Abstract
To assess microbial dynamics during anaerobic digestion (AD) of sewage sludge (SWS) from a municipal Wastewater Treatment Plant (WWTP), a Biochemical Methane Potential (BMP) assay at 37 °C under mono-digestion conditions was conducted. Utilizing the Illumina MiSeq platform, 16S ribosomal RNA (rRNA) gene sequencing unveiled a core bacterial community in the solid material, showcasing notable variations in profiles. The research investigates changes in microbial communities and metabolic pathways to understand their impact on the efficiency of the digestion process. Prior to AD, the relative abundance in SWS was as follows: Proteobacteria > Bacteroidota > Actinobacteriota. Post-AD, the relative abundance shifted to Firmicutes > Synergistota > Proteobacteria, with Sporanaerobacter and Clostridium emerging as dominant genera. Notably, the methanogenic community underwent a metabolic pathway shift from acetoclastic to hydrogenotrophic in the lab-scale reactors. At the genus level, Methanosaeta, Methanolinea, and Methanofastidiosum predominated initially, while post-AD, Methanobacterium, Methanosaeta, and Methanospirillum took precedence. This metabolic transition may be linked to the increased abundance of Firmicutes, particularly Clostridia, which harbor acetate-oxidizing bacteria facilitating the conversion of acetate to hydrogen.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Sewage sludge (SWS) generation and disposal have become one of the greatest sanitary challenges of the twenty-first century (Nascimento et al. 2018). As a by-product of biological wastewater treatment, SWS generation is facing a dramatic increase with the population growth and the continuous improvement in the wastewater treatment facility (Guo et al. 2023). In some regions with insufficient sewerage and wastewater treatment facilities SWS is discharged directly into receiving water bodies. In 2020, the volume of municipal wastewater generated annually worldwide was estimated to be 360–380 cubic kilometres, with prediction of a 24% increase by 2030 and a 51% increase by 2050 (Giacomo and Romano 2022). Feng et al (2023) estimated that the annual global production of SWS may rise from 53 million tons dry solids in 2023 to 160 million tons if the wastewater generated globally is to be treated to a similar level as in the 27 European Union countries/UK. As the production of SWS increases, there is a corresponding rise in the energy demand for its treatment (Ferrentino et al. 2023).
As a renewable energy, biogas has been proved to be a feasible alternative to fossil fuels, since it can contribute to slow down the non-renewable energy exhaustion (Fu et al. 2023) and is considered one of the key environmental technologies that can provide affordable, sustainable, and secure energy (Garlicka et al. 2023). Several investigations have been conducted to improve the SWS anaerobic digestion (AD), specially to: enhance biogas/methane yields (Gu et al. 2020; Nguyen et al. 2021; Tao et al. 2020a, b); for nutrient rich digestate production; inactivating pathogens; decreasing the abundance of antibiotic resistance genes and; degrading emerging contaminants (Liew et al. 2022; Li et al. 2022).
The biochemical methane potential (BMP) assay is defined as a measure of substrate biodegradability determined through the cumulative CH4 production from an organic material anaerobically incubated and monitored over time (Hafner et al. 2020; Holliger et al. 2021; Raposo et al. 2011; VDI 4630 2016). This methodology is widely used to test the degradability of different organic wastes and it is considered a suitable method to compare the degradability of different substrates in bench scale (Lavergne et al. 2018).
It is well-known the complex process of AD is based on close interactions between numerous microorganisms (MO), which degrade organic polymers in a sequence of steps involving hydrolysis, acidogenesis, acetogenesis and methanogenesis, resulting in methane (CH4), carbon dioxide (CO2) and water production (Angelidaki et al. 2018).
Microbial methanogenesis activity has been widely studied in bench scale systems. Methanogenic archaea can produce CH4 in anaerobiosis through different biochemical pathways, but mainly using acetoclastic and hydrogenotrophic metabolisms (Cai et al. 2021). Methanogenic archaea are specialized in using different substrates, such as hydrogen (H2), methanol and acetate, to produce CH4. Each metabolic pathway creates a unique environment able to provide syntrophic relations with fermenters bacteria in a complex microbiome (Zhu et al. 2020). Several MO such as Methanobacterium sp., Methanosarcina sp., Methanococcus sp., Methanosaeta sp., and Methanospirillum were isolated from different ecological systems and identified for their contribution to biogas production (Tao et al. 2020a, b).
In recent decades, culture-independent molecular biological techniques have made considerable contributions to describe the microbial communities involved in biological processes, mainly by targeting the 16S rRNA gene (Walter et al. 2019). Advances in next-generation sequencing (NGS) technologies have revolutionized the field of environmental microbiology. NGS platforms now enable the retrieval of an unprecedent amount of DNA sequence directly from environmental samples, providing a cost-effective and precise representation of microbial diversity (Treu et al. 2018).
This investigation sought to advance our understanding of microbial community dynamics in SWS digestion, particularly in the context of AD processes, which play a crucial role in wastewater treatment plants (WWTPs). While previous studies have indeed explored microbial communities in AD, this research introduces several novel components that contribute to the current scientific knowledge.
Firstly, the study focuses on the specific context of a municipal WWTP in Rio de Janeiro, Brazil, which may harbour unique microbial populations due to regional environmental factors and operational conditions. Understanding microbial communities in diverse geographical locations is essential for developing tailored strategies for wastewater treatment.
Secondly, the investigation employs Illumina MiSeq technology, a state-of-the-art high-throughput sequencing method, to characterize the taxonomic composition of both bacteria and archaea communities.
Thirdly, the study evaluates microbial community dynamics before and after AD in BMP reactors. This temporal analysis provides insights into how microbial communities adapt and evolve in response to AD conditions, shedding light on the ecological processes underlying AD performance and efficiency.
Methodology
BMP configuration and sampling
SWS were taken from an anaerobic digestor at a large municipal WWTP in Rio de Janeiro city/ Brazil. The WWTP has capacity to treat 7,400 m3day−1. The hydraulic retention time (HRT) of the anaerobic reactor is 28 days. It currently treats 2.5 m3s−1 of wastewater and serves a population equivalent to 1.5 million inhabitants.
The BPM assay was carried out according to previous studies by Angelidaki et al (2009), Hafner et al (2020) and the German Guideline for Fermentation of Organic Materials (VDI 2016), to evaluate CH4 production from SWS in bench-scale. The methodology for assembling the BMP and validating the system are found in detail in our previous publication (Rocha et al. 2024).
The experiment was conducted in 3 replicates (n = 3) incubated during 11-days, when daily CH4 production during three consecutive days is < 1% of the accumulated CH4 volume (Holliger et al. 2021) under mesophilic conditions (37 ± 0.1 °C) using water bath and digestion bottles of 250 mL (total volume) and 100 mL (working volume). The reactors were called R1, R2 and R3 before AD and R4, R5, R6 post-AD.
The 3 SWS samples were kept in 1 L Schott bottles and immediately taken to the laboratory to set up the BMP assays (Table 1). SWS initial and final values for the main physicochemical parameters (Table 2) were measured according to Standard Methods Protocol (APHA 2017) as shown in our previous publication (Rocha et al. 2024).
Chemical oxygen demand (COD) analyses were conducted with a Shimadzu UV-1800 UV–VIS Spectrophotometer and the alkalinity through potentiometric titration. The pH was measured with MS Tecnopon model Mpa210 meter and the temperature was recorded with a digital thermometer. The gravimetric method in the analytical scale Gehaka AG200 was used. Total organic carbon (TOC) was analysed using Shimadzu Total Organic Carbon Analyzer TOC 5000A. The SWS used in the assays (100 mL in each assay) played the role of both inoculum and substrate. All parameters were measured in each replicate and the values are presented as mean value and standard deviation.
The BMP bottles were sealed with a silicone stopper. To prevent gas leakage, caps and connectors were sealed with high vacuum grease. To purge the existing O2, N2 gas was flushed into the bottle's headspace for 2 min. The BMP units sealing was checked with the aid of a high-pressure pump, a differential dual port piezoresistive pressure transducer MPX5050DP, a Fluke multimeter and an Arduino data logger. The bottles were gently shaken manually twice a day to prevent particle retention and to avoid clogging in the system.
DNA extraction
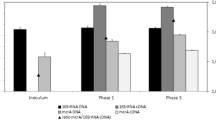
Genomic DNA was extracted from the samples using the PowerSoil™ DNA Isolation Kit (MoBio, Carlsbad, CA, USA), according to the manufacturer’s instructions. The DNA was quantified with a nanodrop ND-1000 spectrophotometer, and its yield and purity were documented (characterized by the absorption ratio of 260/280 nm). To verify the integrity of the extracted DNA, a 5μL aliquot of the sample was subjected to electrophoresis at 80 Volts in agarose gel (0.8%) for two hours. The gel was stained for approximately 15 min in ethidium bromide solution (2 μg/mL) and observed on a transilluminator with ultraviolet light.
Microbial communities in both stages (at the beginning and the end of the 11-days of AD) in the BMP assays were characterized using 16S rRNA marker gene. Sampling replicates were carried out in both stages of the experiment and the results were compared to evaluate adaptation and specialization of the microbial consortia during the AD.
Library preparation and sequencing analysis
The libraries were prepared following the Illumina recommendations. Primers were used for locus-specific amplification of bacteria flank the locus-specific region. Overhang sequence of adapters was included in locus-specific primers. The following Illumina linker sequences (locus-specific sequence) were hybridized to the sequences immobilized on the sequencing slide:
-
i.
forward overhang: 5'-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG-
-
ii.
reverse overhang: 5'-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAG-
Sequencing was conducted using the Illumina Miseq system, resulting in paired end reads of 250 base pairs each. Initially, a polymerase chain reaction (PCR) was carried out to selectively amplify locus-specific regions of 16S rRNA. Following this, AMPure XP beads were employed to purify the PCR products, and the size distribution of the resulting fragments was assessed through agarose gel electrophoresis. Subsequently, a second PCR step was performed to incorporate barcodes from the Nextera XT kit, and additional PCR purification and library validation steps were executed. Next, the libraries were quantified, ensuring that all samples/ libraries were combined in equimolar proportions into a unified pool. To introduce sequencing diversity, a heterogeneous control in the form of the phi-X phage was blended with the amplicon pool. Finally, denaturation of both the libraries and the phi-X control was carried out to facilitate the sequencing process.
Data analysis
Multiplexed reads were assigned on biological samples. The DADA2 program (Callahan et al. 2016), an open-source package implemented in the R language, was used to model and correct amplicons errors. The DADA2 package has a complete pipeline implemented to transform the sequencer's fastq files into inferred, disassembled, and chimera-free sample sequences. Filtering of fastq files was performed to cut the PCR primer sequences and filter the 3' ends of the reads due to quality decay (Q < 30).
After filtering, the reads had a size of 2 × 250 bp, keeping the overlap for later joining the readings and reassembling the fragment. The DADA2 algorithm makes use of a parametric error model, and each amplicon dataset has a different set of error rates. The learnErrors method learns this error model from the data, alternating between estimating error rates and inferring sample composition until they converge on a consistent solution. As with many machine learning problems, the algorithm must start with an initial guess, for which, the maximum possible error rates on that data are used (the error rates if only the most abundant sequence is correct and everything else is errors). For greater accuracy, the error is estimated with component samples from the entire sequencing run. Then, the denoising step is performed to obtain a detailed list of unique sequences and their abundances and produce consensus position quality scores for each unique sequence, averaging the positional qualities of the component reads.
After the initial processing of the sequencing data by DADA2, taxonomies were assigned to each amplicon sequencing variants (ASV) using a DADA2 program implementation of the naive Bayesian classifier method for this purpose. The assignTaxonomy function takes as input a set of sequences (ASVs) to be classified and a training set of reference sequences with known taxonomy and assigns taxonomies. The Silva 138.2 database was used as a reference. The taxonomic classifications, and their quantifications, generated by DADA2 were imported into the Phyloseq program (McMurdie and Holmes 2013), and implemented in R. ASVs that were not classified at least up to the family level were filtered out, and ASVs assigned the same genus were clustered.
Results and Discussion
Physicochemical analysis
In our previous publication (Rocha et al. 2024), we presented the results of the assembly and validation of bench-scale bioreactors with the insertion of experimental parameters to feed a theoretical model for AD (the ADM1 model), and the subsequent mathematical model validation and calibration for SWS AD biogas yield from a WWTP. The bench-scale BMP assays had a methane gas cumulative production of 136 ± 7.5 Nml CH4 and a yield of 124 ± 6.23 mL CH4 g−1 VS−1. Similar results for SWS mono-digestion methane production in experimental assays (138.2, 121, 124.4 mL CH4 g−1 VS−1) are found in scientific publications by Alves et al (2020), Kashi et al (2017) and Pan et al (2019), respectively.
Physicochemical analysis (Table 2) shows pH values (7.43 ± 0.1 in the beginning and 7.60 ± 0.1 after AD) within the range expected due to the growth of MO and biogas production in all reactors without the addition of a buffer solution.
TOC values measured before and after the experiment (895 ± 102 and 789 ± 100 mg L−1 respectively) refers to the organic carbon content in the SWS and indirectly reflects the content of organic matter (OM) (Zhu et al. 2020). The high-temperature combustion applied in the TOC method has been reported to be reliable with higher oxidation rates of various types of OM (Park et al. 2021). The OM in the system may be relatively stable and resistant to degradation or alteration under the experimental conditions. Microbial processes, such as decomposition and assimilation of OM, could have reached an equilibrium, which maintains the overall OM content even if there are fluctuations in the TOC values.
The alkalinity values measured before and after AD (2382 ± 102 and 2332 ± 101 mg CaCO3 L−1 respectively) (Table 2) indicates the sludge capability of buffering the reaction. High values for alkalinity possibilities that the reaction is buffered, so the pH tends to not undergo major changes (Campanaro et al. 2020).
The SWS TCOD values (Table 2) in the beginning and after 11 days of AD (21,903 ± 1,002 mg L−1 and 16,502 ± 598 mg L−1 respectively) could indicate that a significant portion of the OM is converted into gases, primarily CH4 and CO2, which are released from the system.
Serna-García et al (2020) and Xie et al (2020) found similar values for SWS TCOD from WWTPs (22,430 ± 1190 and 20,400 ± 1050 mg L−1, respectively). In relation to TCOD reduction, our study presented 24,66% and other studies with BMP tests (Kashi et al. 2017; Maragkaki et al. 2018 and Zhang et al. 2016) obtained similar values: 16%, 28.6%, and 25.22% respectively.
The stability of TOC and the reduction in COD can coexist if part of the OM in the sample has been transformed into products (such as gases or insoluble compounds) that are not detected by the COD method. This can occur due to the diverse nature of organic compounds in the sample and the possible limited capacity of the COD method to detect all types of oxidizable OM.
Total solid (TS), total volatile solids (TVS) and volatile suspended solids (VSS) presented a reduction after AD from 2.0 ± 0.26 to 1.0 ± 0.13%, 1.1 ± 0.11 to 0.7 ± 0.09%, and 0.8 ± 0.10 to 0.5 ± 0.08%, respectively. These results suggest that there is a greater proportion of organic compounds compared to inorganic compounds in the SWS. The reduction observed in the organic fraction during AD, indicates that a substantial portion of the sludge consists of OM that is susceptible to microbial degradation.
Bacterial biodiversity at phylum level
The microbial community profile of the SWS varied before and after the AD process, which means the microbial structure in these BMP reactors were affected during the process (Fig. 1, Fig. 2 and Supplementary Files). AD needs a complex microbial community and specific nutrients in the substrates to promote methanogenic activity (Yang et al. 2016). The AD assays conducted revealed high bacterial and archaeal diversity.
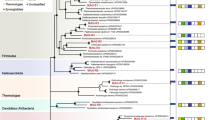
Bacterial Phylum Chord Diagram related to reactors R1, R2 and R3 (before AD) and R4, R5 and R6 (post-AD). The 10 most representative Phylum groups: Firmicutes, Sinergy: Synergistota, Proteob: Proteobacteria, Ba: Bacteroidota, Ca: Caldisericota, Ao: Actinobacteriota, Ai: Acidobacteria, C: Chloroflexi, D: Desulfobacterota and H: Hydrogenedentes
Clear changes were observed in the bacterial diversity after 11 days of the experiment. The most represented phyla in terms of Operational Taxonomic Units (OTU) in the sludge before AD were: Proteobacteria (10,667–15,248 OTU, 52.5%–66.1%) > Bacteroidetes (3544–4943 OTU, 17.2%–21.8%) > Actinobacteriota (1799–2495 OTU, 8.6%–11.1%) > Firmicutes (1082–1619 OTU, 5.5%–6.7%) > Chloroflexi (970–1183 OTU, 4.1%–5.9%). At the end of the BMP assays bacterial community changed the relative abundance and a new dominance community configuration emerged in the system as following: Firmicutes (52,648 OTU–65,368 OTU, 54.3%–78.4%) > Synergistota (6980 OTU–8182 OTU, 7.5%–9.6%) > Proteobacteria (3530 OTU–4010 OTU, 3.6%–4.7%) > Actinobacteriota (2103 OTU–2235 OTU, 2.5%–2.7%) > Desulfobacterota (913–1077 OTU, 1.6%) (Fig. 1). This profile has been consistently reported in previous studies, suggesting there is a general and consistent signature of the AD microbiome (Abendroth et al. 2015; Gao et al. 2022; Goux et al. 2015; Treu et al. 2018).
By analysing the resulting chord diagram (Fig. 2) insights into the connections and patterns among the entities/categories, identify a connection between phylum Firmicutes and Synergistes related with AD process (R4, R5 and R6). Major variety of bacterial phylum was also observed before AD (R1, R2 and R3) with an increase in terms of number of organisms after AD and phylum variety reduction.
At the phylum level, Firmicutes, Bacteroidota, Chloroflexi and Proteobacteria are the main bacteria in WWTP SWS in which, Firmicutes and Bacteroidota are the most common in AD reactors, affecting the degree of substrate fermentation (Gao et al. 2022; Jiang et al. 2022; Nascimento et al. 2018; Schneider et al. 2021). Chloroflexi and Proteobacteria, also corroborate with the complex environment associated with SWS, where microbial diversity adjusts to the characteristics of each reactor type, local climatic conditions, and operational parameters (Al Ali et al. 2020; Saha et al. 2020).
Firmicutes are frequently reported in anaerobic sludge treatment systems (Yang et al. 2014) and are recognized for its metabolic versatility, enabling them to degrade a variety of substrates (Li et al. 2022). In AD, they actively participate in hydrolytic and acidogenic steps (Zhu et al. 2020), producing volatile fatty acids (VFA), an important substrate for AD development (Nascimento et al. 2018). They can produce cellulases, lipases, proteases and other extracellular enzymes that carry out the degradation of several substrates, including protein, lipids, lignin, cellulose, sugars and amino acids (Qin et al. 2021). This group is a well-described fermenting bacterium often developing syntrophic cooperation with methanogens, by degrading butyrate and its analogues (Garcia-Peña et al. 2011). The H2 released in the process is scavenged by methanogens, making the reaction thermodynamically possible for both partners (Walter et al. 2019). Within the phylum Firmicutes, Clostridia is the major class, accounting for 95.5 ± 2.32% of Firmicutes reads.
Bacteriodetes are one of the primary populations participating in hydrolysis and fermentation in SWS carbohydrates. This group is known to play several roles in AD, been reported as sugar fermenters and plant cellulose degraders (Yang et al. 2016). Protein degradation and amino acid fermentation to acetate, propionate and succinate have been documented among species (Kampmann et al. 2012).
Proteobacteria (52.5–66.1% prior to AD and 3.6–4.7% post-AD) is a highly diverse bacteria phylum with significant metabolic capacity, actively participating in the carbon, nitrogen, sulphur, and phosphorus cycles (Meyer et al. 2016). Proteobacteria contributes to all AD steps, producing a broad range of fermentation products (Cai et al. 2016), they also contribute to all metabolic pathways involved in OM degradation (Jiang et al. 2019a, b). The decrease in Proteobacteria post-AD in reactors R4, R5 and R6 could be attributed to the strict anaerobic environmental conditions and microbiological interactions within the system. Some possible explanations refer to microbial competition for available substrates, adaptation to anaerobic conditions in which, some species of Proteobacteria may not be well adapted to strict anaerobic environments and therefore, may decrease in number over time as the system stabilizes anaerobically.
Hydrogenedentes is a recently proposed phylum of bacteria, previously known as NKB19. This phylum increased 7 to 10% post-AD. Genetic analyses of the sequences extracted from the environment suggest these organisms play an important role in nitrogen reduction, sulphite oxidation, sulphate reduction and homoacetogenesis (Momper et al. 2018). They are syntrophic bacteria that can transfer molecular H2 (Dyksma and Gallert 2022) and they can also be related to glycerol and lipids degradation in detrital biomass (Nobu et al. 2015).
The metabolic potential of the phylum Chloroflexi is still unclear, but several studies suggest a role in carbohydrate degradation (Campanaro et al. 2018). Zhou et al (2023) made statistical analysis to uncovers significant correlations between process parameters, dominant bacterial phyla and archaeal genera. The results indicate that Firmicutes exhibit negative correlations with Proteobacteria and Chloroflexi, which make sense in the present study.
Microbial diversity at genus level
At genus level, the most representative groups (Fig. 3) at the end of AD in terms of OTU and relative abundance where Sporanaerobacter (29,013–36,689 OTU, 43%–47%) followed by Clostridium (16,208–20,451 OTU, 19%)—both from Class Clostridia; JGI-0000079-D21 (2785–3166 OTU, 3.4%–4.1%) and Aminiphilus (2380–2874 OTU, 2.9%–3.7%) from Class Synergistia; CI75cm.2.12 from Class γ-Proteobacteria (2421–2619 OTU, 3.1%–3.5%), Rhabdanaerobium (2356–3035 OTU, 3.1%–3.6%), Caldicoprobacter (1454–1839 OTU, 1.9%–2.3%) and Romboutsia (1244–1638 OTU, 1.5%–1.9%) from Class Clostridia, Syner-01 (1081–1237 OTU, 1.3%–1.6%) from Class Synergistia, Gordonia (1102–1133 OUT, 1.3%–1.6%) from Class Actinobacteria.
The abundance of all these groups exponentially increased with AD, some of them in the beginning had an abundance < 100 OTU, such as Sporanaerobacter, Clostridium, JGI-0000079-D21, Aminiphilusfrom, CI75cm.2.12, Rhabdanaerobium, Caldicoprobacter and Syner-01. Two groups (Romboutsia and Gordonia) presented 100 < OTU < 200.
The most abundant genus in the beginning of the operation was Ottowia (2851 OTU), Candidatus Competibacter (1933 OTU), Ellin6067 (1480 OTU) both from Class γ-Proteobacteria. Defluviicoccus (1379 OTU) from Class α-Proteobacteria and OLB8 (1271 OTU) from Class Bacteroidia.
Changes between communities were expected, as each one of them plays a different role in specific functions throughout each stage of AD. The Sporanaerobacter and Clostridium high increase post-AD could be related to competition between α- and γ-Proteobacteria glycogen accumulating organisms for acetate.
Sporanaerobacter is a glucose-to-acetate fermenter that produces H2 and CO2 as the main end products (Hernandez-Eugenio et al. 2002). During the degradation of organic compounds, Sporanaerobacter generates acetate as a metabolic by-product.
Clostridium is represented as Gram-positive bacteria responsible for degradation of organic compounds and is related to the hydrolysis of complex biopolymers (Zhao et al. 2019). High abundance of Clostridium in domestic sludge is expected since it represents 10–40% of human intestinal microbiota (Lopetuso et al. 2013). Clostridium is commonly the most abundant genera in sludges from WWTPs (Arelli et al. 2021).
The genus Aminiphilus is also strictly anaerobic and mesophilic. This genus ferments peptide compounds, amino acids, malate, fumarate, glycerol, pyruvate releasing acetate, propionate and branched-chain fatty acids. Carbohydrates are not used by this group (Díaz et al. 2007).
The Sporanaerobacter and Clostridium presence in the reactors post-AD contributed significantly to the ecosystem robustness and stability. By occupying various functional niches, these generalist microbes ensured that essential metabolic processes continue unabated, thus safeguarding the overall efficiency and resilience of the reactor ecosystem. Consequently, the presence of such generalist microbial communities represents a vital component in ensuring the sustained functionality and adaptability of AD systems. Generalist groups can perform similar ecological functions in a reactor and this redundancy ensures that even when the environmental conditions change, the ecosystem will stay functional, because its functional niches are occupied (Krohn et al. 2022; Zhu et al. 2020).
Archaeal community
The most representative archaeal community at phylum level was expressed in terms of relative abundance (Fig. 4) by Euryarchaeota (15.0% before AD; 76.94 to 93.96% after AD) and Halobacterota (84.0% before AD; 6.0 to 23.0% after AD). Phylum Thermoplasmatota and Crenarchaeota (0.4%, 0.2%, respectively before AD) represent the minority in the beginning of the experiment, with no detection of these groups after AD.
At Genus level (Figs. 5 and 6) the dominance of methanogenic groups before AD was represented by Methanosaeta (32,328–34,497 OTU, 43.12%–54.52%) > Methanolinea (14,747–16,261 OTU, 19.7%–25.7%) > Methanofastidiosum (6467–7032 OTU, 8.6%–11.1%) > Methanospirillum (3277–3987 OTU, 4.4%–6.3%) > Methanosarcina (1521–1782 OTU, 2.0%–2.8%) > Methanobrevibacter (1213–1471 OTU, 1.6%–2.3%). Post-AD the dominance shifted to Methanobacterium (33,995—62,398 OTU, 76.5%–93.7%) > Methanosaeta (1516–2617 OTU, 3.9%—4.2%) > Methanospirillum (497–9382 OTU, 1.5%–17.9%).
Methanogenic community at genus level Chord Diagram related to reactors R1, R2 and R3 (before AD) and R4, R5 and R6 (post-AD). The seven most representative groups were: Methanobacterium; Methanosaeta; Methanolinea; Msum: Methanofastidiosum; Mlum: Methanospirillum; S: Methanosarcina and B: Methanobrevibacter
The chord diagram results (Fig. 6) make clear the connections and patterns between AD and Methanobacterium in these reactors and the dominance of this group above others (Methanosaeta, Methanolinea, Methanosarcina). Therefore, a major variety of archaeal genera (Methanofastidiosum, Methanospirillum, Methanosarcina, Methanobrevibacter, Methanolinea) coexisting before AD was followed by an increase in terms of number of organisms (especially Methanobacterium) after AD, but a reduction observed in the variety of genus.
Methanosaeta is an obligate acetoclastic methanogen, which produces CH4 using acetate (Conklin et al. 2006; Pan et al. 2016). This is a versatile genus that can utilize acetate, methylamines, methanol, and H2/CO2 for methanogenesis (Zhang et al. 2021) and keeps its robustness even when operational conditions are not stable. It is worth to mention that high concentrations of ammonia and salt, as well as changes in temperature are often deleterious for methanogenic activity (Jiang et al. 2019a, b).
The hydrogenotrophic Methanobacterium genus showed a significant increase in abundance in the reactors, with the acetoclastic Methanoaseta following as the second-most abundant group. Similar shifts in the dominant methanogens from Methanosaeta to Methanobacterium and Methanosarcina have been observed in other studies of SWS anaerobic digestion (Cai et al. 2022; Zhang et al. 2019). This shift in microbial community diversity can also result in a change in the metabolic pathway for methane production, from the acetoclastic to the hydrogenotrophic pathway, as observed in other studies (Cai et al. 2022; Pan et al. 2021).
Homoacetogenic bacteria, strict anaerobes that produce acetate as their only metabolic by-product, and acetate-oxidizing syntrophs (SAOB) regulate AD through acetate producing and acetate consuming, cooperating with acidogenesis and with fatty acids oxidizing bacteria (Zeng et al. 2024). They cooperate with acidogenic bacteria and those that oxidize fatty acids, and they establish a syntrophic relationship with acetoclastic methanogenic archaea (Lv et al. 2023). However, CH4 can also be produced through syntrophic interactions between SAOB and hydrogenotrophic methanogenic archaea (HM). The interactions between these functional microbes are quite complex, but the microbial conversions and interactions in an anaerobic digestors offer flexibility to overcome stress under different environment disturbances. The SAO-HM process can replace acetoclastic methanogenesis in some environments where the activity of acetoclastic methanogens is inhibited (Pan et al. 2021; Wang et al. 2022). This could happen due to different possible situations, such as: (i) syntrophic relationships, where certain bacteria oxidize acetate (CH3COO) to H2 and CO2 in syntrophic relationships with HM; this syntrophic acetate oxidation allows for the conversion of CH3COO, a key intermediate in AD pathways, into H2 and CO2, which can then be used by HM to produce CH4; (ii) substrate availability, even when acetoclastic methanogenesis is inhibited, and CH3COO can still be present as a substrate in the environment due to the breakdown of complex OM by other microbial activities; (iii) redox balance, in environments where acetoclastic methanogenesis is inhibited and the SAO-HM process helps maintaining redox balance by facilitating the conversion of CH3COO to CH4 and; (iv) flexibility and adaptability, when the SAO-HM process offers flexibility and adaptability for changing environmental conditions. When acetoclastic methanogenesis is inhibited, MO capable of SAO and HM can thrive and replace acetoclastic methanogens in driving CH4 production. This process is particularly important in environments with high CH3COO concentrations, as it prevents its accumulation, which can inhibit microbial activity and disrupt AD processes (Amin et al. 2021). The SAO-HM process provides an alternative pathway for utilizing acetate as an energy source for methane production, ensuring that this valuable substrate is not wasted (Yadav et al. 2022).
The shift in metabolic route could be related to the increase in Firmicutes, especially the genus Clostridia, that contains acetate-oxidizing bacteria who convert acetate to H2. Full-scale anaerobic digestion bioreactors showed positive correlations between populations of hydrogenotrophic methanogens and Clostridia (Zhang et al. 2019). Ruiz-Sánchez et al. (2019) also reported an increase predominance of syntrophic bacteria, such as Clostridium and Bacteroides, and alternation of acetoclastic to hydrogenotrophic pathway during AD.
Pan et al. (2021) observed that H2-consuming methanogens help to maintain balanced biomass and stabilize pH. Both H2 producers (acetogens) and H2 consumers (methanogens) can only be favoured at a narrow range of H2 concentration. H2 is the most important product in determining the free energy of the reaction because more H2 is produced stoichiometrically than other products. Several studies have found that inoculum dominated by H2-utilizing methanogens exhibited a higher methane production rate than Methanosaeta, and others acetoclastic methanogens (Gao et al. 2022; Qi et al. 2022). Zhou et al. (2023) found that bacteria and archaea exhibit potential competitivity (between syntrophic acetate-oxidizing bacteria and acetoclastic archaea) and syntrophic (between hydrogen-producing bacteria and hydrogenotrophic archaea) relationships.
Future research on biogas-producing microbial communities will certainly help enhancing AD efficiency and stability. Standardized methods and analyses are essential for generating data that can be compared and utilized for the development and enhancement of anaerobic digestion models.
In summary, the results obtained in the present study offer valuable insight into the microbial dynamics during sewage sludge AD and highlight the importance of considering the microbial community if the purpose is optimizing the process to maximize biogas production and minimize environmental impacts. Future research efforts may focus on exploring interactions between different microbial groups and identifying strategies to enhance the efficiency of AD in municipal wastewater treatment facilities.
Conclusions
BMP assays having SWS as feedstock showed a bacterial diversity consistent with the profile expected in an AD process. Firmicutes was the dominant Phylum during AD followed by Synergistota. Sporanaerobacter and Clostridium were the dominant genera in the BMP reactors. The shift of methanogenic archaeal community from Methanosaeta to Methanobacterium might be related to the increase of Firmicutes and Synergistota Phylum in the reactors, affecting the degree of substrate fermentation, that contains acetate-oxidizing bacteria who convert acetate to hydrogen that is scavenged by methanogens, making the reaction thermodynamically possible for these groups. The microbial profiles observed in the present study expand the current knowledge regarding possible syntrophic relationships between hydrogen-producing bacteria and hydrogenotrophic methanogenic archaea in sewage sludge from WWTP. The results obtained help to foresee new horizons for future microbial ecology studies and improvement of biogas production from sewage sludge.
Data availability
This is not applicable.
References
Abendroth C, Vilanova C, Günther T et al (2015) Eubacteria and archaea communities in seven mesophile anaerobic digester plants in Germany. Biotechnol Biofuels 8:87
Al Ali AA, Naddeo V, Hasan SW et al (2020) Correlation between bacterial community structure and performance efficiency of a full-scale wastewater treatment plant. J Water Process Eng 37:101472
Alves IRFS, Mahler CF, Oliveira LB et al (2020) Assessing the use of crude glycerol from biodiesel production as an alternative to boost methane generation by anaerobic co-digestion of sewage sludge. Biomass Bioenergy 143:105831
Amin FR, Khalid H, El-Mashad HM et al (2021) Functions of bacteria and archaea participating in the bioconversion of organic waste for methane production. Sci Total Environ 763:143007
Angelidaki I, Alves M, Bolzonella D et al (2009) Defining the biomethane potential (BMP) of solid organic wastes and energy crops: a proposed protocol for batch assays. Water Sci Technol 59:927–934
Angelidaki I, Treu L, Tsapekos P et al (2018) Biogas upgrading and utilization: current status and perspectives. Biotechnol Advances 36:452–466
APHA (American Public Health Association) (2017) Standard Methods for Evaluation of Water and Wastewater, 23rd edn. American Water Works Association, Washington, DC
Arelli V, Mamindlapelli NK, Begum S et al (2021) Solid state anaerobic digestion of food waste and sewage sludge: impact of mixing ratios and temperature on microbial diversity, reactor stability and methane yield. Sci Total Environ 793:148586
Cai M, Wilkins D, Chen J et al (2016) Metagenomic reconstruction of key anaerobic digestion pathways in municipal sludge and industrial wastewater biogas-producing systems. Front Microbiol 7:1664–2302
Cai C, Zhang X, Wu M et al (2021) Roles and opportunities for microbial anaerobic oxidation of methane in natural and engineered systems. Energy Environ Sci 14:4803–4830
Cai Y, Zheng Z, Wei L et al (2022) The characteristics of multi-substrates (low and high C/N) anaerobic digestion: focus on energy recovery and the succession of methanogenic pathway. Bioresour Technol 343:125976
Callahan BJ, McMurdie PJ, Rosen MJ et al (2016) DADA2: High-resolution sample inference from illumina amplicon data. Nature methods 13:581–583
Campanaro S, Treu L, Panagiotis G et al (2018) Metagenomic binning reveals the functional roles of core abundant microorganisms in twelve full-scale biogas plants. Water Res 140:123–134
Campanaro S, Treu L, Rodriguez-R LM et al (2020) New insights from the biogas microbiome by comprehensive genome-resolved metagenomics of nearly 1600 species originating from multiple anaerobic digesters. Biotechnol Biofuels 13:25
Conklin A, Stensel HD, Ferguson J (2006) Growth kinetics and competition between Methanosarcina and Methanosaeta in mesophilic anaerobic digestion. Water Environ Res 78:486–496
Di Giacomo G, Romano P (2022) Evolution and prospects in managing sewage sludge resulting from municipal wastewater purification. Energies 15:5633
Díaz C, Baena S, Fardeau ML, Patel BKC (2007) Aminiphilus circumscriptus gen. nov., sp. nov., an anaerobic amino-acid-degrading bacterium from an upflow anaerobic sludge reactor. Int J Syst Evol Microbiol 57:1914–1918
Dyksma S, Gallert C (2022) Effect of magnetite addition on transcriptional profiles of syntrophic Bacteria and Archaea during anaerobic digestion of propionate in wastewater sludge. Environ Microbiol Rep 14:664–678
Feng J, Burke IT, Chen X et al (2023) Assessing metal contamination and speciation in sewage sludge: implications for soil application and environmental risk. Rev Environ Sci Biotechnol 22:1037–1058
Ferrentino R, Langone M, Fiori L, Andreottola G (2023) Full-scale sewage sludge reduction technologies: a review with a focus on energy consumption. Water 15:615
Fu J, Yan B, Gui S, Fu Y, Xia S (2023) Anaerobic co-digestion of thermo-alkaline pretreated microalgae and sewage sludge: methane potential and microbial community. J Environ Sci 127:133–142
Gao N, Zhang H, Xu X, Teng J (2022) Mutual effects of CO2 absorption and H2-mediated electromethanogenesis triggering efficient biogas upgrading. Sci Total Environ 818:151732
Garcia-Peña E, Parameswaran P, Kang D et al (2011) Anaerobic digestion and co-digestion processes of vegetable and fruit residues: process and microbial ecology. Bioresour Technol 102:9447–9455
Garlicka A, Umiejewska K, Nielsen PH, Muszyński A (2023) Hydrodynamic disintegration of thickened excess sludge and maize silage to intensify methane production: Energy effect and impact on microbial communities. Bioresour Technon 376:128829
Goux X, Calusinska M, Lemaigre S et al (2015) Microbial community dynamics 524 in replicate anaerobic digesters exposed sequentially to increasing organic loading rate, acidosis, and process recovery. Biotechnol Biofuels 8:122
Gu J, Liu R, Cheng Y, Stanisavljevic N, Li L, Djatkov D, Peng X, Wang X (2020) Anaerobic co-digestion of food waste and sewage sludge under mesophilic and thermophilic conditions: Focusing on synergistic effects on methane production. Bioresour Technol 301:122765
Guo W, Li D, Zhang Z et al (2023) A novel approach for the fractionation of organic components and microbial degraders in ADM1 and model validation based on the methanogenic potential. Water Res 236:119945
Hafner SD, Fruteau de Laclos H, Koch K et al (2020) Improving inter-laboratory reproducibility in measurement of biochemical methane potential (BMP). Water 12:1752
Hernandez-Eugenio G, Fardeau ML, Cayol JL et al (2002) Sporanaerobacter acetigenes gen. nov., sp. nov., a novel acetogenic, facultatively sulfur-reducing bacterium. Int J Syst Evol Microbiol 52:1217–1223
Holliger C, Astals S, de Laclos HF et al (2021) Towards a standardization of biomethane potential tests: a commentary. Water Sci Technol 83:247–250
Jiang Y, McAdam E, Zhang Y et al (2019a) Ammonia inhibition and toxicity in anaerobic digestion: a critical review. J Water Process Eng 32:100899
Jiang J, Wang Y, Liu J et al (2019b) Exploring the mechanisms of organic matter degradation and methane emission during sewage sludge composting with added vesuvianite: Insights into the prediction of microbial metabolic function and enzymatic activity. Bioresour Technol 286:121397
Jiang X, Xie Y, Liu M (2022) Study on anaerobic co-digestion of municipal sewage sludge and fruit and vegetable wastes: methane production, microbial community and three-dimension fluorescence excitation-emission matrix analysis. Bioresour Technon 347:126748
Kampmann K, Ratering S, Kramer I et al (2012) Unexpected stability of bacteroidetes and firmicutes communities in laboratory biogas reactors fed with different defined substrates. Microb Ecol 78:7
Kashi S, Satari B, Lundin M et al (2017) Application of a mixture design to identify the effects of substrates ratios and interactions on anaerobic co-digestion of municipal sludge, grease trap waste, and meat processing waste. J Environ Chem Eng 5:6156–6164
Krohn C, Khudur L, Dias DA et al (2022) The role of microbial ecology in improving the performance of anaerobic digestion of sewage sludge. Front Microbiol 13:1079136
Lavergne C, Jeison D, Ortega V, Chamy R, Donoso-Bravo A (2018) A need for a standardization in anaerobic digestion experiments? Let’s get some insight from meta-analysis and multivariate analysis. J Environ Manag 222:141–147
Li M, Ge Song G, Liu R, Huang X, Liu H (2022) Inactivation and risk control of pathogenic microorganisms in municipal sludge treatment: a review. Front Environ Sci Eng 16(6):70
Liew CS, Yunus NM, Chidi BS et al (2022) A review on recent disposal of hazardous sewage sludge via anaerobic digestion and novel composting. J Hazard Mater 423:126995
Lopetuso LR, Scaldaferri F, Petito V, Gasbarrini A (2013) Commensal clostridia: leading players in the maintenance of gut homeostasis. Gut Pathog 5:23
Lv Z, Pan X, Ye ZL et al (2023) Homoacetogenesis is altering the metabolic pathway of acidogenic microbiome and combating volatile fatty acid accumulation in anaerobic reactors. J Environ Chem Eng 11(4):110224
Maragkaki AE, Fountoulakis M, Kyriakou A et al (2018) Boosting biogas production from sewage sludge by adding small amount of agro-industrial by-products and food waste residues. Waste Manage 71:605–611
McMurdie PJ, Holmes S (2013) Phyloseq: an r package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217
Meyer DD, de Andrade PA, Durrer A et al (2016) Bacterial communities involved in sulfur transformations in wastewater treatment plants. Appl Microbiol Biotechnol 100:10125–10135
Momper L, Aronson HS, Amend JP (2018) Genomic description of ‘candidatus abyssubacteria’, a novel subsurface lineage within the candidate phylum hydrogenedentes. Front Microbiol 9:1993
Nascimento AL, Souza AJ, Andrade PAM et al (2018) Sewage sludge microbial structures and relations to their sources, treatments, and chemical attributes. Front Microbiol 9:1664–2302
Nguyen LN, Kumar J, Minh TV et al (2021) Biomethane production from anaerobic co-digestion at wastewater treatment plants: a critical review on development and innovations in biogas upgrading techniques. Sci Total Environ 765:142753
Nobu MK, Narihiro T, Rinke C et al (2015) Microbial dark matter ecogenomics reveals complex synergistic networks in a methanogenic bioreactor. ISME J 9(8):1710–1722
Pan X, Angelidaki I, Alvarado-Morales M et al (2016) Methane production from formate, acetate and H2/CO2; focusing on kinetics and microbial characterization. Bioresour Technol 218:796–806
Pan Y, Zhi Z, Zhen G et al (2019) Synergistic effect and biodegradation kinetics of sewage sludge and food waste mesophilic anaerobic co-digestion and the underlying stimulation mechanisms. Fuel 253:40–49
Pan X, Zhao L, Li C (2021) Deep insights into the network of acetate metabolism in anaerobic digestion: focusing on syntrophic acetate oxidation and homoacetogenesis. Water Res 190:116774
Park M, Kim N, Jung S et al (2021) Optimization and comparison of methane production and residual characteristics in mesophilic anaerobic digestion of sewage sludge by hydrothermal treatment. Chemosphere 264:128516
Qi X, Jia X, Wang Y (2022) Development of a rapid startup method of direct electron transfer-dominant methanogenic microbial electrosynthesis. Bioresour Technol 358:127385
Qin S, Wainaina S, Liu H (2021) Microbial dynamics during anaerobic digestion of sewage sludge combined with food waste at high organic loading rates in immersed membrane bioreactors. Fuel 303:121276
Raposo F, De La Rubia MA, Fernández-Cegrí V, Borja R (2011) Anaerobic digestion of solid organic substrates in batch mode: an overview relating to methane yields and experimental procedures. Renew Sustain Energy Rev 16:861–877
Rocha ME, Lazarino TC, Oliveira G, Teixeira L, Marques M, Mangiavacchi N (2024) Analysis of biogas production from sewage sludge combining BMP experimental assays and the ADM1 model. PeerJ 12:e16720
Ruiz-Sánchez J, Guivernau M, Fernández B et al (2019) Functional biodiversity and plasticity of methanogenic biomass from a full-scale mesophilic anaerobic digester treating nitrogen-rich agricultural wastes. Sci Total Environ 649:760–769
Saha S, Basak B, Hwang J-H et al (2020) Microbial symbiosis: a network towards biomethanation. Trends Microbiol 28:12
Schneider D, Zühlke D, Poehlein A et al (2021) Metagenome-assembled genome sequences from different wastewater treatment stages in germany. Microbiol Resour Announc 10:e00504-e521
Serna-García R, Borrás L, Bouzas A et al (2020) Insights into the biological process performance and microbial diversity during thermophilic microalgae co-digestion in an anaerobic membrane bioreactor (AnMBR). Algal Res 50:101981
Tao B, Zhang Y, Heaven S, Banks CJ (2020a) Predicting pH rise as a control measure for integration of CO2 biomethanisation with anaerobic digestion. Appl Energy 277:115535
Tao Y, Ersahin ME, Ghasimi DSM et al (2020b) Biogas productivity of anaerobic digestion process is governed by a core bacterial microbiota. Chem Eng J 380:122425
Treu L, Kougias PG, Diego-Díaza B et al (2018) Two-year microbial adaptation during hydrogen-mediated biogas upgrading process in a serial reactor configuration. Bioresour Technol 264:140–147
VDI 4630. (2016) Fermentation of organic materials - Characterization of the substrate, sampling, collection of material data, fermentation tests. Germany
Walter A, Probst M, Franke-Whittle IH et al (2019) Microbiota in anaerobic digestion of sewage sludge with an without co-substrates. Water Environ J 33:214–222
Wang Z, Wang S, Hu Y et al (2022) Distinguishing responses of acetoclastic and hydrogenotrophic methanogens to ammonia stress in mesophilic mixed cultures. Water Res 224:119029
Xie S, Li X, Wang C et al (2020) Enhanced anaerobic digestion of primary sludge with additives: performance and mechanisms. Bioresour Technol 316:123970
Yadav M, Joshi C, Paritosh K et al (2022) Organic waste conversion through anaerobic digestion: a critical insight into the metabolic pathways and microbial interactions. Metab Eng 69:323–337
Yang Y, Yu K, Xia Y et al (2014) Metagenomic analysis of sludge from full-scale anaerobic digesters operated in municipal wastewater treatment plants. Appl Microbiol Biotechnol 98:5709–5718
Yang Z, Xu R, Zheng Y et al (2016) Characterization of extracellular polymeric substances and microbial diversity in anaerobic co-digestion reactor treated sewage sludge with fat, oil, grease. Bioresour Technol 212:164–173
Zeng Y, Zheng D, Li LP et al (2024) Metabolism of novel potential syntrophic acetate-oxidizing bacteria in thermophilic methanogenic chemostats. Appl Environ Microbiol 90(2):e01090-e1123
Zhang Z, Zheng H, Sun Y et al (2016) A combined process of chemical precipitation and flocculation for treating phosphating wastewater. Desalin Water Treat 57(53):25520–25531
Zhang W, Zhang F, Li YX (2019) No difference in inhibition among free acids of acetate, propionate and butyrate on hydrogenotrophic methanogen of Methanobacterium formicicum. Bioresour Technol 294:122237
Zhang L, Gong X, Wang L et al (2021) Metagenomic insights into the effect of thermal hydrolysis pre-treatment on microbial community of an anaerobic digestion system. Sci Total Environ 791:148096
Zhao X, Li L, Wu D et al (2019) Modified anaerobic digestion model No. 1 for modeling methane production from food waste in batch and semi-continuous anaerobic digestions. Bioresour Technol 271:109–117
Zhou B, Wu W, Li X et al (2023) A state-of-the-art review on anaerobic digestion of sewage sludge based on microbial abundance: correlations among microbiota, performance and process parameters. Crit Rev Environ Sci Technol. 1–24
Zhu X, Campanaro S, Treu L et al (2020) Metabolic dependencies govern microbial syntrophies during methanogenesis in an anaerobic digestion ecosystem. Microbiome 8:22
Acknowledgements
We are thankful to the State Company of Water and Wastewater (CEDAE), for their help in collecting wastewater.
Funding
This research was supported by the Department of Innovation of the Rio de Janeiro State University (UERJ). The financial support from the Carlos Chagas Filho Foundation for Supporting Research in the State of Rio de Janeiro (FAPERJ), the National Council for Scientific and Technological Development (CNPq) (Proc. 308335/2017-1), Finep (01.19.0087.00) and Capes are also acknowledged.
Author information
Authors and Affiliations
Contributions
Conceptualization: MER, LT, MM, NM; Writing: MER, LT; Images and tables: MER, LT; Material and Methods: MER, LT; Discussion: MER, LT, MM; Conclusions: MER, LT, MM, NM and Revision: MER, LT, MM, NM.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Ethical approval
This is not applicable.
Consent to participate
All the authors consent to participate in this study.
Consent to publish
All the authors consent to publish in this study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Rocha, M.E., Mangiavacchi, N., Marques, M. et al. Succession from acetoclastic to hydrogenotrophic microbial community during sewage sludge anaerobic digestion for bioenergy production. Biotechnol Lett (2024). https://doi.org/10.1007/s10529-024-03528-6
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10529-024-03528-6