sp. type I), Algimonas ampicilliniresistens (Algimonas sp. type II), and Polaribacter porphyrae were novel species isolated for the first time [24–27]. In previous culture studies of Pyropia spp., Aeromonas, Bacillus, Corynebacterium, Enterobacteriaceae, Escherichia, Flavobacterium, Micrococcus, Phycisphaera, Staphylococcus, and Vibrio have been identified on the basis of phenotypic characteristics [28–30], which differed from those bacteria observed in this study. On the other hand, a culture-independent clone library method also detected the sequences of Alteromonas sp., Marinobacter sp., and Polaribacter sp. from the thalli of a laboratory strain of P. yezoensis [31]. However, this study detected no clone sequences related to α-Proteobacteria isolated in our study. This result could be explained by the finding that the sequences of the 75F primer developed by Namba et al. [31] were different from our α-Proteobacteria target sequences. It was reported that bacterial communities in laboratory blades of Porphyra umbilicalis which were treated with antibiotics differed from those in field samples [32]; therefore, the bacterial floras observed in P. yezoensis laboratory cultures might not fully reflect that in natural environments. However, because the laboratory thalli grew normally in the three algal media, these strains were implicated in a role of increased morphological changes.
Viable bacterial counts of the protoplasts (PGCM, PGAM, and PGNM) were lower than in the algal media and on the thallus surface (Table 1). The reduced viable counts of protoplasts may be attributed to the enzyme treatments and hypertonic solution used for protoplast production. However, the bacterial abundance of the protoplasts differed among three cultivation groups with/without antibiotics. The presence of neomycin caused low-level accumulation of Sphingopyxis sp. (<103 MPN/g) in/on protoplasts (Table 1 and Fig. 2). Our results suggested that even if an antibiotic was added to the algal medium, it was difficult to exclude epiphytic bacteria associated with P. yezoensis. On the other hand, the isolation of protoplasts from thalli incubated in algal medium without antibiotics allowed us to obtain a large number of axenic protoplasts (Table 1, PGCM). Therefore, axenic protoplasts without antibiotics were used as experimental material for the bacterial incubation in this study. Evidence for the consistency of the axenic status of the protoplasts was obtained throughout four replicate experiments of bacterial inoculation. These bacteria-free protoplasts have also been used as a powerful material in genome analysis of P. yezoensis [14].
When protoplasts were cultured in NP medium with and without bacteria, the incubated protoplasts exhibited four surviving morphologies, i.e., normal morphogenetic, irregular morphogenetic, callus-like, and anaplastic cells (Fig. 1). Similar morphologies have been observed during the regeneration of protoplasts from Pyropia and Porphyra [9, 12, 13]. It was reported that callus cells also formed thalli from the edges [13] or produced small cells which germinated into thalli [12]. In most previous studies, thalli were treated with antibiotics prior to protoplast isolation or antibiotics were added to the culture medium for protoplast regeneration; however, the presence of bacteria in the starting materials and bacterial contaminants has not been fully examined. When protoplasts of P. yezoensis were grown under axenic conditions, most of the axenic protoplasts observed were either irregular morphogenetic or anaplastic cells, and the number of normal morphogenetic cells was the lowest (Fig. 4). Our results correspond with those of a previous report showing that axenic conchospores of P. yezoensis failed to attain typical morphology [15].
The effects of 17 phylogenetic groups, including 259 bacterial strains, on the surviving and normal morphologies of protoplasts differed (Fig. 3). The differences among the 17 phylogenetic groups are likely to be attributed to the competition for nutrients between bacteria and protoplasts, or to the inhibition by either organism. Interestingly, the bacterial strains that showed significantly higher levels in the survival or normal morphogenetic rates were classified in the same family, Hyphomonadaceae (the genera Hyphomonas and Algimonas), on the basis of 16S rRNA gene sequences (Fig. 4 and Table 2). In the co-culture experiments, the relationship between bacterial population and normal morphogenetic effect was not observed in 6 weeks incubation, suggesting that the normal morphogenetic activity was specific for Hyphomonas strains. Members of the genus Hyphomonas have been characterized as having a biphasic life cycle consisting of flagellated swarmer cells and prosthecate cells that adhere to surfaces [33]. Other Hyphomonas strains are ubiquitous members of seawater, hydrothermal vents, and shellfish beds [33–35]. Using a culture-independent approach based on the 16S rRNA gene, sequences related to the genus Hyphomonas were detected in blades of Porphyra umbilicalis [32], which were cultured in a field. However, the distribution and function of Hyphomonas strains on aquacultured or wild species of Pyropia in natural environments is not clear.
A few strains that induce morphogenesis of conchospores in P. yezoensis were identified as members of the family Rhodobacteraceae (α-Proteobacteria) [16, 36]. These strains were distinct from the strains identified in our study, although the bacterial isolates associated with the morphogenesis of P. yezoensis in our study also belonged to the class α-Proteobacteria. In contrast, bacterial isolates involved in the morphogenesis of calli or callus-like masses of the green algae Ulva spp. belonged to a variety of taxonomic groups, including α-Proteobacteria, γ-Proteobacteria, Bacteroidetes, and Firmicutes [2, 3]. In this study, the direct inoculation of the Hyphomonas strain into P. yezoensis protoplasts resulted in high levels of survival and morphogenetic activities compared to indirect inoculation. A substance known as “thallusin”, which is produced by a strain of the Cytophaga-Flavobacteria-Bacteroides group, was essential for the morphogenesis of Monostroma cells and the germination of Ulva and Enteromorpha spores [37]; however, culture supernatants of active bacteria did not induce morphogenesis in the callus-like masses of U. pertusa [2]. It is not clear whether the activity of the Hyphomonas strain in P. yezoensis is the same as either of those observed in green algae or is a different phenomenon altogether. Our results imply that each organism may be dependent on the metabolites produced by the other.
Our bacteria belonging to the genera Hyphomonas and Algimonas were originally isolated from algal media and thalli in the laboratory environments of P. yezoensis, and these strains exhibited the survival or normal morphogenetic activities against protoplasts, which were artificially isolated from the thalli and did not present typical morphologies throughout the life cycle of the alga. An endophytic bacterial association with macroalgae is known for a Rhodopseudomonas-like bacterium of α-Proteobacteria isolated from the cytoplasm of rhizoids of the green alga Caulerpa taxifolia that has the potential to fix nitrogen in rhizoids [38]. In the red macroalga Prionitis, endophytic bacteria of α-Proteobacteria are responsible for gall formation (abnormal tissue growth) by overproduction of the phytohormone indole-3-acetic acid [39, 40]. From the results of this study, Hyphomonas strains might have a specific effect on the synthesis of cell walls or initial differentiation of protoplasts, unlike the phenomenon on the surface of thalli. However, to our knowledge, there are no reports on the biological meaning associated with interactions between bacteria and protoplast cultures of marine macroalgae to support this speculation. The mechanism of action by which Hyphomonas strains induce normal morphogenesis of P. yezoensis protoplasts and detailed differences among Hyphomonas strains need to be investigated in future studies.
In conclusion, the normal morphogenesis of axenic protoplasts is entirely dependent on the incubation with bacteria, whereas the addition of Hyphomonas strains significantly influences the development of normal morphology in protoplasts. This is the first report on the isolation of bacteria associated with normal morphogenesis in protoplasts of marine algae; previously, the effects of bacteria on marine algae have been evaluated using spores and calli, and relationships between bacteria and protoplast regeneration of marine algae have been poorly understood. This new technique using P. yezoensis protoplasts and bacteria that induce normal morphogenesis in the alga can be utilized for effective breeding in aquaculture.
References
Provasoli L, Pintner IJ (1980) Bacteria induced polymorphism in an axenic laboratory strain of Ulva lactuca (Chlorophyceae). J Phycol 16:196–201
Nakanishi K, Nishijima M, Nishimura M, Kuwano K, Saga N (1996) Bacteria that induce morphogenesis in Ulva pertusa (Chlorophyta) grown under axenic conditions. J Phycol 32:479–482
Marshall K, Joint I, Callow ME, Callow JA (2006) Effect of marine bacterial isolates on the growth and morphology of axenic plantlets of the green alga Ulva linza. Microbial Ecol 52:302–310
Matsuo Y, Suzuki M, Kasai H, Shizuri Y, Harayama S (2003) Isolation and phylogenetic characterization of bacteria capable of inducing differentiation in the green alga Monostroma oxyspermum. Environ Microbiol 5:25–35
Mumford TF, Miura A (1988) Porphyra as food: cultivation and economics. In: Lembi CA, Waaland JR (eds) Algae and human affairs. Cambridge University Press, Cambridge, UK, pp 87–117
Sahoo D, Tang X, Yarish C (2002) Porphyra—the economic seaweed as a new experimental system. Curr Sci 83:1313–1316
Miura A (1985) Genetic analysis of the variant color types of light red, light green and light yellow phenotypes of Porphyra yezoensis (Rhodophyta, Bangiaceae). In: Hara H (ed) Original and evolution of diversity in plants and plant communities. Academia Scientific Book, Tokyo, pp 270–284
Saga N, Sakai Y (1984) Isolation of protoplasts from Laminaria and Porphyra. Bull Jap Soc Sci Fish 50:1085
Fujita Y, Migita S (1985) Isolation and culture of protoplasts from some seaweeds. Bull Fac Fish Nagasaki Univ 57:39–45 (in Japanese)
Fujita Y, Saito M (1990) Protoplast isolation and fusion in Porphyra (Bangiales, Rhodophyta). Hydrobiologia 204(205):161–166
Mizukami Y, Okauchi M, Kito H, Ishimoto S, Ishida T, Fuseya M (1995) Culture and development of electrically fused protoplasts from red marine algae, Porphyra yezoensis and P. suborbiculata. Aquaculture 132:361–367
Chen LCM (1987) Protoplast morphogenesis of Porphyra leucosticta in culture. Bot Mar 30:399–403
Waaland JR, Dickson LG, Watson BA (1990) Protoplast isolation and regeneration in the marine red alga Porphyra nereocystis. Planta 181:522–528
Nakamura Y, Sasaki N, Kobayashi M et al (2013) The first symbiont-free genome sequence of marine red alga, susabi-nori (Pyropia yezoensis). PLoS One 8:e57122
Yamazaki A, Nakanishi K, Saga N (1998) Axenic tissue culture and morphogenesis in Porphyra yezoensis (Bangiales, Rhodophyta). J Phycol 34:1082–1087
Yamazaki A, Sekida S, Hanzawa N, Okuda K, Saga N (2000) Factors in growth and morphogenesis recovery under axenic conditions in Porphyra yezoensis (Bangiales, Rhodophyta), especially the effect on symbiotic bacteria in conditioned media. Bull Inst Oceanic Res Develop, Tokai Univ 21:57–76 (in Japanese)
Araki T, Aoki T, Kitamikado M (1987) Preparation and regeneration of protoplasts from wild-type of Porphyra yezoensis and green variant of P. tenera. Nippon Suisan Gakkaishi 53:1623–1627 (in Japanese)
Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. Wiley, New York, pp 115–175
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Satomi M, Kimura B, Mizoi M, Sato T, Fujii T (1997) Tetragenococcus muriaticus sp. nov., a new moderately halophilic lactic acid bacterium isolated from fermented squid liver sauce. Int J Syst Bacteriol 47:832–836
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Ludwig W, Strunk O, Klugbauer S, Klugbauer N, Weizenegger M, Neumaier J, Bachleitner M, Schleifer KH (1998) Bacterial phylogeny based on comparative sequence analysis (review). Electrophoresis 19:554–568
Fukui Y, Abe M, Kobayashi M, Ishihara K, Oikawa H, Yano Y, Satomi M (2012) Maritalea porphyrae sp. nov., isolated from a red alga (Porphyra yezoensis), and transfer of Zhangella mobilis to Maritalea mobilis comb. nov. Int J Syst Evol Microbiol 62:43–48
Fukui Y, Abe M, Kobayashi M, Saito H, Oikawa H, Yano Y, Satomi M (2013) Algimonas porphyrae gen. nov., sp. nov., a member of the family Hyphomonadaceae, isolated from the red alga Porphyra yezoensis. Int J Syst Evol Microbiol 63:314–320
Fukui Y, Kobayashi M, Saito H, Oikawa H, Yano Y, Satomi M (2013) Algimonas ampicilliniresistens sp. nov. isolated from the red alga Porphyra yezoensis, and emended description of the genus Algimonas. Int J Syst Evol Microbiol, 63:4407–4412
Fukui Y, Abe M, Kobayashi M, Saito H, Oikawa H, Yano Y, Satomi M (2013) Polaribacter porphyrae sp. nov., isolated from the red alga Porphyra yezoensis, and emended descriptions of the genus Polaribacter and two Polaribacter species. Int J Syst Evol Microbiol 63:1665–1672
Tsukidate J (1971) Microbiological studies of Porphyra plants—II. Bacteria isolated from Porphyra leucosticta in culture. Bull Jap Soc Sci Fish 37:376–379
Duan D, Xu L, Fei X, Xu H (1995) Marine organisms attached to seaweed surfaces in Jiaozhou Bay, China. World J Microbiol Biotechnol 11:351–352
Sunairi M, Tsuchiya H, Tsuchiya T, Omura Y, Koyanagi Y, Ozawa M, Iwabuchi N, Murooka H, Nakajima M (1995) Isolation of a bacterium that causes anaaki disease of the red algae Porphyra yezoensis. J Appl Bacteriol 79:225–229
Namba A, Shigenobu Y, Kobayashi M, Kobayashi T, Oohara I (2010) A new primer for 16S rDNA analysis of microbial communities associated with Porphyra yezoensis. Fish Sci 76:873–878
Miranda LN, Hutchison K, Grossman AR, Brawley SH (2013) Diversity and abundance of the bacterial community of the red macroalga Porphyra umbilicalis: Did bacterial farmers produce macroalgae? PLoS One 8:e58269
Moore RL, Weiner RM, Gebers R (1984) Genus Hyphomonas Pongratz 1957 nom. rev. emend., Hyphomonas polymorpha Pongratz 1957 nom. rev. emend., and Hyphomonas neptunium (Leifson 1964) comb. nov. emend. (Hyphomicrobium neptunium). Int J Syst Bacteriol 34:71–73
Weiner RM, Devine RA, Powell DM, Dagasan L, Moore RL (1985) Hyphomonas oceanitis sp. nov., Hyphomonas hirschiana sp. nov., and Hyphomonas jannaschiana sp. nov. Int J Syst Bacteriol 35:237–243
Weiner RM, Melick M, O'Neill K, Quintero E (2000) Hyphomonas adhaerens sp. nov., Hyphomonas johnsonii sp. nov. and Hyphomonas rosenbergii sp. nov., marine budding and prosthecate bacteria. Int J Syst Evol Microbiol 50:459–469
Hanzawa N, Chiba SN, Miyajima S, Yamazaki A, Saga N (2010) Phylogenetic characterization of marine bacteria that induce morphogenesis in the red alga Porphyra yezoensis. Fish Genet Breeding Sci 40:29–35
Matsuo Y, Imagawa H, Nishizawa M, Shizuri Y (2005) Isolation of an algal morphogenesis inducer from a marine bacterium. Science 307:1598
Chisholm JRM, Dauga C, Ageron E, Grimont PAD, Jaubert JM (1996) ‘Roots’ in mixotrophic algae. Nature 381:382
Ashen JB, Cohen JD, Goff LJ (1999) GC-SIM-MS detection and quantification of free indole-3-acetic acid in bacterial galls on the marine alga Prionitis lanceolata (Rhodopyta). J Phycol 35:493–500
Ashen JB, Goff LJ (2000) Molecular and ecological evidence for species specificity and coevolution in a group of marine algal-bacterial symbioses. Appl Environ Microbiol 66:3024–3030
Acknowledgments
This study was supported by Research Fellowships of the Japan Society for the Promotion of Science for Young Scientists. We would like to thank Dr. J. Bruckner (Desert Research Institute) for discussion of this study.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Fukui, Y., Abe, M., Kobayashi, M. et al. Isolation of Hyphomonas Strains that Induce Normal Morphogenesis in Protoplasts of the Marine Red Alga Pyropia yezoensis . Microb Ecol 68, 556–566 (2014). https://doi.org/10.1007/s00248-014-0423-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-014-0423-4