Abstract
Crop production may benefit from plant growth-promoting bacteria. The knowledge on bacterial communities is indispensable in agricultural systems that intend to apply beneficial bacteria to improve plant health and production of crops such as canola. In this work, the diversity of root bacterial communities associated to two different developmental phases of canola (Brassica napus L.) plants was assessed through the application of new generation sequencing technology. Total bacterial DNA was extracted from root samples from two different growth states of canola (rosette and flowering). It could be shown how bacterial communities inside the roots changed with the growing stage of the canola plants. There were differences in the abundance of the genera, family, and even the phyla identified for each sample. While in both root samples Proteobacteria was the most common phylum, at the rosette stage, the most common bacteria belonged to the family Pseudomonadaceae and the genus Pseudomonas, and in the flowering stage, the Xanthomonadaceae family and the genus Xanthomonas dominated the community. This implies in a switch in the predominant bacteria in the different developmental stages of the plant, suggesting that the plant itself interferes with the associated microbial community.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Crop production is closely related to the soil and rhizosphere bacteria, which interact with plant roots. These interactions can be beneficial, detrimental, or neutral for the plant, and sometimes the effect of a particular bacterium may vary as a consequence of soil conditions [1]. The knowledge on bacterial community is, therefore, indispensable in agricultural systems that intend to make use of beneficial bacteria to improve crop production. It is well known for a long time that soils represent one of the most complex and difficult environments to study and at the same time are of particular interest as they are considered to harbor the most diverse populations of bacteria of any environment on earth [2]. There is evidence that there might be several thousands of microbial species in one soil sample and most (99.5–99.9 %) of the soil bacteria observed by microscopy cannot be isolated and cultured in laboratory media. Isolated bacteria may therefore account for only a minor proportion of the total bacterial diversity in soil [3]. Similar difficulty is faced regarding cultivable bacteria present in the rhizosphere or inside plant tissues, which are thought to represent only a small proportion (0.1–10 %) of the total bacteria in these environments [4].
The application of molecular genetic detection and identification methods greatly aids in clarifying the phylogenetic relationships of the microbial community of rhizosphere-associated bacteria. It is generally accepted that only a combination of methods, including classical cultivation techniques and cultivation-independent techniques, allow a comprehensive insight into the bacterial community in environmental habitats [5, 6]. The ribosomal RNA genes of bacteria, especially those for 16S- and 23S-rRNA, are excellent molecular markers for phylogenetic studies due to their functional constancy, their ubiquitous distribution, and elements rising from highly conserved to highly variable regions within the sequence [7–9]. This molecular phylogenetic approach can be used to identify pure isolates and to assess the diversity of complex communities.
Although many of the recent publications have shown the use of next-generation sequencing technologies, using 16S rRNA amplified regions, as a way to get insights into the diversity of certain bacterial communities such as deep sea [10] and human gut [11, 12], not much work has been done on microbial communities in soil or on root-associated bacterial communities. For studying bacterial communities, pyrosequencing of amplified V9 regions of the 16S rRNA genes present in soil DNA or of regions V3 and V6 of the 16S rRNA genes has been successfully demonstrated [12]. However, in both cases, the obtained sequences showed average sizes of 100–200 base pairs (bp), sometimes even shorter (50 bp). With the increased capability of the new generation of sequencing machines to sequence up to 400 bp or more, it is now possible to span most hypervariable regions, multiple adjacent hypervariable regions, or possibly combinations of nonadjacent hypervariable regions through paired-end sequencing strategies [12]. As a consequence, the reads with purpose of molecular ecology studies can be more useful and accurate than the ones generated by classical capillary sequencing [13].
Canola (B. napus L.) is a very important oil-producing plant. The oil produced by this plant, besides its benefits to human health, with a large amount of vitamins and omega 3, is also very important for biodiesel production worldwide [14]. Canola is used as a partner in crop rotation systems with other important crops, such as wheat and soybean [15]. However, studies regarding the diversity or the isolation of the bacterial community that could interact with canola plants are still very preliminary [15–19].
The aim of this work was to estimate the diversity of microorganisms that interact with the roots of canola plants at two different stages of development. By using next-generation sequencing technology, we were able to show that the bacterial community inside the roots changed between these two growth periods, implying in a shift in the dynamics of bacterial populations.
Methods
Sampling and Location
Roots of canola (B. napus L., variety Hyola 60) were collected in an experimental field from Fundação Estadual de Pesquisa Agropecuária (Fepagro/RS), in the city of Vacaria, south of Brazil (28°30′43″ S, 50°56′02″ W). The first sample, roots of canola at the rosette stage, was collected in September 14, 2009; the second sample, roots of canola at the flowering stage, was collected in October 15, 2009. For each stage, five independent plants at least 2 m away from each other were taken randomly and bulked to obtain a representative composite sample. Roots were separated from the rest of the canola plants. This material was used for bulk DNA extraction.
Total Bacterial DNA Extraction
Bacteria that were inside the roots were isolated by surface disinfection performed by washing the roots in running tap water, followed by a 70 % ethanol wash for 1 min, a sodium hypochlorite solution (4 %, v/v) wash for 2 min, and five serial rinses in sterilized distilled water. Immediately after disinfection, the roots were sliced with a sterile scalpel and macerated. A total of 10 g of root segments for each sample were placed in distinct 500-ml Erlenmeyer sterile flasks containing 90 ml of sterile saline solution (0.85 % NaCl). Samples were incubated at 4 °C under agitation (200 rpm) for 3–4 h. DNA from bacterial communities that were inside the roots of canola plants was extracted according to Soares and co-workers [20]. Briefly, bacterial cells were rinsed with TES buffer (10 mM Tris pH 8.0, 25 mM EDTA, and 150 mM NaCl) and suspended in TE buffer (10 mM Tris pH 8.0, 25 mM EDTA). Cell lyses took place in 20 mg ml−1 lysozyme at 37 °C and 4 % sodium dodecyl sulfate. Extractions with phenol/chloroform and precipitation in ethanol were performed. DNA quality and integrity was checked by electrophoresis on 0.8 % agarose ethidium bromide gel. DNA was quantified by spectrophotometer.
Amplification of the Metagenomic DNA with Primers for the V6 to V8 Region of the 16S rRNA Gene
Amplification was performed by using FastStart High Fidelity PCR system (Roche®). The amount of DNA template was 50 ng per reaction. Primers U968 (AACGCGAAGAACCTTAC) and L1401 (CGGTGTGTACAAGACCC) [21] flanking a region of about 500 base pairs between nucleotides 968 and 1401 of the Escherichia coli 16S rRNA gene, including variable regions V6 to V8 [22], were used. Amplification reactions (25 μl) contained 20 μM of each deoxynucleoside triphosphates, 0.3 μM of each primer U968 and L1401, 2 mM MgCl2, and 2 U High Fidelity Taq polymerase (Roche®) in 1X Taq buffer. The amplifications were performed in a PCR Express Temperature Cycling System (Thermo Hybrid) as follows: an initial denaturation step at 94 °C for 5 min followed by 30 cycles at 94 °C for 45 s, 52 °C for 45 s, 72 °C for 45 s, and one cycle at 72 °C for 10 min for final elongation. Whole PCR reaction was purified by gel extraction (Minelute, Qiagen). PCR template for the next reaction was 1 μl of the purified samples and the specific primers were CCATCTCATCCCTGCGTGTCTCCGACTCAG ACGAGTGCGTAACGCGAAGAACCTTAC and CCTATCCCCTGTGTGCCTTGGCAGTCTCAGCGGTGTGTACAAGACCC for the DNA obtained from bacteria that were inside the roots of canola at the rosette stage; and CCATCTCATCCCTGCGTGTCTCCGACTCAG ACGCTCGACAAACGCGAAGAACCTTAC and CCTATCCCCTGTGTGCCTTGGCAGTCTCAGCGGTGTGTACAAGACCC for the DNA obtained from bacteria that were inside the roots of canola at the flowering stage. The primers are constituted of the Roche adaptors (30 bp, forward is in bold and backward is in italic), Roche multiplex identifiers (10 bp, underlined), and the rDNA specific sequences (17 bp). The underlined bases indicated tags that were later used for the identification of the samples. Amplifications conditions were the same as mentioned above. The PCR probes were purified twice by gel extraction (Minelute, Qiagen).
Sequencing Procedures
DNA sequencing was performed at CeBiTec (University of Bielefeld, Germany), according to the manufacturers’ protocols (Roche/454 Life Sciences, Branford, USA). An aliquot of the DNA preparation served as template DNA for sequencing on the GS FLX Titanium platform. The sequencing library was constructed according to the protocol of the GS Titanium General Library Prep Kit (Roche Applied Science). After titration of the library using the GS Titanium SV emPCR Kit (Roche Applied Science), a full sequencing run was carried out on the GS FLX Titanium platform. Metagenomic sequence data have been submitted to European Bioinformatics Institute (EMBL-EBI) databank for SRA archive (study accession number is ERP001267).
Sequencing Data Analysis
Untrimmed raw pyrosequencing reads were consecutively checked for various kinds of quality features according to standardized practice. Filtering of obvious sequencing errors, i.e. (a) reads with ambiguous call bases (N) and (b) incorrect amplification primers, was done using an in-house developed processing pipeline. Inexact matches of the leading and tailing amplification primers to the raw read with up to two mismatches were allowed by this processing step using a fuzzy pattern matching algorithm. Reads not fulfilling these criteria were filtered out; leftover reads were trimmed of the primer sequences.
Operational taxonomic unit (OTU) clustering, species diversity, and species richness estimations were performed using the ESPRIT algorithm [23] with default parameters except for the preprocessing step where only exact matching sequences were considered to form groups of unique sequences.
A rarefaction analysis was employed to assess the coverage of the microbial community by the datasets based on the OTU clustering results. Rarefaction curves were obtained by plotting the sample sizes versus the estimated number of OTUs.
Taxonomic classification of sequencing reads up to the genus level were performed using the RDP naive Bayesian Classifier [24] with an 80 % confidence threshold.
Results
Canola develops from seeding to rosette, flowering, maturity, postharvest fall stubble, and overwintered stubble stages [18, 25], and it is likely that root-associated bacteria facilitate plant growth during initial stages of plant development, providing the host plants with compounds such as phytohormones that play a critical role in plant growth and development [26, 27]. In this study, the bacterial diversity at the rosette and flowering developmental phases of canola (B. napus L.) plants was assessed through the application of new generation sequencing technology. Total bacterial DNA was extracted from root samples of canola grown in Vacaria, south of Brazil. Samples were identified as the following: R1 consisted of DNA extracted from bacteria that were inside the roots of canola plants at the rosette stage and R2 consisted of DNA extracted from bacteria that were inside the roots of canola plants at the flowering stage. These DNA samples were used in PCR reactions to amplify a specific fragment corresponding to the V6 to V8 region of the 16S rRNA gene using primers with molecular IDs as described in the “Methods” section. Sequencing of the amplicons yielded 64,675 reads for sample R1 and 16,699 reads for sample R2 after the initial filtering step.
Rarefaction curves (Fig. 1), which plot the number of observed OTUs (cluster count) versus the fraction of sample observed to assess coverage, were done for each sample, and all plots approached an asymptote.
Rarefaction analysis of the samples. The curves represent the number of observed OTUs (cluster count) plotted against the fraction of sample. R1, DNA extracted from bacteria that were inside the roots of canola plants at the rosette stage; R2, DNA extracted from bacteria inside the roots of canola plants at flowering stage
In the sample R1, microorganisms that could be identified belonged to several classes, in which Gammaproteobacteria, Flavobacteria, Sphingobacteria, Bacilli, Betaproteobacteria, Actinobacteria, and Alphaproteobacteria were the predominant classes, respectively. The most representative phylum in this sample was Proteobacteria, followed by Bacteroidetes, Firmicutes, and Actinobacteria.
Pseudomonadaceae was the most abundant family in sample R1, followed by Flavobacteriaceae and Enterobacteriaceae families. Xanthomonadaceae, Sphingobacteriaceae, and Paenibacillaceae families were identified as well although less frequently. Other families, like Oxalobacteraceae, Comamonadaceae, Rhizobiaceae, Sanguibacteraceae, and Alcaligenaceae appeared with fewer representatives. Figure 2 shows a phylogenetic tree for the R1 sample up to the family level. About 50 % (51.70) of the sequences of the R1 sample were characterized as belonging to the genus Pseudomonas. Other bacterial genera, such as Chryseobacterium, Sphingobacterium, Erwinia, Saccharibacillus, Stenotrophomonas, Azomonas, Serratia, Riemerella, and Xanthomonas, were also abundant in this sample. In total, 35 bacterial genera were identified in sample R1 (see also Supplementary Material Table S1).
Taxonomic tree of sample R1 showing the most abundant lineages up to the family level. Line thickness indicates the relative abundance of a lineage; the numbers associated with each node give the total number of classified sequences assigned to this taxon; numbers in parentheses, the number of sequences that could not be assigned on a more specific level
Proteobacteria was also the most representative phylum in sample R2, with almost 70 % (69.21) of the sequences that were characterized belonging to it. The predominating group in this sample was also Gammaproteobacteria, followed by Flavobacteria, Alphaproteobacteria, Betaproteobacteria, Sphingobacteria, Actinobacteria (class), Bacilli, Verrucomicrobiae, and Chlamydiae classes. Sequences belonging to Bacteroidetes, Actinobacteria, Firmicutes, Acidobacteria, Verrucomicrobia, and Chlamydiae phyla were also present.
The most abundant family in sample R2 was Xanthomonadaceae, followed by Flavobacteriaceae, Pseudomonadaceae, Rhizobiaceae, Sphingobacteriaceae, Comamonadaceae, and Enterobacteriaceae. Other families were also identified but with fewer representatives. Figure 3 shows a phylogenetic tree for the R2 sample until the family level. In sample R2, 8.53 % of the identified sequences could be assigned to members of the genus Xanthomonas, followed by sequences of members of Rhizobium, Pseudomonas, Flavobacterium, Stenotrophomonas, Chryseobacterium, Pedobacter, Variovorax, Epilithonimonas, Sanguibacter, and Pigmentiphaga genera. In total, 57 bacterial genera were identified in sample R2 (see also Supplementary Material Table S1).
Taxonomic tree of sample R2 showing the most abundant lineages up to the family level. Line thickness indicates the relative abundance of a lineage; the numbers associated with each node give the total number of classified sequences assigned to this taxon; numbers in parentheses, the number of sequences that could not be assigned on a more specific level
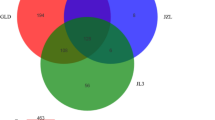
From rosette development stage to flowering stage, we could observe that the bacterial community changed in its composition and abundance. The diversity appeared to increase from rosette to flowering stages. In sample R1, there was a clear predominance of Pseudomonadaceae and Enterobacteriaceae families, whereas in sample R2, Xanthomonadaceae and Flavobacteriaceae families surpass Pseudomonadaceae family, although it was still present. On the other hand, members of the Rhizobacteriaceae family were detected only in the flowering development stage sample (R2). Both of the samples show the presence of members of Sphingobacteriaceae family. Figure 4 summarizes the results concerning to the most abundant families identified for samples R1 and R2.
Discussion
The occurrence and distribution of microbial communities in the soil and rhizosphere is influenced by many factors like root morphology, the stage of plant growth, root exudates, and the physical and chemical properties of the soil [28]. Whereas for several economically important crops the knowledge concerning the diversity of bacterial communities that associate with those plants is growing fast [28, 29], little information is still available for canola plants. In this work, we made use of the 454 deep sequencing technology to unravel the microbial community associated to two developmental stages of canola (B. napus L. v. Hyola 60). This strategy was chosen since it is the most suitable for ecological diversity studies [2]. Roesch and co-workers [2] performed one of the first works applying the deep sequencing methodology to describe microbial diversity in soil samples. In this work, the authors sequenced the V9 region of the 16S rRNA gene amplified by PCR, and they were able to characterize a great amount of bacteria that were not previously accounted for. This work was also important to increase the estimation of microbial diversity in the soil, showing that this environment is of great importance for microbial ecology. In the present work, the V6 to V8 region of the 16S rRNA gene was chosen for the bacterial identification of the bacterial communities present inside canola roots. This region has also proved to be very useful in the identification of bacteria [21] in most cases until the genus level as could be observed in this work and several others [10–12]. In the future, as the size of the reads increased rapidly, next-generation sequencing technology should result in larger reads. This advance will enable researchers to easily identify bacteria until the species level, making the deep sequencing technology even more suitable for ecological diversity studies.
One of the purposes of our research was to investigate whether the stage of development of the plant plays a role in the diversity of the microbial community associated to it. To achieve this goal, two different plant developmental stages, rosette (R1) and flowering (R2) [18], were used for DNA extraction of the bacterial community present inside canola roots. These stages of canola plants were chosen since they are critical for the increase of the leaf area and for grain formation, respectively, which are two important periods for canola development and when the bacterial communities that are interacting with its roots must be well established [30, 31]. The results obtained after the deep sequencing analysis showed that the bacterial communities inside canola roots indeed changed with the growing stage of the plants. There was a difference in the abundance of the genera, family, and even the phyla identified for each sample. In both root samples, Proteobacteria was the most common phylum, but sample R1 presented a lower number of representative phyla, four in total, whereas sample R2 presented seven. In this sample (at rosette stage), the most common bacteria belonged to the genera Pseudomonas, and the family Pseudomonadaceae was the most representative family (Figs. 2 and 4). In the flowering stage (R2), the genus Xanthomonas was the most common, with the Xanthomonadaceae family the most representative family in this sample (Figs. 3 and 4). Also, the number of bacterial genera identified between the rosette stage (R1 = 35) and the flowering stage (R2 = 57) increased. Although root exudation from canola plants has not been assessed, it is likely that the exudation pattern of canola roots changes as plants develop, altering rhizosphere microbial community composition, as reported for maize rhizosphere [32].
Germida and co-workers [15] suggested that plants have a major role in determining the composition of the rhizoplane and endo-rhizosphere bacterial communities among root-associated bacteria. Several studies on many plant species in different locations, using both culturing and nonculturing (molecular) methods, have also indicated that plant type, genotype, root zone, plant age, and plant community composition are, indeed, more important factors influencing the diversity of microbial communities than, for example, soil abiotic parameters [15, 33–37]. Farina et al. [19] isolated cultivable bacteria associated with soil, rhizosphere, and the roots of canola based on their growth on three selective semi-solid media without nitrogen. Although no dominant group of bacteria was identified in their work, strains belonging to Agrobacterium, Burkholderia, Enterobacter, and Pseudomonas genera were the most abundant in all the sampling sites analyzed and, in general, bacteria belonging to the Enterobacteriaceae family were the predominant canola root-associated bacteria. This study also showed that the microbial community structure was influenced by seasonal variation, and in canola rosette stage samples, the diversity of the bacteria associated with the rhizospheric soil was higher than those associated with the roots of canola. Nevertheless, the results obtained in the present work are in agreement with those of Smalla and co-workers [38] that compared the relative abundance of 16S rDNA targets and found that enrichment of bacterial populations associated with canola was most pronounced when canola was at the flowering stage. Interestingly, both the families (Pseudomonadaceae and Xanthomonadaceae) and genera (Pseudomonas and Xanthomonas) identified here were found in both developmental stages but at different proportions. This result reinforces the occurrence of a switch in the predominant bacteria in the different developmental stages of the plant, clearly demonstrating that the plant itself interferes with the associated microbial community.
Finally, our results also showed that the percentage of bacteria belonging to the group of bacteria known as plant growth-promoting rhizobacteria is high inside the plant’s root samples (R1, 52 %; R2, 59 %). Rhizobacteria are better adapted to colonization of roots than bacteria from non-rhizospheric soil [39, 40]. Gram-negative bacteria, especially species of Pseudomonas genus, are by far the most common rhizobacteria [41], and they have been the most extensive group of bacteria interacting with plants studied, as they are readily isolated from plant tissues, easily handled, and amenable to genetic approaches. The commonly used culture-dependent isolation methods mainly detect Pseudomonas and other gram-negative genera [42]. Marilley and Aragno [43] found that the rhizosphere, which is a relatively nutrient-rich niche for bacteria, has a positive selection for the Proteobacteria and reduced the percentage of the Acidobacterium division.
References
Lynch JM (1990) Longevity of bacteria—considerations in environmental release. Curr Microbiol 20:387–389
Roesch LF, Fulthorpe RR, Riva A, Casella G, Hadwin AKM et al (2007) Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J 1:283–290
Torsvik V, Goksoyr J, Daae FL (1990) High diversity in DNA of soil bacteria. Appl Environ Microbiol 56:782–787
Duineveld BM, Rosado AS, van Elsas JD, van Veen JA (1998) Analysis of the dynamics of bacterial communities in the rhizosphere of the chrysanthemum via denaturing gradient gel electrophoresis and substrate utilization patterns. Appl Environ Microbiol 64:4950–4957
Hartmann A, Assmus B, Kirchhof G, Schloter M (1997) Direct approaches for studying soil microbes. In: van Elsas JD, Trevors JT, Wellington EMH (eds) Modern soil microbiology. Dekker, New York
Liesack W, Janssen PH, Rainey FA, Ward-Rainey NL, Stackebrandt E (1997) Microbial diversity in soil: the need for a combined approach using molecular and cultivation techniques. In: van Elsas JD, Trevors JT, Wellington EMH (eds) Modern soil microbiology. Dekker, New York
Amann RI, Ludwig W, Schleifer KH (1995) Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev 59:143–169
Ludwig W, Strunk O, Klugbauer S, Klugbauer N, Weizenegger M et al (1998) Bacterial phylogeny based on comparative sequence analysis. Electrophoresis 19:554–568
Rappe MS, Giovannoni SJ (2003) The uncultured microbial majority. Annu Rev Microbiol 57:369–394
Sogin ML, Morrison HG, Huber JA, Mark Welch D, Huse SM et al (2006) Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc Natl Acad Sci U S A 103:12115–12120
Dethlefsen L, Huse S, Sogin ML, Relman DA (2008) The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol 6:e280
Huse SM, Dethlefsen L, Huber JA, Welch DM, Relman DA, et al (2008) Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. Plos Genetics 4
Huse SM, Huber JA, Morrison HG, Sogin ML, Mark Welch D (2007) Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biology 8
Tomm GO (2000) Situação atual e perspectivas da canola no Brasil. Comunicado Técnico On-line, 58. Passo Fundo: Embrapa Trigo
Germida JJ, Siciliano SD, de Freitas JR, Seib AM (1998) Diversity of root-associated bacteria associated with held-grown canola (Brassica napus L.) and wheat (Triticum aestivum L.). FEMS Microbiol Ecol 26:43–50
Kloepper JW, Lifshitz R, Schroth MN (1988) Pseudomonas inoculants to benefit plant-production. Isi Atl Sci-Anim Pl Sci 1:60–64
Siciliano SD, Goldie H, Germida JJ (1998) Enzymatic activity in root exudates of Dahurian wild rye (Elymus dauricus) that degrades 2-chlorobenzoic acid. J Agric Food Chem 46:5–7
Dunfield KE, Germida JJ (2003) Seasonal changes in the rhizosphere microbial communities associated with field-grown genetically modified canola (Brassica napus). Appl Environ Microbiol 69:7310–7318
Farina R, Beneduzi A, Ambrosini A, de Campos SB, Lisboa BB et al (2012) Diversity of plant growth-promoting rhizobacteria communities associated with the stages of canola growth. Appl Soil Ecol 55:44–52
Soares RA, Roesch LFW, Zanatta G, Camargo FAO, Passaglia LMP (2006) Occurrence and distribution of nitrogen fixing bacterial community associated with oat (Avena sativa) assessed by molecular and microbiological techniques. Appl Soil Ecol 33:14
Felske A, Engelen B, Nubel U, Backhaus H (1996) Direct ribosome isolation from soil to extract bacterial rRNA for community analysis. Appl Environ Microbiol 62:4162–4167
Brosius J, Palmer ML, Kennedy PJ, Noller HF (1978) Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc Natl Acad Sci U S A 75:4801–4805
Sun Y, Cai Y, Liu L, Yu F, Farrell ML et al (2009) ESPRIT: estimating species richness using large collections of 16S rRNA pyrosequences. Nucleic Acids Res 37:e76
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267
Lancashire PD, Bleiholder H, Vandenboom T, Langeluddeke P, Stauss R et al (1991) A uniform decimal code for growth-stages of crops and weeds. Ann Appl Biol 119:561–601
Costacurta A, Vanderleyden J (1995) Synthesis of phytohormones by plant-associated bacteria. Crit Rev Microbiol 21:1–18
Khalid A, Arshad M, Zahir ZA (2004) Screening plant growth-promoting rhizobacteria for improving growth and yield of wheat. J Appl Microbiol 96:473–480
Beneduzi A, Peres D, da Costa PB, Bodanese Zanettini MH, Passaglia LM (2008) Genetic and phenotypic diversity of plant-growth-promoting bacilli isolated from wheat fields in southern Brazil. Res Microbiol 159:244–250
Beneduzi A, Peres D, Vargas LK, Bodanese-Zanettini MH, Passaglia LMP (2008) Evaluation of genetic diversity and plant growth promoting activities of nitrogen-fixing bacilli isolated from rice fields in South Brazil. Appl Soil Ecol 39:311–320
McClinchey SL, Kott LS (2008) Production of mutants with high cold tolerance in spring canola (Brassica napus). Euphytica 162:51–67
Chavarria G, Tomm GO, Muller A, Mendonca HF, Mello N et al (2011) Leaf area index of canola under varying row spacing and plant density of sowing. Ciencia Rural 41:2084–2089
Di Cello F, Bevivino A, Chiarini L, Fani R, Paffetti D et al (1997) Biodiversity of a Burkholderia cepacia population isolated from the maize rhizosphere at different plant growth stages. Appl Environ Microbiol 63:4485–4493
Grayston SJ, Wang SQ, Campbell CD, Edwards AC (1998) Selective influence of plant species on microbial diversity in the rhizosphere. Soil Biol Biochem 30:369–378
Kaiser O, Puhler A, Selbitschka W (2001) Phylogenetic analysis of microbial diversity in the rhizoplane of oilseed rape (Brassica napus cv. Westar) employing cultivation-dependent and cultivation-independent approaches. Microb Ecol 42:136–149
Marschner P, Yang CH, Lieberei R, Crowley DE (2001) Soil and plant specific effects on bacterial community composition in the rhizosphere. Soil Biol Biochem 33:1437–1445
Kowalchuk GA, Buma DS, de Boer W, Klinkhamer PG, van Veen JA (2002) Effects of above-ground plant species composition and diversity on the diversity of soil-borne microorganisms. Antonie Van Leeuwenhoek 81:509–520
Mitchell RJ, Hester AJ, Campbell CD, Chapman SJ, Cameron CM et al (2010) Is vegetation composition or soil chemistry the best predictor of the soil microbial community? Plant Soil 333:417–430
Smalla K, Wieland G, Buchner A, Zock A, Parzy J et al (2001) Bulk and rhizosphere soil bacterial communities studied by denaturing gradient gel electrophoresis: plant-dependent enrichment and seasonal shifts revealed. Appl Environ Microbiol 67:4742–4751
Miller HJ, Henken G, Vanveen JA (1989) Variation and composition of bacterial-populations in the rhizospheres of maize, wheat, and grass cultivars. Can J Microbiol 35:656–660
Hozore E, Alexander M (1991) Bacterial characteristics important to rhizosphere competence. Soil Biol Biochem 23:717–723
Kloepper JW (1993) Plant growth-promoting rhizobacteria as biological control agents. Marcel Dekker, New York
Francis I, Holsters M, Vereecke D (2010) The Gram-positive side of plant-microbe interactions. Environ Microbiol 12:1–12
Marilley L, Aragno M (1999) Phylogenetic diversity of bacterial communities differing in degree of proximity of Lolium perenne and Trifolium repens roots. Appl Soil Ecol 13:127–136
Acknowledgments
We thank R. Luís Mayer Weber for his valuable scientific comments. The work from Brazilian side was financed by a grant and fellowships from the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior, and Instituto Nacional de Ciência e Tecnologia da Fixação Biológica do Nitrogênio. Work in the lab of VFW was supported in part by grant BRA 09/008 from the IB-BMBF Germany.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Table S1
Sequencing data for samples R1 and R2 (PDF 61 kb)
Rights and permissions
About this article
Cite this article
de Campos, S.B., Youn, JW., Farina, R. et al. Changes in Root Bacterial Communities Associated to Two Different Development Stages of Canola (Brassica napus L. var oleifera) Evaluated through Next-Generation Sequencing Technology. Microb Ecol 65, 593–601 (2013). https://doi.org/10.1007/s00248-012-0132-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-012-0132-9