Abstract
Mine wastes have been considered as a source of heavy metal (HM) contamination in the environment and negatively impact many important ecosystem services provided by soils. Plants like Miscanthus, which tolerate high HM concentrations in soil, are often used for phytoremediation and provide the possibility to use these soils at least for the production of energy crops. However, it is not clear if plant growth at these sites is limited by the availability of nutrients, mainly nitrogen, as microbes in soil might be affected by the contaminant. Therefore, in this study, we investigated in a greenhouse experiment the response of ammonia-oxidizing microbes in the root–rhizosphere complex of Miscanthus × giganteus grown in soils with different levels of long-term arsenic (As) and lead (Pb) contamination. Quantitative PCR of the ammonia monooxigenease gene (amoA) was performed to assess the abundance of ammonia-oxidizing bacteria (AOB) and archaea (AOA) at two different points of plant growth. Furthermore, bulk soil samples before planting were analyzed. In addition, terminal restriction fragment length polymorphism (T-RFLP) analysis was used to investigate the diversity of archaeal amoA amplicons. Whereas high concentrations of As and Pb in soil (83 and 15 g/kg, respectively) resulted independent from plant growth in a clear reduction of AOA and AOB compared to the control soils with lower HM contents, in soils with contamination levels of 10 g/kg As and 0.2 g/kg Pb, only AOB were negatively affected in bulk soil samples. Diversity analysis of archaeal amoA genes revealed clear differences in T-RFLP patterns in response to the degree of HM contamination. Therefore, our results could clearly prove the different response patterns of AOA and AOB in HM-contaminated soils and the development of archaeal amoA phylotypes which are more tolerant towards HMs in soil samples from the areas that were impacted the most by mining waste, which could contribute to functional redundancy of ammonia-oxidizing microbes in soils and stability of nitrification pattern.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The mining industry produces large amounts of waste contaminated by metals and metalloids [1], leading to the accumulation of toxic elements in the environment. Whereas heavy metal (HM)-contaminated soils are unsuitable for food production, energy crops (e.g., Miscanthus × giganteus, a perennial grass with large annual biomass production potential) can allow the commercial exploitation of these soils by establishing biofuel feedstock production systems. In addition, the cultivation of these plants offers opportunities for site stabilization and phytoremediation of contaminated soils [2, 3].
However, macronutrients deficiency is one of the growth-limiting factors of plant cultures on contaminated soils [4]. It is therefore of key importance to investigate the response of the functional microbial biomass involved in nutrient turnover to contamination. In this respect, nitrogen (N) cycling is of major concern as contaminated sites are usually not fertilized, and N available to plants is closely linked to N mineralization from dead biomass and subsequent transformation. Several authors have postulated that nitrification responds at a very sensitive level to different types of contamination [5, 6] due to low functional diversity compared to other steps in the N cycle [7]. Nitrification is a two-step process consisting of: (1) oxidation of NH +4 to NO −2 by ammonia-oxidizing bacteria (AOB) and ammonia-oxidizing archaea (AOA) [8, 9] and (2) oxidation of NO −2 to NO −3 by nitrite-oxidizing bacteria (NOB), the first step being assumed as rate limiting [10]. While HM contamination can affect AOB communities [11–13], little is known about how AOA is affected by contamination. Although AOA are thought to be more tolerant to chronic stress conditions than bacteria [14, 15], the influence of HMs on AOA is discussed controversially in literature [16–18]. Furthermore, the few studies assessing the response of both AOA and AOB to HM contaminations were mostly performed using spiked soils [12, 16–21], which do not allow a prediction of the AOA and AOB dynamics in soils with a long history of HM contamination due to adaptation processes of the microflora on the one hand and changes in the amount of bioavailable HMs on the other hand [22, 23].
Therefore, in this study, we investigated in a greenhouse experiment the response of ammonia-oxidizing microbes in the root–rhizosphere complex (RRC) of Miscanthus × giganteus grown in soils with different levels of long-term combined arsenic (As) and lead (Pb) contamination. We analyzed the abundance of both archaeal and bacterial amoA genes in the bulk soils before planting and 6 and 12 weeks, respectively, after planting of the bioenergy crop. Furthermore, we studied the diversity of the amoA genes of AOA. We postulated that AOA will be less affected by HM contamination in soil than AOB due to a shift in the AOA diversity towards more HM-resistant phylotypes over time.
Materials and Methods
Experimental Design
Soils classified as Anthroposol Artificiel according to the French classification or Technosol (WRB nomenclature, FAO 2006) from the former gold mining area at La Petite Faye (Limousin, France, 01°34′23″ E, 46°08′37″ N) were chosen for the experiments. This site, which has been abandoned since 1964, presents zones with different As and Pb concentrations and has been colonized by local vegetation (e.g., grasses, ferns, horsetails, birches). Three plots were selected for soil sampling, reflecting zones with severe (S), medium (M), and low (L) levels of HM contamination based on total Pb and As values. The values for Pb ranged from 15,200 mg/kg at plot S to values smaller than 500 mg/kg at plots M and L. Arsenic values ranging from 83,000 mg/kg at plot S to 1,700 mg/kg at plot L were measured. Soils were sampled in January 2010 up to 20 cm soil depth after removing the organic horizon. Sieved soil (2 mm) was transferred into plastic bags and placed into pots (21 × 21 × 21 cm) at a bulk density of 1.0 and equilibrated for 1 month at 20 °C and constant water content. Major chemical and physical soil parameters are summarized in Table 1.
After an equilibration phase of 7 days at 50 to 60 % of field water capacity at 20 °C, one pre-grown Miscanthus × giganteus rhizome (Novabiom, France) was introduced per pot. Miscanthus × giganteus plants were grown in a phytotron for 3 months, without receiving additional nutrients. The plants were subjected to a photoperiod of 16 h light at 350 μmol m−2 s−1 and 8 h of darkness at temperatures of 23 and 18 °C, respectively, under constant soil water content (field capacity). Samplings were performed before planting the rhizome (t 0) and after 6 (t 1) and 12 (t 2) weeks. The experiment was conducted in a randomized block design with four independent replicates per soil and per sampling time point. Bulk soil (at t 0) and composite samples of roots with attached soil (at t 1 and t 2) were sampled from each pot. After shaking the roots vigorously, the roots and attached soil were treated as one compartment called the RRC. One part of the samples was immediately shock-frozen in liquid nitrogen and stored at −80 °C for nucleic acid extraction; the other part was directly extracted with 0.01 M CaCl2 for determination of water-extractable As and Pb, organic carbon (WEOC), ammonium–N (NH +4 –N), and nitrate–N (NO −3 –N). Bulk soil samples were directly treated by lithium metaborate/tetraborate fusion and nitric acid digestion for HM determination.
Soil Parameters
Samples consisting of bulk soil or RRC were shaken overhead for 45 min in 0.01 M CaCl2 for determination of water-extractable As and Pb (soil/solution ratio 1:10), WEOC, NH +4 –N and NO −3 –N (soil/solution ratio 1:4). After filtration, Pb and As were measured by graphite furnace atomic absorption spectrometry (SpectrAA 880 Z, Varian, CA, US) equipped with a Zeeman background correction. NH +4 –N and NO −3 –N were determined using a continuous-flow analyzer (SA 20/40, Skalar Analytical, The Netherlands). WEOC was measured using a TOC analyzer (TOC-5050A, Shimadzu Corporation, Japan).
Plant Parameters
The dry weight of rhizome, stem, and leaves of Miscanthus plants, respectively, were measured 12 weeks after planting (t 2). As and Pb contents were determined in each organ at t 2 by ICP-MS after nitric acid digestion (Acme Analytical Laboratories Ltd., Canada).
DNA Extraction
DNA of each of 12 samples (four replicates × three time points) was extracted from 0.5 g of bulk soil and RRC, respectively, after a bead beater lysis step (Bertin Technologie, France) using the FastDNA SPIN kit for soil (MP biomedicals, Germany) according to the manufacturer’s instructions. DNA concentration was measured by using a microvolume spectrophotometer (NanoDrop, PeqLab, Germany).
Quantitative PCR Assay
Quantitative PCR (qPCR) was used to determine the abundance of functional communities involved in ammonium oxidation by targeting amoA genes (encoding the ammonia monooxygenase) in both AOB and AOA using a SYBR® GreenI-based detection system (Applied Biosystems, Germany). Absolute quantification of investigated target genes was carried out in 25-μL samples in triplicate on the ABI Prism 7300 Cycler (Applied Biosystems). The reaction mixture contained 15 μg bovine serum albumin (Sigma-Aldrich, Germany), 0.2 μM of each primer for amoA AOA, and 0.3 μM of each primer for amoA AOB amplification, respectively (Metabion, Germany), 1X Power SYBR Green PCR master mix (Applied Biosystems), and 40 ng DNA template. PCR conditions and primers used are shown in Table 2. The specificity of the amplification products was confirmed by melting curve analysis and agarose gel electrophoresis. No template controls served as null value. Samples were diluted tenfold as no inhibitory effects on the PCR amplification were detected when known amounts of standard (AOB and AOA) were spiked with tenfold diluted environmental DNA samples (data not shown). Dilution series of plasmids containing cloned DNA of the amoA gene from Nitrosomonas multiformis ATCC25196 for AOB and of the fosmid clone 54d9 [9] for AOA, respectively, were used to generate standard curves ranging from 101 to 106 gene copies per microliter. The calculated efficiencies for qPCRs of AOA and AOB were comprised between 90 and 95 %.
Terminal Restriction Fragment Length Polymorphism Fingerprinting
Diversity analysis of archaeal amoA gene was carried out by terminal restriction fragment length polymorphism (T-RFLP). Archaeal amoA gene amplicons were generated by two successive PCRs using the primers described for qPCR assay, with forward primer labeled with 5′-carboxyfluorescein. The first PCR reaction (50 μL) contained ~100 ng of template DNA, 0.2 μM of each primer, 0.2 mM dNTPs (Fermentas, Germany), 60 μg of BSA (Sigma-Aldrich, Germany), 1 U Top Taq and 1 × PCR buffer (Qiagen, Germany), and nuclease-free water (Promega, Germany). The PCR reaction was achieved according to the following thermal profile: 5 min at 95 °C, followed by 30 cycles of 45 s at 94 °C, 45 s at 55 °C, 45 s at 72 °C, and finally 5 min at 72 °C. Four microliters of PCR products was amplified subsequently in the same conditions as previously described in a final volume of 100 μL. Generated amplicons were checked by standard agarose gel electrophoresis and ethidium bromide staining and purified with the QIAquick PCR purification kit (Qiagen) prior enzymatic digestion with the restriction enzyme MwoI (Fermentas, Germany) according to the manufacturer’s protocol. The restriction enzyme was selected based on in silico T-RFLPs using the program REPK (Restriction Endonuclease Picker) [24]. Digested amplicons (~50 ng in 10 μL) were subsequently purified with the MinElute Reaction cleanup kit (Qiagen). Desalted digests (1 μL) were mixed with 13 μL of Hi-Di formamide (Applied Biosystems) containing an 800-fold dilution of a 6-carboxy-X-rhodamine-labeled MapMarker 1000 ladder (Bio-Ventures, TN, US), denatured (3 min at 95 °C), and cooled on ice. Electrophoresis was performed as described previously [25] using an ABI 3730 DNA analyzer (Applied Biosystems). Electropherogram evaluation was performed using the GeneMapper 5.1 software (Applied Biosystems).
Statistical Analysis
Prior to analysis, gene abundance data were ln-transformed to achieve normal distribution. Data were analyzed by one-way ANOVA with soil (L, M, S) as factor at a given time point for both bacterial and archaeal amoA genes with significance level set to α = 0.05. Soil parameters data were analyzed similarly. Statistical tests were calculated in SPSS 11.5 (SPSS, Inc., IL, USA). T-RFLP data were imported into T-REX [26] and a data matrix based on peak area was generated. Peaks were inactivated for lack of length (<50 bp) and aligned using clustering threshold of 1 bp. Fragments with relative abundance of less than 1 % were considered as background noise. The data matrix was exported for analysis with the ADE4 package [27] within the R software environment (www.R-project.org) using between-group analysis (BGA) based on correspondence analysis using the function dudi.coa followed by bca. Because correspondence analysis is only the first step in the between-group analysis, no detrended form with downweighting of variables was used. The between-groups inertia percentage was used for a global test of any difference between the groups. Based on 999 permutations, a P value was calculated. In case of significant results (P < 0.05), pairwise tests were performed; the P values were adjusted for multiple comparisons by the method of Hommel [28].
Results
Physical and Chemical Characterization of Bulk Soil, RRC, and Plants
The CaCl2-extractable fraction of As and Pb in all soil samples was lower than 1 % of the total amount of the respective HM and ranged from 10.4 to 2.1 mg/kg for As and from 134.2 to 1.6 mg/kg for Pb, respectively, at t 0. Surprisingly, the clear gradient in total HM in soil samples from S, M, and L was not reflected in the amount of CaCl2-extractable As and Pb. Over the experimental period, the values for the As CaCl2-extractable fraction did not change significantly in all soil samples; for Pb in soil samples from plot S and L, a clear reduction was visible (Table 3).
Initial NO −3 –N and NH +4 –N concentrations in bulk soil were independent of the degree of metal and metalloid contamination (Table 3). Higher NO −3 –N and NH +4 –N concentrations were found in all samples at t 0. At t 1, significantly higher NO −3 –N concentrations and lower NH +4 –N concentrations were measured in RRC samples from soil L compared to soil S. At t 2, however, no differences in NO −3 –N and NH +4 –N concentrations were apparent. The amount of WEOC was not influenced by the degree of HM contamination and was, as expected, higher in the RRC than in bulk soil. Between t 1 and t 2, no changes in WEOC amount were observed.
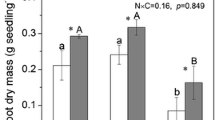
Biomass of plant leaves, steam, and rhizomes was not affected by the different amounts of HM present in soil (Table 4) at the last sampling time point t 2. All plants accumulated As over time. At t 2, highest As concentrations were measured in the rhizomes independent from the degree of contamination in soil. Surprisingly, the highest accumulation of As was found in plants grown in soil M with total As concentrations of 282.7 mg/kg. For soil S and L, the accumulation of As in plants was comparable (56.2–72.0 mg/kg). The accumulation of Pb in the plants was also observed, and a higher accumulation of this element in leaves and stem was observed in soil S compared to As. Greater Pb accumulations were found at t 2 in plants grown in soil S and M (103.2–121.5 mg/kg). Pb contents in plants harvested from soil L were lower than 10 mg/kg.
Quantification of Bacterial and Archaeal Ammonia Oxidizers
In bulk soil at t 0, a clear influence of the HM contamination on the abundance of AOA and AOB was visible. Whereas amoA copy number in the control soil L was, for both AOA and AOB, in the range of 2.9–4.8 × 105 copies/g soil, in the severely impacted soil S only 2.2–5.2 × 104 copies/g were measured. In soil M, AOA was obviously not influenced by HM contamination and values were comparable to soil L; for AOB, a clear influence of contamination was visible and values were similar to soil S (Fig. 1).
Quantification of amoA (AOB and AOA) in bulk soil (t 0) and in root–rhizosphere complex of Miscanthus × giganteus after 6 (t 1) and 12 (t 2) weeks in three acidic soils contaminated with heavy metals (S, M, and L). Significant differences between the three soils at a particular time point are indicated by letters. Error bars represent standard deviation of mean (n = 4). RC RRC root-rhizosphere complex
At t 1 in the RRC of plants grown in soil L, as expected, AOB outcompeted AOA by almost one order of magnitude (2.4 × 105 and 4.0 × 104 amoA copies/g RRC, respectively). In contrast, in the RRC of plants from soil M, AOA copy numbers were higher (5.7 × 105 amoA copies/g) compared to those from soil L, whereas no significant differences in AOB copy numbers between plants from soil L and M were visible. In the RRC from plants obtained from soil S, as expected, amoA copy numbers for AOA and AOB showed similar response pattern and were significantly lower compared to plants from the other soils (9.9 × 103—1.7 × 104 amoA copies/g).
At t 2, similar copy numbers for amoA could be measured. Only in the RRC of plants from soil L were the differences observed at t 1 between AOA and AOB no longer visible and values for both groups of ammonia oxidizers were in the range of 1.5 × 105 amoA copies/g.
Diversity Analysis of Archaeal amoA genes
T-RFLP analysis of the archaeal amoA gene resulted in a detectable T-RF number/sample ranging from two (t 0) to six (t 1) in soil S, from two (t 1, t 2) to three (t 0) in soil M, and from two (t 0) to nine (t 1) in soil L (Fig. 2). T-RF-162 was dominant in bulk soil samples (t 0) for all soils and in the RRC samples (t 1, t 2) for soil S and M, ranging from 66 to 97 % of the total relative community. In the RRC of soil L, T-RFs-162 and -253 had a similar level of relative abundance, contributing for 33 and 30 % of the total amoA AOA community at t 1 and 43 and 51 % at t 2, respectively. The contribution of T-RF-253 to the community richness in soil L was comparable at the different time points. However, T-RF-253 was absent in soil S. An increased number of T-RFs was found at t 1 and, to a lower extent, at t 2 compared to t 0 in soil S and L. Statistical evaluation of overall AOA diversity by BGA revealed a clustering according to the different soils at a given time point, with the exception of soils S and M at t 2 which clustered together (Fig. 3; Tables 5 and 6). BGA also indicated significant differences for all soils between t 0 and t 1, as well as between t 0 and t 2 for soil S and t 1 and t 2 for soil M.
Contributions of T-RFs to total amoA AOA gene fragment diversity in bulk soil (t 0) and in root–rhizosphere complex of Miscanthus × giganteus after 10 (t 1) and 20 (t 2) days in three acidic soils contaminated with heavy metals (S, M, and L). Significant differences between the three soils at a particular time point are indicated by letters
Between-group analysis based on correspondence analysis of the T-RFLP data set for amoA AOA gene fragments. The first two axes explain 61 % of variance. Symbols illustrate the four replicates for each soil (S, M, and L) at each time point (t 0, t 1, and t 2). Ellipses surround the four replicates for each soil, showing that they cluster together
Discussion
The objective of this study was to assess the impact of different degrees of As and Pb contamination in soil and the planting of the energy crop Miscanthus × giganteus on the functional microbial biomass involved in the oxidation of ammonia in soil. Therefore, soils with different long-term As and Pb contamination were used for plant growth. By targeting the marker gene amoA, we quantified ammonia-oxidizing bacteria and archaea in bulk soil as well as in the RRC at two different time points during plant development and described the diversity pattern of archaeal ammonia oxidizers. As shown by Nicol and coworkers [29], there is good evidence that amoA diversity well reflects the phylogeny of ammonia-oxidizing bacteria and archaea. Furthermore, Schauss et al. [30] calculated the maximum number of amoA genes per cell for AOA and AOB and estimated for both groups of ammonia oxidizers a maximum of three operons per cell; thus, amoA has been considered as a good proxy for the total number of ammonia-oxidizing microbes.
Bioavailability of Pb and As in Soils with a Long History of Contamination
In this study, the investigated soils showed different degrees of As and Pb contamination. Remarkably, the total Pb and As contents and the CaCl2-extractable fractions, respectively, did not follow the same trend. This might be related to the pH values of the different soils and the difference in the HM speciation to mineralogy (e.g., respective amount of As- and Pb-bearing phases). Similarly, Cui and colleagues [31] observed an increased availability of As and Pb with higher and lower soil pH, respectively. Total HM concentrations are a poor indicator of the actual concentration in the soil solution to which soil microbes are exposed, and there is no universally acceptable method to assess bioavailable soil metal concentrations [32].
As and Pb as Drivers for Abundance and Diversity of Ammonia-Oxidizing Microbes
In our study, AOA and AOB amoA copy numbers in bulk soil, ranging from 4.4 × 103 to 4.8 × 105, were lower than what has been previously reported [16–19, 21] (Fig. 1). However, the investigated soils in this study were of mining waste origin, presumably hostile for microbial populations, low in organic matter content, and not of agricultural provenance. In soil S, amoA copy numbers for both groups of ammonia oxidizers were significantly reduced compared to soil M and L, which could be interpreted as a negative impact of the high HM concentration in this soil. These results may indicate low in situ turnover rates of ammonia in response to high HM contamination, as observed in our experiment by the increased NH +4 –N and reduced NO −3 –N concentrations at t 1 in soil S compared to soil L. However, potentially lower transformation rates of ammonia into nitrate in soil S might be partly compensated by higher dentrification activities in soil L.
At t 0, while amoA AOA gene abundance was significantly higher in soil M than in soil S, no significant difference between these two soils was observed in amoA AOB abundance, indicating a higher sensitivity of AOB than AOA towards Pb and As. Similarly, a higher tolerance of AOA than AOB in soils contaminated by Zn [17] and Cu [21] has been suggested. However, other studies showed contrasting results and ascribed tolerance development in ammonia-oxidizing communities to AOB rather than AOA populations [16, 18]. Nevertheless, as most of these results are based on soils spiked with HMs, a direct comparison to the data presented in this study is not possible as bioavailability of HMs is different in soils with artificially added HMs, and the time of adaptation of microbes in response to the stressor is missing [22].
Mainly, the latter argument has been proven to be of high importance in our study as different AOA phylotypes showing differences in HM tolerance were observed. For example, T-RF-253 was below the detection limit in soil S and relatively less abundant in soil M than L, possibly indicating the sensitivity of this genotype to high HM contamination (Fig. 2). Vice versa, T-RF-162 was dominating AOA diversity patterns in soil S and M at all time points, indicating the importance of this phylotype in soils highly contaminated by HMs and possibly its increased resistance against HMs.
Although Mertens and colleagues [13] demonstrated that AOB populations from long-term contaminated soil samples were able to tolerate higher Zn concentrations than AOB populations from uncontaminated soil samples and microbial HM, resistance mechanisms for AOB including Nitrosomonas europaea [33, 34] have been described in literature in our study; obviously, only a very low tolerance level towards Pb and As of AOB was observed. This might be related to the contamination with two HM in combination with other stressors which affect AOB, like low pH. In contrast, detailed data on the resistance development towards HM of AOA are still missing as the first cultures of AOA from soil, which may form the basis for such studies, have been isolated only recently [35].
Other Factors Driving the Abundance and Diversity of Ammonia-Oxidizing Microbes
When plots for this study were selected, much care was taken on identifying soils which only differ in their As and Pb concentrations. Therefore, soil parameters like soil texture, total C, and total N content were comparable. However, soils differed in their pH values. Whereas soil S and M had a comparable pH (3.5), soil L was less acidic with a pH of 5.3. Soil pH is known to shape the distribution and activity of archaeal and bacterial ammonia oxidizers [29, 36] and is a driver for selecting different bacterial and archaeal communities [37–40]. Furthermore, nitrification has been suggested to be driven by AOA in highly acidic soils [36, 40]. Therefore, the effects observed in this study may be partly assigned to differences in soil pH.
Although no clear influence of the degree of HM contamination on plant growth was observed, as plant biomass values were comparable from all soils (Table 4), differences were observed in the amount of accumulated As and Pb. Roots are known to shape microbial communities and activity because of the wide variety of organic compounds that they provide [41, 42]. Accumulated HM change overall plant physiology and consequently alter the quantity and quality of root exudates. Taking into account that mainly AOA, as autotrophic microbes, which exhibit a rather oligotrophic lifestyle [43], are often outcompeted by heterotrophic microbes in the rhizosphere due to (1) the increased amounts of available carbon and (2) the competition between plants and microbes for ammonia, resulting in the release of nitrification-inhibiting substances by the plant [44], the change in exudation patterns may explain the shifts in the relative abundance of AOA and AOB, mainly in plants grown in soil M.
Conclusion
Based on the data of this study, it can be postulated that selected phylotypes of AOA tolerate higher concentrations of Pb and As in soil and RRC compared to AOB. However, abundance of a functional group cannot be directly linked to the activity of these microbes. Therefore, it remains unclear if, mainly in soil M, AOA can substitute AOB and if functional redundancy between both groups of ammonia-oxidizing microbes exists under the given conditions. Based on the ammonium levels measured, which are similar between soil L and M, this might be speculated; nitrate concentrations between both soils differ significantly though. Thus, in situ data cannot answer this question in total as it is not clear how nitrite oxidation is influenced by As and Pb. To address these points in detail, further studies are needed, including (1) analysis of mRNA, (2) using 15 N-labeled ammonium, and (3) studying the effects of HM on nitrite oxidation.
References
Alloway BJ (1990) Heavy metals in soils. Blackie, Glasgow
Chaney RL, Malik M, Li YM, Brown SL, Brewer EP, Angle JS, Baker AJM (1997) Phytoremediation of soil metals. Curr Opin Biotechnol 8:279–284
Vangronsveld J, Herzig R, Weyens N, Boulet J, Adriaensen K, Ruttens A, Thewys T, Vassilev A, Meers E, Nehnevajova E, van der Lelie D, Mench M (2009) Phytoremediation of contaminated soils and groundwater: lessons from the field. Environ Sci Pollut Res 16:765–794
Tordoff GM, Baker AJM, Willis AJ (2000) Current approaches to the revegetation and reclamation of metalliferous mine wastes. Chemosphere 41:219–228
Dalzell DJB, Alte S, Aspichueta E, de la Sota A, Etxebarria J, Gutierrez M, Hoffmann CC, Sales D, Obst U, Christofi N (2002) A comparison of five rapid direct toxicity assessment methods to determine toxicity of pollutants to activated sludge. Chemosphere 47:535–545
Broos K, Mertens J, Smolders E (2005) Toxicity of heavy metals in soil assessed with various soil microbial and plant growth assays: as comparative study. Environ Toxicol Chem 24:634–640
Verstraete W, Mertens B (2004) The key role of soil microbes. In: Doelman P, Eijsackers HJP (eds) Vital soil: function, value, and properties, vol 29. Elsevier, Amsterdam, pp 127–157
Kowalchuk GA, Stephen JR (2001) Ammonia-oxidizing bacteria: a model for molecular microbial ecology. Annu Rev Microbiol 55:485–529
Leininger S, Urich T, Schloter M, Schwark L, Qi J, Nicol GW, Prosser JI, Schuster SC, Schleper C (2006) Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442:806–809
Prosser JI (1989) Autotrophic nitrification in bacteria. Adv Microb Physiol 30:125–181
Stephen JR, Chang Y-J, Macnaughton SJ, Kowalchuk GA, Leung KT, Flemming CA, White DC (1999) Effect of toxic metals on indigenous soil beta-subgroup proteobacterium ammonia oxidizer community structure and protection against toxicity by inoculated metal-resistant bacteria. Appl Environ Microbiol 65:95–101
Gremion F, Chatzinotas A, Kaufmann K, Von Sigler W, Harms H (2004) Impacts of heavy metal contamination and phytoremediation on a microbial community during a twelve-month microcosm experiment. FEMS Microbiol Ecol 48:273–283
Mertens J, Springael D, De Troyer I, Cheyns K, Wattiau P, Smolders E (2006) Long-term exposure to elevated zinc concentrations induced structural changes and zinc tolerance of the nitrifying community in soil. Environ Microbiol 8:2170–2178
Schleper C, Jurgens G, Jonuscheit M (2005) Genomic studies of uncultivated archaea. Nat Rev Microbiol 3:479–488
Valentine DL (2007) Adaptations to energy stress dictate the ecology and evolution of the Archaea. Nat Rev Microbiol 5:316–323
Ruyters S, Mertens J, Springael D, Smolders E (2010) Stimulated activity of the soil nitrifying community accelerates community adaptation to Zn stress. Soil Biol Biochem 42:766–772
Xia Y, Zhu Y-G, Gu Q, He J-Z (2007) Does long-term fertilization treatment affect the response of soil ammonia-oxidizing bacterial communities to Zn contamination? Plant Soil 301:245–254
Mertens J, Broos K, Wakelin SA, Kowalchuk GA, Springael D, Smolders E (2009) Bacteria, not archaea, restore nitrification in a zinc-contaminated soil. ISME J 3:916–923
Mertens J, Wakelin SA, Broos K, McLaughlin MJ, Smolders E (2010) Extent of copper tolerance and consequences for functional stability of the ammonia-oxidizing community in long-term copper-contaminated soils. Environ Toxicol Chem 29:27–37
Frey B, Pesaro M, Rüdt A, Widmer F (2008) Resilience of the rhizosphere Pseudomonas and ammonia-oxidizing bacterial populations during phytoextraction of heavy metal polluted soil with poplar. Environ Microbiol 10:1433–1449
Li XF, Zhu YG, Cavagnaro TR, Chen MM, Sun JW, Chen XP, Qiao M (2009) Do ammonia-oxidizing archaea respond to soil Cu contamination similarly as ammonia-oxidizing bacteria? Plant Soil 324:209–217
Diaz-Ravina M, Baath E (1996) Development of metal tolerance in soil bacterial communities exposed to experimentally increased metal levels. Appl Environ Microbiol 62:2970–2977
Giller KE, Witter E, McGrath SP (1998) Toxicity of heavy metals to microorganisms and microbial processes in agricultural soils: a review. Soil Biol Biochem 30:1389–1414
Collins RE, Rocap G (2007) REPK: an analytical web server to select restriction endonucleases for terminal restriction fragment length polymorphism analysis. Nucleic Acids Res 35:W58–W62
Lueders T, Kindler R, Miltner A, Friedrich MW, Kaestner M (2006) Identification of bacterial micropredators distinctively active in a soil microbial food web. Appl Environ Microbiol 72:5342–5348
Culman SW, Bukowski R, Gauch HG, Cadillo-Quiroz H, Buckley DH (2009) T-REX: software for the processing and analysis of T-RFLP data. BMC Bioinforma 10:10
Thioulouse J, Chessel D, Doledec S, Olivier JM (1997) ADE-4: a multivariate analysis and graphical display software. Stat Comput 7:75–83
Hommel G (1988) A stagewise rejective multiple test procedure based on a modified Bonferroni test. Biometrika 75:383–386
Nicol GW, Leininger S, Schleper C, Prosser JI (2008) The influence of soil pH on the diversity, abundance and transcriptional activity of ammonia oxidizing archaea and bacteria. Environ Microbiol 10:2966–2978
Schauss K, Focks A, Leininger S, Kotzerke A, Heuer H, Thiele-Bruhn S, Sharma S, Wilke BM, Matthies M, Smalla K, Munch JC, Amelung W, Kaupenjohann M, Schloter M, Schleper C (2009) Dynamics and functional relevance of ammonia-oxidizing archaea in two agricultural soils. Environ Microbiol 11:446–456
Cui Y, Du X, Weng L, Van Riemsdijk WH (2010) Assessment of in situ immobilization of lead (Pb) and arsenic (As) in contaminated soils with phosphate and iron: solubility and bioaccessibility. Water Air Soil Pollut 213:95–104
Giller KE, Witter E, McGrath SP (2009) Heavy metals and soil microbes. Soil Biol Biochem 41:2031–2037
Haferburg G, Kothe E (2007) Microbes and metals: interactions in the environment. J Basic Microbiol 47:453–467
Park S, Ely RL (2008) Genome-wide transcriptional responses of Nitrosomonas europaea to zinc. Arch Microbiol 189:541–548
Tourna M, Stieglmeier M, Spang A, Konneke M, Schintlmeister A, Urich T, Engel M, Schloter M, Wagner M, Richter A, Schleper C (2011) Nitrososphaera viennensis, an ammonia oxidizing archaeon from soil. Proc Natl Acad Sci U S A 108:8420–8425
Gubry-Rangin C, Nicol GW, Prosser JI (2010) Archaea rather than bacteria control nitrification in two agricultural acidic soils. FEMS Microbiol Ecol 74:566–574
De Boer W, Kowalchuk GA (2001) Nitrification in acid soils: micro-organisms and mechanisms. Soil Biol Biochem 33:853–866
Lehtovirta LE, Prosser JI, Nicol GW (2009) Soil pH regulates the abundance and diversity of Group 1.1c Crenarchaeota. FEMS Microbiol Ecol 70:367–376
Nugroho RA, Roling WFM, Laverman AM, Verhoef HA (2007) Low nitrification rates in acid scots pine forest soils are due to pH-related factors. Microb Ecol 53:89–97
Yao H, Gao Y, Nicol GW, Campbell CD, Prosser JI, Zhang L, Han W, Singh BK (2011) Links between ammonia oxidizer community structure, abundance, and nitrification potential in acidic soils. Appl Environ Microbiol 77:4618–4625
Brimecomb MJ, de Leij FA, Lynch JM (2001) The effect of root exudates on rhizosphere microbial populations. In: Pinton R, Varanini Z, Nannipieri P (eds) The rhizosphere: biochemistry and organic substances at the soil–plant interface. Marcel Dekker, New York, pp 95–140
Marschner P, Yang CH, Lieberei R, Crowley DE (2001) Soil and plant specific effects on bacterial community composition in the rhizosphere. Soil Biol Biochem 33:1437–1445
Martens-Habbena W, Berube PM, Urakawa H, de la Torre JR, Stahl DA (2009) Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature 461:976–U234
Ollivier J, Töwe S, Bannert A, Hai B, Kastl E-M, Meyer A, Su MX, Kleineidam K, Schloter M (2011) Nitrogen turnover in soil and global change. FEMS Microbiol Ecol 78:3–16
Rotthauwe JH, Witzel KP, Liesack W (1997) The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl Environ Microbiol 63:4704–4712
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ollivier, J., Wanat, N., Austruy, A. et al. Abundance and Diversity of Ammonia-Oxidizing Prokaryotes in the Root–Rhizosphere Complex of Miscanthus × giganteus Grown in Heavy Metal-Contaminated Soils. Microb Ecol 64, 1038–1046 (2012). https://doi.org/10.1007/s00248-012-0078-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-012-0078-y