Abstract
Microbes are key components of the soil environment, playing an important role in maintaining soil health, sustainability, and productivity. The composition and structure of soil bacterial communities were examined in winter wheat–rice (WR) and winter wheat–maize (WM) cropping systems derived from five locations in the Low-Middle Yangtze River plain and the Huang-Huai-Hai plain by pyrosequencing of the 16S ribosomal RNA gene amplicons. A total of 102,367 high quality sequences were used for multivariate statistical analysis and to test for correlation between community structure and environmental variables such as crop rotations, soil properties, and locations. The most abundant phyla across all soil samples were Proteobacteria, Acidobacteria, and Bacteroidetes. Similar patterns of bacterial diversity and community structure were observed within the same cropping systems, and a higher relative abundance of anaerobic bacteria was found in WR compared to WM cropping systems. Variance partitioning analysis revealed complex relationships between bacterial community and environmental variables. The effect of crop rotations was low but significant, and interactions among soil properties, locations, and crop rotations accounted for most of the explained variation in the structure of bacterial communities. Soil properties such as pH, available P, and available K showed higher correlations (positive or negative) with the majority of the abundant taxa. Bacterial diversity (the Shannon index) and richness (Chao1 and ACE) were higher under WR than WM cropping systems.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Winter wheat (Triticum aestivum L.) is one of the most important cereal crops worldwide, and the Low-Middle Yangtze River (LMYR) plain and the Huang-Huai-Hai (HHH) plain are the two main winter wheat growing areas in China, with winter wheat–rice (WR) rotation in the LMYR plain and winter wheat–maize (WM) rotation in the HHH plain. With the increasing food demand in China, high productivity and sustainable food production is becoming increasingly important.
Soil microorganisms are important components of sustainable agro-ecosystems [1] as they mediate many processes that are crucial to soil agricultural productivity, including recycling of soil nutrients, maintenance and development of soil structure, degradation of agrochemicals, and the control of pests and plant pathogens [2–5]. Bacteria are the most abundant and diverse group of microorganisms in soil [6], and there are estimated to be as many as 109–1010 bacteria and 6,000–50,000 bacterial species per gram of soil [7, 8]. Bacteria are major drivers for nearly all biogeochemical cycles in terrestrial ecosystems and participate in maintaining the health and productivity of soil in agricultural systems [9, 10].
It is well known that management practices (e.g., application of fertilizer or fungicide, periodic flooding, and crop rotations), plant species, soil type, geographic distance, etc. largely determine the structure of bacterial communities [11–16]. Crop rotation is an important agricultural practice for increasing crop yield and disease suppression [17, 18]. It may also result in shifts in the microbial communities as different plant species secrete different types of root exudates, which can shape soil bacterial community structure [14, 19]. Significant differences in composition, abundance, and diversity were observed following different crop rotation systems [17, 20, 21]. However, in some cases, crop rotation appeared to have smaller effects than other management practices such as organic and conventional management of the cropping system [22].
Understanding the soil microbial community and its response to the various land use and management systems is important for selecting suitable management practices to improve ecosystem service and soil function [21, 23, 24]. Therefore, experiments examining the effects of soil properties, locations, and crop rotations on the microbial community should help reveal what drives changes in microbial populations. In the present study, we employ 16S ribosomal RNA (16S rRNA) pyrosequencing to reveal the bacterial diversity and community structure under two main winter wheat cropping systems in China. The large amount of data generated by this method provides detail information on the soil bacterial assemblages, allows us to make comparative analyses of bacterial diversity and community composition, and thus makes it possible to characterize the majority of variation within the soil microbial community.
The objectives of this study were (1) to determine the abundance, taxonomic diversity, and composition of the bacterial community between the two winter wheat cropping systems and (2) to determine the contributions of environmental variables that correlated to changes in the structure of bacterial communities.
Materials and Methods
Site Descriptions and Soil Sampling
Five study sites were selected in the LMYR plain and HHH plain. Three sites were in the LMYR plain with a winter wheat–rice rotation system: Changshu (CS), Jintan (JT), and Zhangjiagang (ZJG). The other two sites were in the HHH plain with a winter wheat–maize rotation system: Dezhou (DZ) and Quzhou (QZ). The sites differed in location, soil types, and cropping systems (Table 1; Table S1). Management practices and fertilization were in accordance with the standard of each site. Irrigation was routinely used in the rice season of the winter wheat–rice region to maintain the shallow standing water during seedling stage to maturity stage, while wheat and maize were watered when necessary for agriculture. Herbicides and pesticides were applied when necessary.
Soil samples were collected randomly from the plough layer (0–20 cm) at each site using a 2.5-cm diameter soil auger soon after the winter wheat crops were harvested in June 2011. Triplicate samples were collected at each site, and each sample was a mixture of 10 cores (2.5 cm diameter × 20 cm deep) within an area of ∼100 m2. All samples were packed on ice upon collection and transported to the laboratory. The soil was sieved to 2 mm and thoroughly homogenized; one portion was air-dried for soil properties analysis and the other for DNA extraction.
Soil Characteristics Analysis
Soil pH was determined with a glass electrode (soil/water = 1:5). Available K in the soil was extracted with ammonium acetate and determined by flame photometry. Available P in the soil was extracted with sodium bicarbonate and then determined using the molybdenum blue method. Soil organic C (SOC) and total N (TN) were determined by an Elementar Vario EL III (Germany). Soil particle size distributions were determined by a Beckman Coulter LS 230 (USA) following the manufacturer’s recommendations.
DNA Extraction, PCR amplification, and 454 Pyrosequencing
Total genomic DNA was extracted from 1.0 g subsamples using a PowerSoil DNA Isolation Kit (Mo Bio Laboratories Inc., Carlsbad, CA, USA) according to the manufacturer’s instructions. Three successive DNA extractions of each sample were pooled before PCR (to minimize DNA extraction bias). The concentration and quality (ratio of A 260/A 280) of the DNA were determined with a spectrophotometer (NanoDrop, ND2000, ThermoScientific, Wilmington, DE, USA).
PCR amplification of the bacterial 16S rRNA genes from the genomic DNA of the 15 soil samples was conducted using a highly conserved universal bacterial primer set as described by Dethlefsen et al. [25] and Huse et al. [26] The 27F (5′-GCCTTGCCAGCCCGCTCAG-TC-AGAGTTTGATCCTGGCTCAG-3′) and 533R fusion primers (5′-GCCTCCCTCGCGCCATCAG-AC-NNNNNNNNNN-TTACCGCGGCTGCTGGCAC-3′) were employed. For each sample, this fusion primer included Roche-454 A/B adapters (shown in italics) and a 2-bp linker sequence (shown in bold) followed by a unique, error-correcting barcode sequence (Ns) and the 16S rRNA primer. The region amplified by this primer set is well suited for accurate phylogenetic placement of bacterial sequences [27].
All PCR reactions for each sample were performed in triplicate (including two negative control reactions) with 2 μM of each primer, 0.25 μM dNTPs (Takara), 4 μL of 5× FastPfu Buffer (TransGen, TransGen Biotech Co., Ltd., Beijing, China), 1 U of FastPfu DNA polymerase (2.5 U/μL, TransGen), and 20 ng of soil DNA template with a final volume of 20 μL. The PCR conditions were an initial denaturation step of 95 °C for 2 min followed by 25 cycles of denaturation at 95 °C for 30 s, annealing at 55 °C for 30 s, extension at 72 °C for 30 s, and a final elongation at 72 °C for 5 min. PCR products were visualized on 2 % agarose gels.
PCR products of each sample were pooled by volume (to minimize PCR bias), purified with the PCR Purification Kit (Axygen Bio, USA), and quantified with PicoGreen® (Promega, USA). Next, the amplicons from each sample were pooled in equimolar concentrations into a single tube, and an emulsion PCR was carried out to make the single strands on beads as required for 454 pyrosequencing. Pyrosequencing was performed on a 454 GS-FLX Titanium System (Roche, Switzerland) by Majorbio Bio-pharm Technology Co., Ltd (Shanghai, China).
Processing of Pyrosequencing Data
Pyrosequencing data were processed using Mothur (version 1.25.1) [28] following the Schloss standard operating procedure [29]. In brief, sequences having a minimum flow length of 450 flows were denoised using the Mothur based reimplementation of the PyroNoise algorithm [30] with the default parameters. Denoised sequences with one or more mismatches to the barcode, two or more mismatches to the primer, any ambiguous base call, a homopolymer longer than 8 bp or a length shorter than 200 bp were eliminated, and then the filtered sequences were assigned to soil samples based on unique 10-bp barcodes. After removing the barcode and primer sequences, the unique sequences were aligned against the Silva 106 database [31]. Through screening, filtering, preclustering processes, and chimera removal, the retained sequences were used to build a distance matrix with a distance threshold of 0.2. Using the average neighbor algorithm with a cutoff of 97 % similarity, bacterial sequences were clustered to operational taxonomic units (OTUs), and the most abundant sequence in each OTU was selected as the representative sequence. Representative sequences were taxonomically classified using a Ribosomal Database Project naive Bayesian rRNA classifier [32] with a confidence threshold of 80 %. Relative abundance of a given phylogenetic group was set as the number of sequences affiliated with that group divided by the total number of sequences per sample. To correct for sampling effort, we used a randomly selected subset of 5,940 sequences per sample for further analysis. To compare the effect of the crop rotations, we averaged the relative abundances of OTUs from each field site of each rotation.
Statistical Analysis
An OTU-based analysis was performed to calculate the richness, diversity, evenness, and coverage with a cutoff of 3 % dissimilarity. Richness indices, the Chao1 estimator [33], and the abundance-based coverage estimator (ACE) [34] were calculated to estimate the number of observed OTUs that were present in the sampling assemblage. The diversity within each individual sample was estimated using the nonparametric Shannon diversity index [35]. Good’s nonparametric coverage estimator [36] was used to estimate the percentage of the total species that were sequenced in each sample. Rarefaction curves generated using Mothur [28] were used to compare relative levels of bacterial OTU diversity across all soil samples. Significant differences in the rarefaction curves were considered when there was no overlap between the 95 % confidence intervals.
To compare bacterial community structures across all samples based on the OTU composition and examine the relationship between relative abundances of abundant phyla (proteobacterial classes) and environmental variables, principal component analysis (PCA) of sites and redundancy analysis (RDA) were carried out using CANOCO for Windows [37]. Bray–Curtis indices were calculated and represented in a heat map format with hierarchical cluster analysis to depict the similarity and dissimilarity between bacterial communities. Variance partitioning analysis (VPA) was also used to determine the contributions of soil properties, locations, and crop rotations, as well as interactions between them on the variation in a bacterial community with Hellinger-transformed data. Heat map and VPA were performed in R (Version 2.15.0) [38] with the gplots [39] and vegan [40] packages. Pearson correlation coefficients between soil geochemical characteristics and Spearman’s rank correlations between abundant phyla (proteobacterial classes) and soil properties were calculated using PASW Statistics 18 (SPSS Inc.). We also used two sample t test analyses to compare the alpha-diversity indices and soil properties between two cropping systems. Duncan’s multiple range tests were carried out to compare the soil properties and relative abundance of bacterial communities among sampling sites.
Sequence Accession Numbers
The pyrosequencing-generated nucleotide sequences have been deposited in the NCBI Sequence Read Archive (SRA) database with accession number SRA059577.
Results
Soil Physicochemical Characteristics
The geographical and selected physicochemical characteristics of the analyzed soil samples are summarized in Table 1. The two cropping systems showed significant differences with respect to edaphic properties such as soil pH, available K, available P, TN, and the C/N ratio. The soil SOC in the winter wheat–rice cropping system ranged from 13.46 to 19.16 g/kg, and the SOC in the winter wheat–maize cropping system ranged from 13.74 to 16.91 g/kg.
Bacterial Community Composition
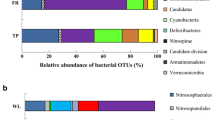
Across all soil samples, we obtained 102,367 quality sequences in total and 5,940–7,511 sequences per sample (mean= 6,824) and were able to classify 77.9 % of those sequences at the phylum level. The dominant phyla (proteobacterial classes) across all samples were Alphaproteobacteria, Betaproteobacteria, Deltaproteobacteria, Gammaproteobacteria, Acidobacteria, Chloroflexi, Bacteroidetes, and Actinobacteria (relative abundance> 4 %), accounting for 62.8 % of the bacterial sequences (Fig. 1). In addition, Verrucomicrobia, Nitrospirae, Gemmatimonadetes, Firmicutes, Planctomycetes, and Armatimonadetes were present in all of the samples with low abundance, and nine other more rare phyla were identified (Table S2). Similar patterns of bacterial community structure were observed within the WR cropping systems, while it was differed within the WM cropping systems (Table S3). The phylum distribution varied under different cropping systems. Betaproteobacteria accounted for 13.37 % of the total bacterial communities under the WR cropping systems, while it was only 6.78 % under WM. In contrast, Alphaproteobacteria made up 11.12 % of the total bacterial communities under WM cropping systems but were present in lower percentages (3.42 %) under WR. Phylum Actinobacteria was also richer under WM (9.38 %) compared to WR (2.52 %), while phylum Chloroflexi showed the opposite pattern, being higher under WR (6.70 %) than WM (2.21 %). Other phyla did not vary much between winter wheat–rice and winter wheat–maize cropping systems. Within WM cropping system, Acidobacteria accounted for 20.73 % of the total bacterial communities in DZ, significantly higher than in QZ (15.17 %). On the contrary, phylum Actinobacteria was striking higher in QZ (14.05 %) than in DZ (4.70 %). Other phyla had smaller differences within the WM cropping systems. Interestingly, a higher diversity (number of taxa) and relative abundance of anaerobic bacteria were observed under WR than WM cropping systems (Table 2). Two anaerobic related phyla Chlorobi and Spirochaetes were only detected under WR, although their relative abundances were low. Likewise, some other anaerobic representatives had a similar pattern. In contrast, relative abundances of some aerobic bacteria were richer under WM than WR.
Relative abundance of the dominant bacteria phyla (proteobacterial classes) in all samples combined and in each cropping system. Relative abundances are based on the proportional frequencies of those DNA sequences that could be classified at the phylum (proteobacterial class) level. WR winter wheat–rice, WM winter wheat–maize
Bacterial α-Diversity
Bacterial diversity and richness of different cropping systems were calculated based on the samples belonging to them, and each sample was calculated based on 5,940 sequences (Table 3). Statistically significant differences (P< 0.05) in richness and diversity were observed for Chao1, ACE, Shannon, and coverage under different cropping systems. The WR cropping system had a higher richness for Chao1 (3,513 ± 259, n= 9) and ACE (4,916 ± 165, n= 9) and a higher diversity for Shannon (6.72 ± 0.07, n= 9) in comparison to the WM cropping system. However, WM had a higher percentage of coverage (0.88 ± 0.01, n= 6) than WR (0.83 ± 0.01, n= 9). In addition, the number of observed OTUs was higher in WR (1,833 ± 51, n= 9) than WM (1,474 ± 50, n= 6). The rarefaction curve (Fig. 2) also indicated that the number of observed OTUs for winter wheat–rice cropping systems was significantly higher than for winter wheat–maize cropping systems, with ZJG having the highest number of observed OTUs at the same sample effort.
Bacterial Community Structure
To compare the similarity and dissimilarity between all samples, we generated a heat map with hierarchical cluster based on Bray–Curtis distance (Fig. 3). Hierarchical cluster analysis showed that different patterns of community structure were observed between WR and WM cropping systems, while they were similar within the same cropping systems. The Bray–Curtis indices showed that a higher similarity of bacterial communities was found in the WR sampling sites than in the WM.
The samples contained 1,432–1,883 OTUs per 5,400 sequences; 19.8–25.4 % of the OTUs were unique to each sample, 20.6–52.1 % of the OTUs overlapped in any pair of samples, indicating that a high percentage of OTUs were shared between each pair of samples (Table S4). In addition, triplicate samples and samples within the same cropping systems shared more OTUs than samples between the two cropping systems, indicating that the community structure was different between WR and WM.
Concomitantly, PCA clearly showed variations between these two cropping systems (Fig. 4). The first two principal components can explain 77.05 % of the total bacterial community variations among the individual samples. Bacterial community compositions shift greatly between WR and WM cropping systems along the second principal component axis. Moreover, bacterial community structures of WR cropping system are still grouped well even in the third principal component axis, while the QZ and DZ are separated with each other (Fig. S1), indicating that WR cropping systems shape a more similar bacterial assemblages than WM cropping systems.
Linking Bacterial Communities to Soil Properties, Location, and Crop Rotation
RDA was performed to study the effect of environmental variables (soil properties, location, and crop rotation) on abundant phyla (proteobacterial classes) (Fig. 5). The results showed significant correlation between environmental variables and abundant bacterial phyla (P= 0.002, Monte Carlo test). The first two axes of RDA explain 69.6 and 16.0 %, respectively, of the total variation in the data. Abundant phyla of the WR were more alike and related to higher SOC, TN, and available P contents, in addition to the similar longitude and same rotation, as shown by their close grouping and by the vectors. On the other hand, bacterial communities of DZ formed a separate group associated with higher available K contents and lower TN and available P contents, while the bacterial communities of QZ separated from others with lower SOC contents. The main abundant phyla differed within WM cropping system were Acidobacteria, Gammaproteobacteria, Verrucomicrobia, and Actinobacteria (Table S3; Fig. 5). Because some environmental variables were highly correlated with each other (Table S5), we also used Spearman’s rank-order correlation to evaluate relationships between abundant phyla (proteobacterial classes) and soil edaphic properties (Table 4). We found that the relative abundance of Betaproteobacteria and Chloroflexi was positively correlated with soil available P and TN contents and negatively correlated with soil pH, available K contents, and the C/N ratio. The relative abundance of Gammaproteobacteria and Bacteroidetes was positively correlated with soil pH, available K contents, and the C/N ratio and negatively correlated with available P and TN (only Bacteroidetes) contents. The relative abundance of Alphaproteobacteria and Actinobacteria was negatively correlated with soil available K contents and positively correlated with soil available P contents. In addition, the relative abundance of Deltaproteobacteria was positively correlated with soil available P and TN contents and negatively correlated with available K contents. We also found that Acidobacteria had no significant correlation with all soil properties, and soil SOC contents showed no correlations with any of the abundant phyla and classes.
To quantify the relative contributions of soil properties, location, and crop rotation to the total bacterial community based on the OTU composition, VPA was carried out, and the variation in bacterial community structure was partitioned among soil properties, crop rotation, and location, as well as interactions between them. These variables explained 46.1 % of the observed variation, leaving 53.9 % of the variation unexplained (Fig. 6). Location, soil properties, and crop rotation explained small portions of the observed variation alone, which accounted for 4.2 % (P= 0.09), 2.7 % (P= 0.01), and 1.3 % (P= 0.03), respectively. The variation was mostly explained by interactions between soil properties and location and soil properties and crop rotation, which accounted for 30.2 and 12.4 %. The interactions between location and crop rotations accounted for only 1.9 % of the variation.
Variation partitioning analysis (VPA) of the effects of soil properties (S), locations (L), crop rotation (C), and interactions between them on the bacterial community structure. Circles on the edges of the triangle show the percentage of variation explained by each factor alone. The percentage of variation explained by interactions between two or three of the factors is shown as squares on the sides and as a circle at the center of the triangle. The unexplained variation is depicted in square on the bottom
Discussion
Winter wheat–rice and winter wheat–maize cropping systems are the two main cropping systems for winter wheat production in China. In this study, pyrosequencing was used to investigate the effect of cropping systems, locations, and soil properties on the total bacterial community. The sequence analyses reveal that phyla Proteobacteria, Acidobacteria, and Bacteroidetes were the most abundant phyla in all of the samples. These results were consistent with several other studies demonstrating that Proteobacteria, Acidobacteria, and Bacteroidetes are dominant soil bacterial taxa using 16S rRNA gene clones or pyrosequencing [41, 42]. Significant differences in soil bacterial composition were also observed in these two cropping systems. In particular, high relative abundances of Betaproteobacteria and Deltaproteobacteria were observed in WR, while Alphaproteobacteria and Gammaproteobacteria were richer in WM (Table S1). To explain why some bacterial phyla are more abundant in soils than other, some researchers have proposed the concept of copio- and oligotrophic bacteria [43, 44]. Betaproteobacteria was considered as copiotrophic (fast growing) and always associated with large amounts of available nutrients [44]. In our study, we found that soil available P and TN contents were significantly higher in WR cropping systems than WM, which may explain why the higher relative abundance of Betaproteobacteria was found in WR. Chu et al. [45] and Rousk et al. [46] have reported that the relative abundances of Alphaproteobacteria and Gammaproteobacteria increase with higher pH, which appears the same in our study (Fig. 1 and Table 1). In fact, ZJG has a higher pH but results in a low abundance of these two subgroups (Table S2). Therefore, we hypothesize that there are also some other factors affecting these groups in soil.
Many studies have shown that environmental factors shape community structure [46–48]. In our study, we found that the proportions of abundant phyla and proteobacterial classes were highly correlated with soil pH. Our results were consistent with several other studies that demonstrated that soil pH was the strongest factor in structuring bacterial communities [45, 46, 49]. The relative abundances of Betaproteobacteria and Chloroflexi were negatively correlated with soil pH, while the abundances of Gammaproteobacteria and Bacteroidetes were positively correlated with soil pH. In addition, the abundances of Alphaproteobacteria and Actinobacteria were poorly correlated with soil pH. Our observations were in contrast to several other studies that showed that Betaproteobacteria and Alphaproteobacteria increase with higher soil pH and that Acidobacteria was sensitive to soil pH [46, 49]. Soil pH in our study varied from just 7.31 to 8.39, resulting in a nonsignificant change in Acidobacteria. This finding is in contrast to other studies in which the soil pH varied from 3 to 8. Furthermore, we observed that several other soil properties such as soil available P, available K, TN, and the C/N ratio were significantly correlated with abundant phyla and proteobacterial classes (Fig. 5). For instance, Betaproteobacteria and Deltaproteobacteria were positively correlated with soil available P contents and negatively correlated with soil available K contents. However, Alphaproteobacteria and Gammaproteobacteria had an opposite pattern. This finding may be a good explanation for the higher percentage of Betaproteobacteria and Deltaproteobacteria in WR, while Alphaproteobacteria and Gammaproteobacteria were richer in WM. We also observed that Actinobacteria was positively correlated with soil available K contents and negatively correlated with soil available P contents. In contrast, Nacke et al. [10] found that Actinobacteria was positively correlated with TN. Thus, we hypothesize that soil properties such as soil available P, available K, TN, SOC, etc., can also drive bacterial communities in some specific situations. Moreover, we found that anaerobic bacteria were more abundant in WR compared to WM, and two anaerobic phyla were only detected in WR. To the best of our knowledge, periodic flooding in the rice season for crop production resulted in an anaerobic environment, which may affect the soil bacterial community. Kikuchi et al. [50] and Noll et al. [51] found that bacterial community structure and diversity in a paddy soil can be affected by flooded/upland conditions and oxygen gradient. Therefore, we hypothesize that soil oxygen is also a factor in shaping bacterial communities and will have an effect for a period of time. We also observed that the abundant phyla were very similar within WR, while they differed between QZ and DZ within WM. We also hypothesize that anaerobic environment in WR help to shape the very similar bacterial community structure.
The bacterial communities were grouped well according to the cropping systems, not only in the alpha diversity but also in community structure (Figs. 2, 3, and 4). Further analysis showed that crop rotation could only explain smaller variation (1.3 %) in the total bacterial community, even though it is significant (P= 0.03) in determining the bacterial community. Most of the variation can be explained by the interactions among soil properties, location, and crop rotation. The variation that cannot be explained may be caused by some other factors, such as cropping history, fertilization, and stochastic factors. This study was consistent with some other studies that showed that crop rotation had a significant effect on the total bacterial community [17, 20]. In addition, locations and soil properties also affect the bacterial community in this study.
Soil microbial diversity is considered to be critical to the integrity, function, and long-term sustainability of soil ecosystems [9], which is usually reduced by agricultural perturbation [52, 53]. Recently, an increasing number of studies have demonstrated that greater biodiversity can enhance the ecosystem and the stability of microbial functions [54, 55]. Hence, maintenance and restoration of microbial biodiversity are becoming key issues in sustainable agricultural systems. In the present study, a more diverse bacterial community was detected in WR compared to WM, indicating that winter wheat–rice cropping systems may have a more stable ecosystem, which may contribute toward sustainable crop production.
Finally, it is important to acknowledge that we used DNA-based deep sequencing to reveal the bacterial diversity and community structure. Detecting DNA from dead bacteria is a common criticism of using DNA-based molecular methods to characterize microbial communities. This potential bias may prevent us from gaining real insights into the bacterial community. In the present study, we cannot be sure whether the anaerobic bacteria are currently active or dormant in the no flooding period of wheat season. Strategies exist to reduce or eliminate nucleic acids from dead bacteria, such as incorporation of DNA digestion steps prior to cell lysis or performing RNA-based analysis. We also acknowledge that choosing the V1–V3 region as a target for deep sequencing may lead to unexpected bias. However, the V1–V3 region was suitable for phylogenetic analysis and distinguishing most bacterial species from the phylum level to the genus level [56, 57]. Therefore, even if some bacterial communities were missed or overestimated, we could still investigate the difference between the different cropping systems.
Conclusions
Cropping systems, locations, and soil properties and the interactions among them are correlated with changes in soil bacterial community. The interactions between these environmental variables and managements can explain the main explicable variation of the community. Similar patterns of bacterial community structure were found in WR, while the WM cropping systems shape different bacterial assemblages. Higher diversity and richness were observed in the winter wheat–rice cropping system and may help maintain a stable and sustainable system.
References
Lupwayi NZ, Rice WA, Clayton GW (1998) Soil microbial diversity and community structure under wheat as influenced by tillage and crop rotation. Soil Biol Biochem 30:1733–1741
Burges HD (1981) Microbial control of pests and plant diseases 1970–80. Academic, London
Kowalchuk GA, Stephen JR (2001) Ammonia-oxidizing bacteria: a model for molecular microbial ecology. Annu Rev Microbiol 55:485–529
Lens PNL, Kuenen JG (2001) The biological sulfur cycle: novel opportunities for environmental biotechnology. Water Sci Technol 44:57–66
Tiedje JM (1988) Ecology of denitrification and dissimilatory nitrate reduction to ammonium. In: Zehnder AJB (ed) Biology of anaerobic microorganisms. Wiley, New York, pp 179–244
Gans J, Wolinsky M, Dunbar J (2005) Computational improvements reveal great bacterial diversity and high metal toxicity in soil. Science 309:1387–1390
Claire Horner-Devine M, Leibold MA, Smith VH, Bohannan BJM (2003) Bacterial diversity patterns along a gradient of primary productivity. Ecol Lett 6:613–622
Curtis TP, Sloan WT, Scannell JW (2002) Estimating prokaryotic diversity and its limits. Proc Natl Acad Sci 99:10494–10499
Kennedy A, Smith K (1995) Soil microbial diversity and the sustainability of agricultural soils. Plant Soil 170:75–86
Nacke H, Thürmer A, Wollherr A, Will C, Hodac L, Herold N, Schöning I, Schrumpf M, Daniel R (2011) Pyrosequencing-based assessment of bacterial community structure along different management types in German forest and grassland soils. PLoS One 6:e17000
Marschner P, Yang CH, Lieberei R, Crowley DE (2001) Soil and plant specific effects on bacterial community composition in the rhizosphere. Soil Biol Biochem 33:1437–1445
Wu T, Chellemi D, Graham J, Martin K, Rosskopf E (2008) Comparison of soil bacterial communities under diverse agricultural land management and crop production practices. Microb Ecol 55:293–310
Sun B, Dong Z-X, Zhang X-X, Li Y, Cao H, Cui Z-L (2011) Rice to vegetables: short- versus long-term impact of land-use change on the indigenous soil microbial community. Microb Ecol 62:474–485
Marschner P, Crowley D, Yang CH (2004) Development of specific rhizosphere bacterial communities in relation to plant species, nutrition and soil type. Plant Soil 261:199–208
Costa R, Götz M, Mrotzek N, Lottmann J, Berg G, Smalla K (2006) Effects of site and plant species on rhizosphere community structure as revealed by molecular analysis of microbial guilds. FEMS Microbiol Ecol 56:236–249
Xiong J, Liu Y, Lin X, Zhang H, Zeng J, Hou J, Yang Y, Yao T, Knight R, Chu H (2012) Geographic distance and pH drive bacterial distribution in alkaline lake sediments across Tibetan Plateau. Environ Microbiol 14:2457–2466
Xuan D, Guong V, Rosling A, Alström S, Chai B, Högberg N (2012) Different crop rotation systems as drivers of change in soil bacterial community structure and yield of rice, Oryza sativa. Biol Fertil Soils 48:217–225
Peters RD, Sturz AV, Carter MR, Sanderson JB (2003) Developing disease-suppressive soils through crop rotation and tillage management practices. Soil Tillage Res 72:181–192
el Zahar Haichar F, Marol C, Berge O, Rangel-Castro JI, Prosser JI, Balesdent J, Heulin T, Achouak W (2008) Plant host habitat and root exudates shape soil bacterial community structure. ISME J 2:1221–1230
Yin C, Jones KL, Peterson DE, Garrett KA, Hulbert SH, Paulitz TC (2010) Members of soil bacterial communities sensitive to tillage and crop rotation. Soil Biol Biochem 42:2111–2118
Acosta-Martínez V, Dowd S, Sun Y, Allen V (2008) Tag-encoded pyrosequencing analysis of bacterial diversity in a single soil type as affected by management and land use. Soil Biol Biochem 40:2762–2770
Li R, Khafipour E, Krause DO, Entz MH, de Kievit TR, Fernando WD (2012) Pyrosequencing reveals the influence of organic and conventional farming systems on bacterial communities. PloS One 7:e51897
Singh BK, Campbell CD, Sorenson SJ, Zhou J (2009) Soil genomics. Nat Rev Microbiol 7:756–756
Qiu M, Zhang R, Xue C, Zhang S, Li S, Zhang N, Shen Q (2012) Application of bio-organic fertilizer can control Fusarium wilt of cucumber plants by regulating microbial community of rhizosphere soil. Biol Fertil Soils 48:807–816
Dethlefsen L, Huse S, Sogin ML, Relman DA (2008) The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol 6:e280
Huse SM, Dethlefsen L, Huber JA, Welch DM, Relman DA, Sogin ML (2008) Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genet 4:e1000255
Liu Z, Lozupone C, Hamady M, Bushman FD, Knight R (2007) Short pyrosequencing reads suffice for accurate microbial community analysis. Nucleic Acids Res 35:e120
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541
Schloss PD, Gevers D, Westcott SL (2011) Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PloS One 6:e27310
Quince C, Lanzen A, Davenport RJ, Turnbaugh PJ (2011) Removing noise from pyrosequenced amplicons. BMC Bioinforma 12:38
Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glöckner FO (2007) SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35:7188–7196
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naïve bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267
Chao A (1984) Nonparametric estimation of the number of classes in a population. Scand J Stat 11:265–270
Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA (2005) Diversity of the human intestinal microbial flora. Science 308:1635–1638
Washington H (1984) Diversity, biotic and similarity indices: a review with special relevance to aquatic ecosystems. Water Res 18:653–694
Bunge J, Fitzpatrick M (1993) Estimating the number of species: a review. J Am Stat Assoc 8:364–373
Etten EV (2005) Multivariate analysis of ecological data using CANOCO. Austral Ecol 30:486–487. doi:10.1111/j.1442-9993.2005.01433.x
R Development Core Team (2012) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. http://www.r-project.org/
Warnes G, Bolker B, Lumley T gplots: Various R programming tools for plotting data. R package version 2.6.0
Dixon P (2003) VEGAN, a package of R functions for community ecology. J Veg Sci 14:927–930
Roesch LFW, Fulthorpe RR, Riva A, Casella G, Hadwin AKM, Kent AD, Daroub SH, Camargo FAO, Farmerie WG, Triplett EW (2007) Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J 1:283–290
Janssen PH (2006) Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl Environ Microbiol 72:1719–1728
Meyer O (1994) 4 Functional groups of microorganisms. Biodiversity and ecosystem function: Proceedings of the Symposium Held Near Bayreuth, Germany, October 1–4, 1991, Springer, p 67
Fierer N, Bradford MA, Jackson RB (2007) Toward an ecological classification of soil bacteria. Ecology 88:1354–1364
Chu H, Fierer N, Lauber CL, Caporaso J, Knight R, Grogan P (2010) Soil bacterial diversity in the Arctic is not fundamentally different from that found in other biomes. Environ Microbiol 12:2998–3006
Rousk J, Baath E, Brookes PC, Lauber CL, Lozupone C, Caporaso JG, Knight R, Fierer N (2010) Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J 4:1340–1351
Brockett BFT, Prescott CE, Grayston SJ (2012) Soil moisture is the major factor influencing microbial community structure and enzyme activities across seven biogeoclimatic zones in western Canada. Soil Biol Biochem 44:9–20
Lauber CL, Strickland MS, Bradford MA, Fierer N (2008) The influence of soil properties on the structure of bacterial and fungal communities across land-use types. Soil Biol Biochem 40:2407–2415
Lauber CL, Hamady M, Knight R, Fierer N (2009) Pyrosequencing-based assessment of soil ph as a predictor of soil bacterial community structure at the continental scale. Appl Environ Microbiol 75:5111–5120
Kikuchi H, Watanabe T, Jia Z, Kimura M, Asakawa S (2007) Molecular analyses reveal stability of bacterial communities in bulk soil of a Japanese paddy field: estimation by denaturing gradient gel electrophoresis of 16S rRNA genes amplified from DNA accompanied with RNA. Soil Sci Plant Nutr 53:448–458
Noll M, Matthies D, Frenzel P, Derakshani M, Liesack W (2005) Succession of bacterial community structure and diversity in a paddy soil oxygen gradient. Environ Microbiol 7:382–395
Saggar S, Yeates G, Shepherd T (2001) Cultivation effects on soil biological properties, microfauna and organic matter dynamics in Eutric Gleysol and Gleyic Luvisol soils in New Zealand. Soil Tillage Res 58:55–68
Yao H, Jiao X, Wu F (2006) Effects of continuous cucumber cropping and alternative rotations under protected cultivation on soil microbial community diversity. Plant Soil 284:195–203
Chaer G, Fernandes M, Myrold D, Bottomley P (2009) Comparative resistance and resilience of soil microbial communities and enzyme activities in adjacent native forest and agricultural soils. Microb Ecol 58:414–424
Garbeva P, Postma J, Van Veen J, Van Elsas J (2006) Effect of above‐ground plant species on soil microbial community structure and its impact on suppression of Rhizoctonia solani AG3. Environ Microbiol 8:233–246
Kim M, Morrison M, Yu Z (2011) Evaluation of different partial 16S rRNA gene sequence regions for phylogenetic analysis of microbiomes. J Microbiol Methods 84:81–87
Jeraldo P, Chia N, Goldenfeld N (2011) On the suitability of short reads of 16S rRNA for phylogeny‐based analyses in environmental surveys. Environ Microbiol 13:3000–3009
Acknowledgments
This research was financially supported by the Chinese Ministry of Science and Technology (2013AA102802 and 2011BAD11B03) and the Agricultural Ministry of China (201103004 and 2011-G27). R.Z and Q.S were also supported by the 111 Project (B12009) and the Priority Academic Program Development (PAPD) of Jiangsu Higher Education Institutions. We would like to thank Dr. Xiaoyu Yong, Nanjing University of Technology, and Joshua Kendall, Ohio State University, for their careful comments on this manuscript and Dr. Benli Chai of Michigan State University for his help submitting the data. We also thank Majorbio Bio-pharm Technology Co., Ltd (Shanghai, China) for their help with the pyrosequencing experiments.
Author information
Authors and Affiliations
Corresponding author
Additional information
J. Zhao and R. Zhang contributed equally to this paper.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOC 167 kb)
Rights and permissions
About this article
Cite this article
Zhao, J., Zhang, R., Xue, C. et al. Pyrosequencing Reveals Contrasting Soil Bacterial Diversity and Community Structure of Two Main Winter Wheat Cropping Systems in China. Microb Ecol 67, 443–453 (2014). https://doi.org/10.1007/s00248-013-0322-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-013-0322-0