Abstract
Studying shifts in microbial communities under different land use can help in determining the impact of land use on microbial diversity. In this study, we analyzed four different land-use types to determine their bacterial and archaeal diversity and abundance. Three natural ecosystems, that is, wetland (WL), grassland (GL), and forest (FR) soils, and one agricultural soil, that is, tea plantation (TP) soil, were investigated to determine how land use shapes bacterial and archaeal diversity. For this purpose, molecular analyses, such as quantitative polymerase chain reaction (Q-PCR), 16S rRNA gene sequencing, and terminal restriction fragment length polymorphism (T-RFLP), were used. Soil physicochemical properties were determined, and statistical analyses were performed to identify the key factors affecting microbial diversity in these soils. Phylogenetic affiliations determined using the Ribosomal Database Project (RDP) database and T-RFLP revealed that the soils had differing bacterial diversity. WL soil was rich in only Proteobacteria, whereas GR soil was rich in Proteobacteria, followed by Actinobacteria. FR soil had higher abundance of Chloroflexi species than these soils. TP soil was rich in Actinobacteria, followed by Chloroflexi, Acidobacteria, Proteobacteria, and Firmicutes. The archaeal diversity of GL and FR soils was similar in that most of their sequences were closely related to Nitrososphaerales (Thaumarchaeota phylum). In contrast, WL soil, followed by TP soil, had greater archaeal diversity than other soils. Eight different archaeal classes were found in WL soil, and Pacearchaeota class was the richest one. The abundance of bacterial and archaeal 16S rRNA gene copies in WL and GL soils was significantly higher than that in FR and TP soils. Redundancy analysis showed that bacterial diversity was influenced by abiotic factors, e.g., total organic carbon and pH, whereas total nitrogen, pH, and cation exchange capacity (CEC) significantly affected archaeal community composition. Pearson correlation analysis showed that bacterial and archaeal 16S rRNA gene abundance had the highest correlation with clay content (r > 0.905, P < 0.01), followed by total-P, CEC, pH, and silt (%). These results will lead to more comprehensive understanding of how land use affects microbial distribution.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Soil is a spatially structured, heterogeneous, discontinuous and dynamic biological system although generally poor in nutrients and energy sources (Nannipieri et al. 2003). Bacteria are the most abundant microorganisms in soil (Roesch et al. 2007), and their activity, ecology, and population dynamics are driven by the physical and chemical properties of soils (Fierer and Jackson 2006) and land-use types, e.g., wetland, forestland, and cropland (Nacke et al. 2011).
Most studies have focused on temperate grasslands and forests, although tropical forest and wetland soils are some of the most endangered ecosystems in the world. These ecosystems are important because of their potential response to current and future climate change, and as potential sites for agriculture (Balser et al. 2010). Approximately half of the world’s wetland areas are found in the tropics and wetlands have been estimated to cover 5–7% of the world’s surface (Scharpenseel 1993). Natural forests are recognized as sites of high biodiversity, where complex relationships among fauna, flora, and microflora are maintained because of the structural richness of the habitat (Hackl et al. 2004). Tea (Camellia sinensis) plantations dominate much of the agricultural landscape in Asia. Tea is usually grown as a monoculture in acidic soils and requires considerable amounts of fertilizers; tea plantations have high amounts of root exudates and leaf litter (Çakmakç et al. 2010). Grassland soils typically have greater soil organic matter (SOM) content and labile C pools. In addition, agricultural and grassland soils support distinct microbial communities that are correlated with factors that define soil quality, suggesting that land-use history and the associated soil quality influence microbial community composition (Steenwerth et al. 2003).
Land-use management, one of the most important aspects of anthropogenic disturbance to terrestrial ecosystems, has exerted extensive impacts on soil biogeochemical cycling and inhabitant microorganisms (Shen et al. 2013). Although various previous studies focused on the microbial diversity in different soils (Cunliffe et al. 2008; Keshri et al. 2013; Suna et al. 2014), comparative studies on the distribution of bacterial and archaeal populations in different ecosystems and how land use alter the diversity of bacteria and archaea in these ecosystems are largely unavailable. Therefore, studying shifts in microbial communities under different ecosystems can help to identify the impact of land use.
The main objective of this study was to investigate how land use shapes the bacterial and archaeal diversities in soils from four different ecosystems using molecular analyses, such as quantitative-PCR (Q-PCR), sequencing analysis, and terminal restriction fragment length polymorphism (T-RFLP). Comparison of bacterial communities by T-RF analysis has been proven to provide information consistent with that obtained from analysis of clone libraries (Hackl et al. 2004). Soil physicochemical properties were determined, and statistical analyses were performed to identify the key factors affecting microbial diversities in these four different ecosystems.
Materials and methods
Soil information
Soil samples were collected in four different ecosystems encompassing different land-use types, i.e., wetland (WL), grassland (GL), forest (FR), and tea plantation (TP), in southern China. The dominant plant species in WL soils was Spartina alterniflora, whereas the dominant plant species in GL were Stipa baicalensis, Stipa sareptana var. krylovii, Cleistogenes squarrosa, Agrostis gigantea, Bromus inermi, and Deyeuxia langsdorffii. In FR soil, the dominant plant species were trees (e.g., Castanopsis carlesii and Elaeocarpus decipiens), shrubs (e.g., Itea chinensis and Adinandra dichotama), and grasses (Gahnia tristis, Dicranopteris dichotama, and Iles pubescens). TP soil was planted with tea for 5 years. Full site details and soil properties are given in Table S1.
Soil sampling and pre-preparation
Four independent samples from the surface Ah horizon (0–20 cm) were used in the experiments. Field moist soil from each replicate sample was hand-sorted to remove visible plant residues, and then sieved (<5 mm mesh). Prior to use, the wetland soils (WL) were flooded with distilled water, whereas the other soils (GL, FR, and TP) were adjusted to 45% of the field water holding capacity. To accommodate subsequent changes in microbial activity (Butterly et al. 2011), one set of microcosms of the four soils, each with four replicates, was left to equilibrate prior to analysis for 2 weeks after flooding or rewetting.
Soil characteristics
Soil pH and electronic conductivity (EC) were determined at a soil:water ratio of 1:2.5 using a Delta 320 pH meter (Mettler-Toledo Instruments Co., China). Soil moisture content was determined after oven-drying at 105 °C overnight to constant weight. Cation exchange capacity (CEC) was determined by titration (Rhoades 1982; Müller and Höper 2004). Total nitrogen (TN) and total phosphate (TP) were determined by Kjeldahl digestion (Page et al. 1982), and by NaOH fusion and colorimetric analysis (Olsen and Somers 1982), respectively. Soil texture was determined using a laser particle analyzer (Mastersizer 2000, UK).
Microbial DNA extraction from soil samples and PCR
DNA was extracted from 500 mg of soil, in duplicates from each sample, using the FastDNA Spin Kit (MPbio, USA) according to the manufacturer’s protocol. The integrity and yield of extracted nucleic acids were checked by agarose gel electrophoresis and UV quantification (NanoDrop ND-1000, Germany).
Cloning and sequencing of bacterial, archaeal 16S rRNA
For bacterial 16SrRNA identification, Eub-8f /926r primers were used (Lu et al. 2003), and for archaea, Arc-109f/934r primers were used (Aschenbach et al. 2013). PCR products from four replicates were pooled, purified, and cloned. Randomly selected clones were then screened for positive inserts by PCR performed using the SP6 and T7 primers (Sambrook and Russell 2001) and were sequenced by Invitrogen, China. After chimera removal, the similarity of sequences from desired gene clones was checked using MOFFAT (http://www.ebi.ac.uk/Tools/msa/mafft) and a sequence identity of >97% was defined as an operational taxonomic unit (OTU). Phylogenetic affiliations and taxonomical hierarchy based on 16S rRNA were determined with 80% confidence levels using the CLASSIFIER tool (http://rdp.cme.msu.edu) of the Ribosomal Database Project (RDP)-II database (Wang et al. 2007).
Nucleotide sequence accession numbers
The sequences were deposited in the NCBI GenBank database under accession numbers KT460206–KT460753 for bacterial 16S rRNA gene sequences and KT460975–KT461150 for archaeal 16S rRNA gene sequences.
T-RFLP analysis
The community composition of the bacteria and archaea in four different soils was estimated by T-RFLP to support the clone library analysis. T-RFLP was performed using the restriction enzyme HhaI (TAKARA, Japan). The same PCR programs and primer sets were used for clone library analysis. The other procedures were the same as those described by Yuan et al. (2013). Only T-RFs with an average value of Ra > 1% from four replicates were analyzed. Peaks with a Ra above 10% were regarded as dominant T-RFs. To compare bacterial and archaeal diversity under different land uses, Shannon–Wiener and evenness indices were calculated as previously described by Yuan et al. (2013).
Quantification of 16S rRNA genes
Prior to Q-PCR analyses (ABI 7900, Foster City, CA, USA), all DNA extracts were diluted to approximately 5 ng μl− 1 with sterilized ddH2O to reduce inhibition by co-extracted substances. For the bacterial and archaeal 16S rRNA genes, Q-PCRs were performed using the two universal primer pairs Eub-8f/Eub-926r and Arc109f/Arc934r, respectively. The amplifications were performed using three technical replicates per sample. The thermal protocol for both genes was an initial denaturation for 30 s at 95 °C; followed by 40 cycles each of 15 s at 95 °C and 30 s at 60 °C. The standard curve preparation and other procedures were performed as described by Yuan et al. (2012).
Statistical analysis
One way analysis of variance (ANOVA) and multiple comparisons of significant differences were performed using Duncan’s test (P < 0.05). Correlation analysis of soil abiotic properties and bacterial and archaeal abundance was performed using SPSS 16.0 (SPSS Inc., Chicago, IL), with significance defined as P < 0.05 unless otherwise stated. Rare-faction curves for bacterial and archaeal 16S rRNA were generated by plotting the number of OTUs observed against the number of clones sequenced (http://strata.uga.edu/software/Software.html) to assess the sampling effort. The effects of environmental variables on microbial diversity were analyzed by redundancy analysis (RDA) with CANOCO 5.0 for Windows (Monte Carlo permutation test, P = 0.001; Microcomputer Power, USA).
Results
Soil characteristics
The results for basic soil parameters significantly differed for the four soil types (Table 1). Compared to the three natural soils, the TP soil showed the highest TN, TOC, and clay content and the lowest pH. GL soils had the highest pH and sand (%) and the lowest CEC. The highest total-P, CEC, and silt (%) were found in WL soil, whereas the lowest TOC, total-P, and clay (%) were observed in FR soil. According to the soil particle analysis, WL soil was silt clay loam, GL soil was sierozems, and FR and TP soil were sandy clay loam.
Cloning library analysis of bacterial and archaeal 16S rRNA
For bacterial clone libraries, 154 clones from WL, 140 clones from GL, 163 clones from FR, and 163 clones from TP soils were obtained after trimming chimera. Then, 92 to 135 bacterial OTUs were observed with a sequence identity of >97% by MOFFAT. Phylogenetic affiliations determined using the RDP database revealed that bacterial diversity of four different soils was different (Fig. 1a). The high bacterial diversity in WL included a high relative abundance of Proteobacteria (72.59%), followed by Chloroflexi (8.8%) (Fig. 1a). In GR soil, Proteobacteria had the greatest abundance (31.8%), followed by Actinobacteria (27.3%). In TP soils, Actinobacteria had the greatest abundance (27.2%), followed by Chloroflexi (23.9%) and Acidobacteria (20.7%) (Table S2). The rare-faction curves for the bacterial 16S rRNA gene clone library showed that the number of sequenced clones from each library was insufficient to cover the bacterial diversity of four different soils (Fig. 2a).
Relative abundance of different bacterial phylogenetic groups in a bacterial and b archaeal 16S rRNA gene clone libraries from four different soils. Analysis of amplified 16S rRNA gene sequences was performed in comparison with the RDP database at the 80% confidence level. The percentages of the phylogenetically classified sequences are plotted on the X axis. FR forest, GL grassland, TP tea plantation, WL wetland
The phylogeny of the archaeal 16S rRNA gene library comprised 131 clones and 90 OTUs from WL soil, 102 clones and 15 OTUs from GL soil, 103 clones and 14 OTUs from FR soil, and 91 clones and 16 OTUs from TP soil (Table S3). The archaeal diversity of GL and FR soils was similar in that most of their sequences were closely related to Nitrososphaerales under the Thaumarchaeota phylum, except that one of the FR OTUs was closely related to Methanomicrobia. In contrast, the archaeal diversity of WL soil followed by TP soil was higher than that of other soils (Fig. 1b). Eight different types of archaeal classes were found in WL soil; Pacearchaeota was the richest, followed by Crenarchaeota, Nitrososphaerales, and Methanomicrobia. In TP soil, Nitrososphaerales was the richest followed by Nitrosopumilales and Methanomicrobia. Rare-faction analysis showed that the number of sequenced clones from each library was sufficient to cover the archaeal diversity of the soils, except for WL soil (Fig. 2b).
T-RFLP analyses of the bacterial and archaeal community compositions
Thirty-six T-RFs were identified and used to compare bacterial communities using the restriction enzyme HhaI (Fig. 3a). Different land use was found to affect both the presence and relative abundance of different bacterial 16S rRNA T-RFs. For WL soil, the dominant T-RFs had length of 61-, 92-, and 211-bp, whereas TR-Fs with lengths of 37-, 61-, 82-, and 208-bp were the major components in GL soil. In FR soil, 199-, 202-, 215-, and 219-bp T-RFs predominated, whereas T-RFs with lengths of 163- and 215-bp were the dominants in TP soil. In silico, analysis revealed that the predominant 37-, 61-, 82-, 92-, 163-, 199-, 202-, 208-, 211-, 215-, and 219-bp T-RFs were most closely related to Gaiella occulta, Dehalogenimonas alkenigignens, Sphingomonas sp. JS5, Nisaea nitritireducens, Acidobacteriaceae bacterium A2-4c, Mizugakiibacter sediminis, Ktedonobacter racemifer DSM 44963, Thioalkalivibrio sulfidiphilus, Methylomicrobium alcaliphilum 20Z, K. racemifer DSM 44963, and Vicinamibacter silvestris (Table S4).
Average relative abundance of T-RFs in four different soils determined by endonuclease digestion with HhaI for bacteria (a) and for archaea (b). The relative abundance of T-RFs is given as a percentage of the total peak height fragment sizes. Bars indicate the standard error of the mean (n = 4), FR forest, GL grassland, TP tea plantation, WL wetland
Comparison of 19 archaeal 16S T-RFLP profiles (Fig. 3b) showed that five T-RFs (240-, 242-, 247-, 263-, and 267-bp) represented the main community sequence types in the soils studied. The 247-bp T-RF (36–81.7% of the total profile) dominated in the three natural soils, with the highest levels detected in GL and FR soils. In comparison, the 263-bp fragment (30.5%) of the total profile was dominant only in WL soil. The 240-bp TR-F was dominant only in FR soil, whereas 242- and 267-bp T-RFs were dominant in TP soil. In silico comparison of fragment lengths showed that the 247-, 263-, and 267-bp T-RFs were most closely related to Nitrososphaera viennensis, Thermofilum carboxyditrophus 1505, and Candidatus Nitrososphaera gargensis Ga9.2 (Table S5).
To extrapolate the similarities and differences between the community compositions of bacteria and archaea in response to different land use, biodiversity analyses were performed on the basis of T-RFLP results. The Shannon–Wiener and evenness values (Table 2) indicated that the bacterial 16S rRNA diversity did not significantly differ among soils. However, for archaeal 16S rRNA, the Shannon–Wiener and evenness values for WL and TP soils were higher than those for GL and FR soils.
Abundance of bacterial and archaeal 16S rRNA genes
The copy number of bacterial 16S rRNA genes ranged from 7.2 × 109 to 1.61 × 1011 copies g− 1 dry soil for four different soils. For the archaeal 16S rRNA gene, the copy number ranged from 1.98 × 106 to 6.3 × 109 copies g− 1 dry soil. Among four different soils, the abundance of bacterial and archaeal 16S rRNA gene copies for WL was the highest and that of GL was the second highest. Bacterial 16S rRNA gene abundance was the lowest in FR soil and archaeal 16S rRNA gene abundance was the lowest in TP soil (Table 3).
Correlation and RDA analyses of bacterial and archaeal abundance and diversity
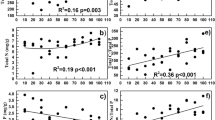
The relationship between soil physicochemical parameters and bacterial and archaeal 16S rRNA gene abundance and diversity in four different soils was evaluated by correlation analysis (Table 4). Soil physiochemical properties were highly correlated with bacterial and archaeal 16S rRNA gene abundance, with the highest correlation being observed for clay (%) (r > 0.905, P < 0.01) followed by total-P, CEC, pH and silt (%). Sand (%) was negatively correlated with gene abundance (r > 0.672, P < 0.01).
To determine the effect of soil abiotic properties on bacterial and archaeal community composition, T-RFLP profiles were subjected to multivariate analysis. The RDA showed that the bacterial communities in the four different soils were clearly distinct from each other (Fig. 4a). The samples were divergent along the first ordination axis, which explained 41.81% of the variation. It was found that clay (%), TOC, and pH were significantly correlated with bacterial community composition (P = 0.001 by Monte Carlo permutation test).
The two-dimensional plot of multivariate statistics (derived from RDA), where the first two axes displayed 57.94 and 29.57% variations, respectively, showed that the archaeal community compositions of FR and TP were similar, although they were clearly distinct from WL and GL soils (Fig. 4b). TN, pH, and CEC were significantly correlated with the archaeal community composition (P = 0.001 by the Monte Carlo permutation test).
Discussion
Although a large amount of knowledge regarding the response of soil bacterial communities to land-use types and soil properties has been assembled (Nacke et al. 2011), the present understanding of how the interaction among land use and soil type affects the microbial community from different ecosystems is still poor. Bacteria are the most abundant and diverse group of soil microorganisms and play multiple important key roles in soil. Any modifications in the microbial community caused by land use change might contribute to changes in ecosystem function and maintenance of soil quality (Konopka 2009). We focused on connecting variations in community composition to differences in soil edaphic characteristics and land use. The present analyses indicate that the microbial communities in the four land uses were different, although land use did not affect the richness and evenness of soil bacterial diversity in these four different ecosystems.
In WL and GR soils, Proteobacteria, especially Alphaprotebacteria, were most abundant followed by Actinobacteria and Chloroflexi (Fig. 1). In TP soils, Actinobacteria were the most abundant, followed by Chloroflexi and Acidobacteria. Proteobacteria and Actinobacteria were dominant in these soils as they are prevalent in the soil of various ecosystems (Zhang and Xu 2008). A high content of Proteobacteria was detected in the pH range of 5.6–7.8 (Long et al. 2015). Alphaprotebacteria prefer nutrient rich environments (Goldfarb et al. 2011), whereas Betaproteobacteria are negatively influenced by clay content indicating that coarse-textured soils are more favorable habitats for this taxon (Rousk et al. 2010). Acosta-Martı´nez et al. (2008) reported that non-disturbed systems under grasses (i.e., Conservation Reserve Program and pastures) had higher Actinobacteria counts as well as microbial biomass and nutrient cycling enzyme activities than crop land. Acidobacteria has also been identified as one of the most common phyla in soil (Zhang et al. 2014); they are slow-growing bacteria that can grow in nutrient-limited environments, such as pristine forest soils (Ward et al. 2009).
However, the bacterial abundance in FR soil was different from that in other soils; Chloroflexi, especially Ktedonobacteria was most dominant, followed by Actinobacteria. This finding was in accordance with that reported by Kim et al. (2014), as the pH of the FR and TP soils was lower than that of WL and GR soil. Thus, the abundance of Ktedonobacteria of Chloroflexi decreased with an increase in soil pH. Chloroflexi accounts for 33–54% of the bacterial sequences in low-pH soils (Long et al. 2015). Within our literature review, not much information was found regarding the ecological role of Ktedonobacteria; however, a few studies noted the occurrence of Chloroflexi phyla in systems with extreme environmental conditions, such as extremely high temperatures (Bjo’rnssona et al. 2002). Bach et al. (2010) also found that vegetation types play an important role in the structure of soil bacterial communities; this was consistent with the findings for different types of soils in our study with different types of plant species. Although the TOC content of TP soil was the highest, the abundance and diversity of bacteria and archaea in this soil were lower than those of other soils. Thus, the findings indicate that microbial diversity depends not only on soil nutrients but also on other abiotic factors such as pH and CEC.
The highest copy number of the 16S rRNA gene in WL soil provides evidence that this ecological system is suitable for microbes, for example, moderate pH and flooding can also provide advantages for anaerobic bacteria as compared to other soil types. The data on 16S rRNA gene copies revealed that the bacteria were much more abundant than archaea in all four soil types. Rare-faction analysis and diversity indices showed the presence of a greater number of species of bacteria than archaea, indicating that this soil is capable of supporting higher density and diversity of bacteria. Bacterial 16S rRNA gene copies were found to be two orders of magnitude more abundant than archaeal 16S rRNA gene copies. A similar ratio of bacterial and archaeal abundance was found in a previous study (Ye et al. 2009). The rare-faction curves indicated that the number of sequenced clones from each library was insufficient to cover the bacterial diversity; however, the curves were near saturation for archaeal diversity.
The phylogenetic analysis showed that the majority of the archaeal clones belonged to the phylum Thaumarchaeota. Similar archaeal diversities have been found in the native forest soil in Amazon and different types of Brazilian soils (Pacchioni et al. 2014; Tupinambá et al. 2016). Bates et al. (2011) found that the Thaumarchaea group appeared to be the most dominant and ubiquitous group in soil; these species were more abundant in acidic soils, such as temperate acidic forest soil (Kemnitz et al. 2007) and acidic red soils (Ying et al. 2010). Members of Thaumarchaeota are now considered to play a major role in the global nitrogen cycle (Stieglmeier et al. 2014). In WL soils, Pacearchaeota were the most dominant species which was compatible with the previous studies describing the wide distribution of Pacearchaeota in saline aquatic environments, where they were present in both marine and inland waters, mainly in microbial mats and sediments (Pachiadaki et al. 2011), followed by hot springs and fresh water. However, their ecological and biological interactions are not known(Kan et al. 2011).
16S rRNA analysis has shown that soil type is also responsible for changes in archaeal communities in soils under different types of land use (native grassland, native forest, eucalyptus and acacia plantations, and soybean and watermelon fields) (Lupatini et al. 2013). The wide range of origin of phylotypes from varying environments might explain the high diversity of bacteria and archaea, although their eco-physiological roles are yet to be investigated.
To accurately evaluate a microbial ecosystem, it is necessary to integrate the influences of biotic and abiotic factors on the diversity and community composition of microbes. Among the soil properties, pH, total-P, CEC, and soil textures were significantly correlated with bacterial and archaeal abundances. The Monte Carlo permutation test showed that pH, TOC, and clay content were significantly correlated with bacterial community composition, and pH, TN, and CEC were significantly correlated with archaeal community composition. All these variables are related to nutrient availability, which has obvious implications for microbial and plant growth. pH imposes significant and direct physiological stress on bacterial cells, resulting in the selection of the best-adapted cells (Fierer and Jackson 2006; Shen et al. 2013). This finding was consistent with those of the previous studies on wetlands of the Qinghai-Tibetan Plateau (Deng et al. 2014). Characterization of abiotic properties is important for understanding the factors that affect bacterial and archaeal diversity and for obtaining a clearer view of how microbial communities change (Faoro et al. 2010). The study of soil bacterial diversity using molecular biological techniques has expanded the current understanding of major bacterial and archaeal groups in soil that contribute to essential soil processes in nutrient cycling (Lynch and Bragg 1985; Acosta-Martı´nez et al. 2010). Future studies such as high-throughput sequencing and gene expression analysis might enhance the understanding of the ecological role of bacterial and archaeal diversity in different ecosystems.
Conclusion
Our data highlight that land use has strong effects on soil microbes, with consequences pertaining to microbial abundance, diversity, and community composition. WL soil showed the highest abundance of bacterial and archaeal 16S rRNA gene copies and also the greatest diversity, as an environment in equilibrium and TP soil showed the lowest bacterial diversity because of the acidic conditions. 16S rRNA and T-RFLP analyses showed that land use was responsible for changes in bacterial and archaeal communities in four different ecosystems. RDA analysis showed that pH, TOC, TN, and CEC played a significant role in shaping the microbial diversity in these ecosystems. Thus, these results provide insights into the microbial community composition in these ecosystems and identify the main factors shaping this composition, which will lead to a more comprehensive understanding of microbial distribution in different ecosystems.
References
Acosta-Martı´nez V, Dowd S, Sun Y, Allen VG (2008) Tag-encoded pyrosequencing analysis of bacterial diversity in a single soil type as affected by management and land use. Soil Biol Biochem 40:2762–2770
Acosta-Martı´nez V, Dowd SE, Sun Y, Wester D, Allen V (2010) Pyrosequencing analysis for characterization of soil bacterial populations as affected by an integrated livestock-cotton production system. Appl Soil Ecol 45:13–25
Aschenbach K, Conrad R, Rˇeháková K, Doležal J, Janatková K, Roey Angel R (2013) Methanogens at the top of the world: occurrence and potential activity of methanogens in newly deglaciated soils in high-altitude cold deserts in the Western Himalayas. Front Microbiol 4:359. doi:10.3389/fmicb.2013.00359
Bach L, Grytnes J, Halvorsen R, Ohlson M (2010) Tree influence on soil microbial community structure. Soil Biol Biochem 42:1934–1943
Balser TC, Wixon D, Moritz LK, Lipps L (2010) The microbiology of natural soils. In: Dixon GR TE (ed) Soil microbiology and sustainable crop production. Springer, Heidelberg, pp 27–58
Bates ST, Berg-Lyons D, Caporaso JG, Walters WA, Knight R, Fierer N (2011) Examining the global distribution of dominant archaeal populations in soil. ISME J 5:908–917
Bjo’rnssona L, Hugenholtz P, Tyson GW, Blackall LL (2002) Filamentous Chloroflexi (green non-sulfur bacteria) are abundant in wastewater treatment processes with biological nutrient removal. Microbiology 148:2309–2318
Butterly CR, McNeill AM, Baldock JA, Marschner P (2011) Rapid changes in carbon and phosphorus after rewetting of dry soil. Biol Fertil Soils 47:41–50
Çakmakç R, Dönmez MF, Ertürk Y, Erat M, Haznedar A, Sekban R (2010) Diversity and metabolic potential of culturable bacteria from the rhizosphere of Turkish tea grown in acidic soils. Plant Soil 332:299–318. doi:10.1007/s11104-010-0295-4
Cunliffe M, Schafer H, Harrison E, Cleave S, Upstill-Goddard R, Murrell JC (2008) Phylogenetic and functional gene analysis of the bacterial and archaeal communities associated with the surface microlayer of an estuary. ISME J 2:776–789
Deng Y, Cui X, Herna´ndez M, Dumont MG (2014) Microbial diversity in Hummock and Hollow soils of three wetlands on the Qinghai-Tibetan plateau revealed by 16S rRNA pyrosequencing. PLoS One 9:e103115. doi:10.1371/journal.pone.0103115
Faoro H et al (2010) Influence of soil characteristics on the diversity of bacteria in the southern Brazilian Atlantic forest. Appl Environ Microbiol 76:4744–4749
Fierer N, Jackson RB (2006) The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci USA 103:626–631
Goldfarb KC et al (2011) Differential growth responses of soil bacterial taxa to carbon substrates of varying chemical recalcitrance. Front Microbiol 22:1–10
Hackl E, Zechmeister-Boltenstern S, Bodrossy L, Sessitsch A (2004) Comparison of diversities and compositions of bacterial populations inhabiting natural forest soils. Appl Environ Microbiol 70:5057–5065
Kan J et al (2011) Archaea in Yellowstone Lake. ISME J 5:1784–1795
Kemnitz D, Kolb S, Conrad R (2007) High abundance of Crenarchaeota in a temperate acidic forest soil. FEMS Microbiol Ecol 60:442–448
Keshri J, Mishra A, Jha B (2013) Microbial population index and community structure in saline–alkaline soil using gene targeted metagenomics. Microbiol Res 168:165–173
Kim HM et al (2014) Bacterial community structure and soil properties of a subarctic tundra soil in Council, Alaska. FEMS Microbiol Ecol 89:465–475
Konopka A (2009) What is microbial community ecology? ISME J 3:1223–1230
Long XE, Yao HY, Wang J, Huang Y, Singh B, Zhu YG (2015) Community structure and soil pH determine chemoautotrophic carbon dioxide fixation in drained paddy soils. Environ Sci Technol 49:7152–7160
Lu J, Idris U, Harmon B, Hofacre C, Maurer JJ, Lee MD (2003) Diversity and succession of the intestinal bacterial community of the maturing broiler chicken. Appl Environ Microbiol 69:6816–6824
Lupatini M, Jacques RJS, Antoniolli ZI, Suleiman AKA, Fulthorpe RR, Roesch LFW (2013) Land-use change and soil type are drivers of fungal and archaeal communities in the Pampa biome. World J Microbiol Biotechnol 29:223–233
Lynch JM, Bragg E (1985) Microorganisms and soil aggregate stability. Adv Soil Sci 2:133–171
Müller T, Höper H (2004) Soil organic matter turnover as a function of the soil clay content: consequences for model applications. Soil Biol Biochem 36:877–888. doi:10.1016/j.soilbio.2003.12.015
Nacke H, Thu¨rmer A, Wollherr A, Will C, Hodac L et al (2011) Pyrosequencing-based assessment of bacterial community structure along different management types in German forest and grassland soils. PLos One. 6:e17000
Nannipieri R, Ascher J, Ceccherini MT, Landi L, Pietramellara G, Renella G (2003) Microbial diversity and soil functions. Euro. J Soil Sci 54:655–670
Olsen SR, Somers LE (1982) Phosphorus. In: K De Page AL (ed) Methods of soil analysis, vol 2. Soil Science Society of America, Madison, pp 403–448
Pacchioni RG, Carvalho FM, Thompson CE et al (2014) Taxonomic and functional profiles of soil samples from Atlantic forest and Caatinga biomes in northeastern Brazil. Microbiol Open. 3:299–315
Pachiadaki MG, Kallionaki A, Dählmann A, De Lange GJ, Kormas KA (2011) Diversity and spatial distribution of prokaryotic communities along a sediment vertical profile of a deep-sea mud volcano. Microb Ecol 62:655–668
Page A, Miller R, Keeney D (1982) Total carbon, organic carbon, and organic matter. In: K De Page AL (ed) Methods of soil analysis, vol 2. Soil Science Society of America, Madison, pp 539–579
Rhoades JD (1982) Cation exchangeable capacity. In: M. R. In Pace AL KD (ed) Methods of soil analysis, part 2: chemical and microbiological properties, 2nd edn. American Society of Agronomy, Inc., Soil Science Society of America, Madison, pp 149–165
Roesch LFW et al (2007) Pyrose-quencing enumerates and contrasts soil microbial diversity. ISME J 1:283–290
Rousk J et al (2010) Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J 4:1340–1351
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Scharpenseel HW (1993) Major carbon reservoirs of the pedosphere; source-sink relations; potential of D14C and δ13C as supporting methodologies. Water Air Soil Pollut 70:431–442
Shen JP, Cao P, Hu HW, He JZ (2013) Differential response of archaeal groups to land use change in an acidic red soil. Sci Total Environ 461:742–749
Steenwerth KL, Jackson LE, Caldero´n FJ, Stromberg MR, Scow KM (2003) Soil microbial community composition and land use history in cultivated and grassland ecosystems in coastal California. Soil Biol Biochem 35:489–500
Stieglmeier M et al (2014) Nitrososphaera viennensis gen. nov., sp. nov., an aerobic and mesophilic, ammonia-oxidizing archaeon from soil and a member of the archaeal phylum Thaumarchaeota. Int J Syst Evol Microbiol 64:2738–2752
Suna H, Terhonena E, Koskinenb K, Paulinb L, Kasanena R, Asiegbua FO (2014) Bacterial diversity and community structure along different peat soilsin boreal forest. Appl Soil Ecol 74:37–45
Tupinambá DD et al (2016) Archaeal community changes associated with cultivation of Amazon Forest soil with oil palm. Archae 2016:1–14
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naïve bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267
Ward NL et al (2009) Three genomes from the phylum Acidobacteria provide insight into the lifestyles of these microorganisms in soils. Appl Environ Microbiol 75:2046–2056
Ye G et al (2009) Distribution and diversity of bacteria and archaea in marine sediments affected by gas hydrates at Mississippi canyon in the Gulf of Mexico. Geomicrobiol J 26:370–381
Ying JY, Zhang LM, He JZ (2010) Putative ammonia-oxidizing bacteria and archaea in an acidic red soil with different land utilization patterns. Environ Microbiol Rep 2(2):304–312
Yuan HZ, Ge T, Chen CY, O’Donnell AG, Wu JS (2012) Significant role for microbial autotrophy in the sequestration of soil carbon. Appl Environ Microbiol 78:2328–2336
Yuan HZ et al (2013) Effect of land use on the abundance and diversity of autotrophic bacteria as measured by ribulose-1,5-biphosphate carboxylase/oxygenase (RubisCO) large subunit gene abundance in soils. Biol Fertil Soils 49:609–616
Zhang L, Xu ZH (2008) Assessing bacterial diversity in soil. J Soil Sediment 8:379–388
Zhang Y et al (2014) Community structure and elevational diversity patterns of soil Acidobacteria. J Environ Sci 26:1717–1724. doi:10.1016/j.jes. 2014.06.012
Acknowledgements
This study was financially supported by the National Natural Science Foundation of China (41522107; 41503081), National Key Research and Development Program (2016YFE0101100), the CHINA-ASEAN Talented Young Scientist Visiting Program (2014–2015), and NSFC Research Fund for International Young Scientists (41450110432). We thank Public Service Technology Center, Institute of Subtropical Agriculture, Chinese Academy of Sciences for technical assistance and Biotechnology Research Department, Myanmar.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Erko Stackebrandt.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Lynn, T.M., Liu, Q., Hu, Y. et al. Influence of land use on bacterial and archaeal diversity and community structures in three natural ecosystems and one agricultural soil. Arch Microbiol 199, 711–721 (2017). https://doi.org/10.1007/s00203-017-1347-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-017-1347-4