Abstract
Nitrosomonas europaea, a Gram-negative obligate chemolithoautotroph, participates in global nitrogen cycling by carrying out nitrification and derives energy for growth through oxidation of ammonia. In this work, the physiological, proteomic, and transcriptional responses of N. europaea to zinc stress were studied. The nitrite production rate and ammonia-dependent oxygen uptake rate of the cells exposed to 3.4 μM ZnCl2 decreased about 61 and 69% within 30 min, respectively. Two proteins were notably up regulated in zinc treatment and the mRNA levels of their encoding genes started to increase by 1 h after the addition of zinc. A total of 27 genes were up regulated and 30 genes were down regulated. Up-regulated genes included mercury resistance genes (merTPCAD), inorganic ion transport genes, oxidative stress genes, toxin-antitoxin genes, and two-component signal transduction systems genes. merTPCAD was the highest up-regulated operon (46-fold). Down-regulated genes included the RubisCO operon (cbbO), biosynthesis (mrsA), and amino acid transporter.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Nitrosomonas europaea (ATCC19718) is an obligate chemolithoautotroph that obtains energy and reductant for growth from the oxidation of ammonia to nitrite through a two-step process (Hyman and Arp 1995). The two steps are carried out by ammonia monooxygenase (AMO) and hydroxylamine oxidoreductase (HAO), respectively, as follows:
The four electrons derived from hydroxylamine oxidation support both AMO activity and ATP synthesis. N. europaea belongs to the betaproteobacteria and contains a single circular chromosome of 2,812,094 bp with 2,460 predicted genes. Most of the genes in the 285 open reading frames related to active transport in N. europaea encode transporters of inorganic ions (Chain et al. 2003).
Zinc is a vital catalytic component of seven enzymes, such as zinc metallopeptidase and zinc transporters, and of two zinc-based domains (termed “zinc fingers”) in N. europaea. However, industrial developments have elevated zinc concentrations in some environments and zinc is regarded as a permanent contaminant because of its non-degradability (Pomposiello et al. 2001). Excessive zinc in wastewater can accumulate inside cells and may cause zinc cytotoxicity in two ways: (1) by binding with SH groups of proteins involved in electron transport systems and (2) by generating reactive oxygen species (HO• or HOO••) (Nies 1999; Outten and O’Halloran 2001).
Heavy metals can enter into the cytosol of prokaryotes through specific or non-specific transporters (Nies 1999). The specific transporters import the metal ions into the cells under requirement, starvation, or specific metabolic conditions. In contrast, the non-specific transporters bring metals into the cells by a diffusion gradient across the cytoplasmic membrane. In this case, metal ions can be imported even after the required micronutrient concentrations are met, causing toxic effects in the cells (Choudhury and Srivastava 2001; Blencowe and Morby 2003).
Bacteria display several resistance mechanisms against metal toxicity. For example, permeable walls allow metals to diffuse out of the cells and active export systems can remove metal ions via efflux pumps. In addition, metal ions can be physically sequestered by periplasmic proteins, cytoplasmic proteins, or ligands like polyphosphate granules (Gilotra and Srivastava 1997; Choudhury and Srivastava 2001). Although physiological and transcriptional responses of N. europaea under starvation and toxic conditions by chloroform and chloromethane have been determined (Wei et al. 2006a, b; Gvakharia et al. 2007) the responses of the cells under zinc stress have not been studied. In this work, we determined the whole-genome transcriptional change of N. europaea exposed to 3.4 μM ZnCl2 for 1 h using cDNA microarrays and by quantitative reverse transcriptase-PCR (q-PCR).
Materials and methods
Cell growth and experimental design (batch reactor)
Nitrosomonas europaea cells were cultured in medium containing 25 mM (NH4)2SO4 as previously described (Ensign et al. 1993; Stein and Arp 1998) and harvested in mid to late exponential phase. At harvest, 1 l of cells (OD600 ≅ 0.07) was washed twice with 40 mM NaHPO4 (pH 7.8) to remove residual ammonia and resuspended in 1 l of 50 mM HEPES buffer (pH 7.8). The cells (OD600 ≅ 0.07) were equally separated into two gas-tight 1.67 l reactor vessels (Wheaton Double-sidearm Cell stir). HEPES buffer was used to prevent zinc precipitates and complexes that can form in phosphate or carbonate buffers. (NH4)2SO4 (2.5 mM) was injected in the reactor vessels and the cultures were stirred with a magnetic stirring bar rotating at 700 rpm. Results from preliminary experiments (data not shown) were used to establish desired zinc concentrations, a preferred monitoring strategy, and appropriate time points and intervals for the physiological, genomic, and proteomic observations (described below). Preliminary experiments also compared performance of cells in HEPES buffer with their performance in the phosphate/carbonate-buffered N. europaea growth media, showing their performance in HEPES buffer to be indistinguishable from their performance in phosphate/carbonate-buffered growth media over the 4-h observation periods used in these experiments (data not shown).
Oxygen uptake measurement and nitrite assay
Nitrite concentration and oxygen uptake rate were determined every 30 min for 4 h. A 1 ml aliquot of the cells was centrifuged immediately and 10 μl of supernatant was analyzed to determine nitrite concentration spectrophotometrically (Hyman and Arp 1995). Ammonia- and hydrazine-dependent specific O2 uptake activities were measured in a 1.8 ml glass, water-jacketed reaction vessel at 30°C using a heated circulating water bath (Ely et al. 1995). The hydrazine-dependent oxygen uptake rate was determined by blocking ammonia-dependent oxygen uptake with 100 μM allylthiourea, followed by the addition of 750 μM hydrazine, an alternative substrate for HAO. The specific oxygen uptake rate (SOUR) of the cells was calculated based on the saturated oxygen concentration in the water (Ely et al. 1995). After a constant nitrite production rate and SOUR were detected over three consecutive time points, where the cells were considered to have reached quasi-steady state (Hyman et al. 1995), the zinc solution was added to the treatment reactor. In separate experiments to determine the recovery of N. europaea activities after zinc stress, cells that had been incubated for 1 h with 3.4 μM ZnCl2 were washed six times with HEPES buffer. The residual zinc concentration was determined by inductively coupled plasma-mass spectrometry (ICP-MS). The washed cells were then resuspended in HEPES buffer with (NH4)2SO4 (5 mM) and nitrite production rate was determined. ICP-MS measurements were also used to confirm that zinc concentrations did not change substantially during the quasi-steady-state reactor experiments.
2-D gel electrophoresis
After 3 h of zinc exposure, total proteins, including membrane proteins, were extracted from the cells using a ReadyPrep Sequential Extraction Kit (BIO-RAD, Hercules, CA, USA). Extracted protein (100 μg) was treated with Benzonase (Sigma-Aldrich, St Louis, MS, USA) and incubated for 60 min to remove residual nucleic acid. The solution was then homogenized for 5 min for complete lysis and centrifuged (10 min at 8,000×g). ReadyStrip Immobilized pH Gradient (IPG) Strips (BIO-RAD), with non-linear pH gradient from 3 to 10, were used for isoelectric focusing (IEF). After IEF, the IPG strips and 3 μl unstained protein standard (BIO-RAD) were placed on 12.5% SDS-polyacrylamide gel (BIO-RAD) to separate the proteins. Proteins were fixed in 30% methanol/7.5% acetic acid and stained with 150 ml SYPRO Ruby protein gel stain (Cambrex Bio science, Rockland, ME, USA) overnight in the dark. The proteins were viewed under fluorescent light and protein spots between control- and zinc-treated conditions were compared. Proteins appearing to be related to zinc stress were excised using a pipette tip. The proteins were identified by nano/LC/MS/MS from Midwest Bio Services, LLC (Overland, KS, USA).
Total RNA preparation
After 60 min of zinc exposure, 180 ml of N. europaea (OD600 ≅ 0.07) was harvested and washed with sterilized 40 mM NaHPO4 (pH 7.8). Total cellular RNA was extracted using 1 ml of Trizol (Ambion Inc., Austin, TX, USA) following the manufacturer’s instructions. The extracted RNA was purified with an RNeasy Mini kit (Qiagen Inc., Valencia, CA, USA) and treated with RNase-free DNase Ι (Qiagen) to digest residual chromosomal DNA. The concentration of purified RNA was determined by using a Nanodrop spectrophotometer (Nanodrop Technologies, Rockland, DE, USA) and RNA quality was checked by the A260/A280 ratio and RNA 6000 Nano LabChip kit on the Agilent Bioanalyzer 2100 (Agilent Technologies Inc., Palo Alto, CA, USA).
DNA microarray construction and microarray experiment
The custom Nimble express™ Made-to-Order array for N. europaea was manufactured by NimbleGen Systems, Inc. for Affymetrix based on the published genome sequence (AL954747; Chain et al. 2003). Identified genes in N. europaea are represented on the array by the probe sets with 24 pairs of perfect match/mismatch oligo probes. Microarray analyses were performed in triplicate (three controls and treatments), using RNA samples extracted from independent experiments. cDNA synthesis, labeling, and hybridization were performed by the Center for Genome Research and Biocomputing Core Lab at Oregon State University, Corvallis OR, USA. GeneSpring software was used to analyze gene expression data obtained from Affymetrix GeneChip Operating Software by applying filters to identify the genes that matched with user defined criteria, and evaluating triplicate samples. GC-RMA (Robust Multi-chip Average, with GC-content background correction) was applied to normalize and summarize probe-level intensity measurements from Affymetrix GeneChips (Irizarry et al. 2003). The two normalized groups were filtered using a detected twofold change as a minimum for up or down regulation. The variation of filtered up- or down-regulated genes was evaluated by using unpaired two sample t-tests with a cutoff P value of 0.05. Biological functions of up-regulated or down-regulated hypothetical genes were searched by BLASTX.
Quantitative reverse transcriptase-PCR
The total RNA from control and treatment samples was used to synthesize cDNA with the IScript™ cDNA Synthesis kit (BIO-RAD, Hercules, CA, USA). The synthesized cDNA was amplified with primers (Invitrogen, Carlsbad, CA, USA) designed by using Primer 3 software (Rozen and Skaletsky 2000) and IQ™ SYBR Green Supermix (BIO-RAD). q-PCR was performed with an ABI 7500 instrument (Applied Biosystems, Foster City, CA, USA). The q-PCR efficiency for each gene was checked through standard curves created by serial dilution of RNA samples. The fold changes of genes showing ideal amplification efficiency were calculated using the formula 2−ΔΔCt (Peirson et al. 2003). Using the 2−ΔΔCt method, the data are presented as fold changes in gene expression normalized to a housekeeping gene and relative to the control condition. Twelve genes representing the most up-regulated genes from each of 12 operons were selected for q-PCR validation of the change of mRNA expression level detected from Affymetrix microarrays.
ArrayExpress accession number
The microarray data for this research is available at the Gene Expression Omnibus database (http://www.ncbi.nih.gov/geo) under accession number GSE7552.
Results
Physiological responses to zinc
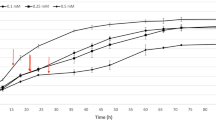
Nitrosomonas europaea in quasi-steady state exposed to 3.4 μM ZnCl2 showed a 61% decrease in the rate of nitrite production (Fig. 1a) and a 69% decrease in the ammonia-dependent specific oxygen uptake rate (AMO-SOUR) (Fig. 1b) within 30 min of exposure and remained at these levels of activity throughout the experiment. Cells in quasi-steady state exposed to 5 μM ZnCl2 showed a 96% decrease in their nitrite production rate (Fig. 1a) and a 94% decrease in the AMO-SOUR (Fig. 1b). The AMO-SOUR remained constant (≅0.35 mM O2/min-OD600) before injection of zinc and in the control condition, but the uptake rate was reduced after 30 min of exposure to zinc. However, the hydrazine-dependent specific O2 uptake rate (HAO-SOUR) did not change after exposure to zinc (data not shown), suggesting that zinc inhibition of ammonia oxidation targets AMO specifically rather than HAO or other metabolic processes. The inhibition pattern of zinc in N. europaea for nitrite production rate and AMO activity was consistent with that of Nitrosococcus mobilis, a halophilic ammonia oxidizing bacteria (Radniecki and Ely, in press). Exposure of batch-cultured N. mobilis to 10 μM ZnCl2 resulted in 100% inhibition in nitrite production, 71% inhibition in AMO-SOUR, and 32% inhibition in HAO-SOUR in 30 min. Also, in our experiments the decreases in nitrite production rate and AMO activity of the cells exposed to 3.4 μM ZnCl2 for 1 h were not recovered when cells were washed to remove residual zinc and placed in fresh HEPES buffer with 5 mM ammonia; 50% inhibition continued for the 4 h observation period (data not shown). ICP-MS analyses confirmed that residual zinc concentration in cell suspensions washed and resuspended, as described above, was essentially zero.
Physiological responses of N. europaea exposed to 3.4 μM ZnCl2 or 5 μM ZnCl2 for 4 h. Nitrite production rate normalized by cell density (a). Ammonia-dependent oxygen uptake rate normalized by cell density (b). Open circles represent no zinc; filled circles represent 3.4 μM ZnCl2; filled triangles represent 5 μM ZnCl2; lines connect data points. Vertical dashed lines indicate the injection time (60 min) of zinc
Analysis of protein expression changes
2-D SDS-PAGE of the soluble protein fractions of N. europaea treated with 3.4 μM ZnCl2 for 3 h showed two notably up-regulated proteins (Fig. 2). The proteins were identified as an outer membrane protein (NE2548) and a S1 RNA binding domain (NE0760; Fig. 2). The transcription levels of the proteins were determined by using q-PCR before the injection of zinc and at 20, 60, and 180 min after the injection of zinc. No substantial up regulation in corresponding genes of these two proteins, as indicated by mRNA levels, was apparent after 20 min of zinc exposure but major up regulation was evident in the 60-min RNA samples (data not shown). Based on this observation RNA samples for corresponding microarray analyses were taken after 60 min of incubation with 3.4 μM ZnCl2.
Transcriptional changes
In total, of the 2,460 predicted genes of the N. europaea genome, 27 genes showed statistically significant (>2-fold) up regulation while 30 genes displayed statistically significant (>2-fold) down regulation following treatment with zinc. The up- and down-regulated genes were categorized by function according to the N. europaea genome database (http://genome.ornl.gov; Fig. 3). The up-regulated genes were involved in various functions such as mercury resistance, inorganic ion transport, oxidative stress, toxin–antitoxin (TA) functions, two-component signal transduction system (TCS), DNA repair, and translation. The largest group of up-regulated genes (11 genes) was related to inorganic transport, while only three were related to DNA repair or translation. When cells in exponential growth are exposed to a high concentration of zinc, the cells import zinc non-specifically into the periplasm, where it causes heavy metal toxicity (Nies 1999). Therefore, genes that encode inorganic transporters and heavy metal binding proteins may be up regulated to sequester the free zinc imported non-specifically. The down-regulated genes in our experiments are involved in transcription (cspD2, NE1312), multicopper oxidase (mnxG, NE0315), biosynthesis (mrsA, NE0530), and amino acid transport (gcvH1, NE0608). In addition, genes for signal transduction mechanisms (cheYZ, NE1923/4) and universal stress protein (yxiE, NE2292) showed statistically significant down regulation. NE1918/9 [cbbOQ, Ribulose bisphosphate carboxylase/oxygenase (RubisCO)] and NE0668/0670 [outermembrane efflux protein (MtrC)] were also down regulated. Since transcript of genes in the RubisCO operon have been observed to increase under low CO2 and decrease under higher CO2, it might indicate that cells in our system were not carbon-limited (Wei et al. 2004). The RubisCO operon for N. europaea also has been shown to be expressed only under non-limited energy conditions (Wei et al. 2004). Hence, the down regulation of cbbOQ may suggest that N. europaea under zinc stress may conserve energy by down regulating transcription of the RubisCO operon.
Differentially expressed genes grouped by functional classification according to the N. europaea genome database (http://genome.ornl.gov). Columns: 1 inorganic transport and membrane permeability, 2 general function prediction, 3 signal transduction system, 4 carbohydrate transport, 5 amino acid transport, 6 DNA repair, 7 translation, 8 transcription, 9 hypothetical proteins, 10 intergenic region
Discussion
Mercury-resistance operon
Mercury scavenger-like transporters that might be involved in heavy metal tolerance of N. europaea (Chain et al. 2003) were members of the highest up-regulated operon detected under zinc stress. The operon (Fig. 4) includes merTPCAD, with merE being adjacently located and merR regulating expression of the operon (Stein et al. 2007). These genes may play an important role in the zinc resistance pathway of N. europaea; that is, once zinc is imported into the periplasm through unspecific transport proteins under a zinc-ample condition, merTPC may transport zinc into the cytoplasm to prevent toxic effects in the periplasm (Nies 1999). The merA is related to glutathione reductase and may reduce the oxidized bisglutathione (GS-SG) formed by the metal cations (Nies 1999). Transcriptional levels of the five genes in merTPCAD were different as shown in Table 1, perhaps because of premature termination of transcription, rapid mRNA processing or mRNA degradation. In a BLASTX search, full-length alignments of NE0837 yielded a best hit (E value, 10−127) to urf-2 (a putative mercury resistance gene) of Pseudomonas sp. A19-1. NE2575 (merE), located between NE0837 and NE0838, may have caused the signal observed from NE0837, which encodes a transposition function, possibly due to the immediate proximity of the two genes or read-through of the region (Stein et al. 2007).
Up regulation of genes involved in metal transport and membrane permeability
NE2124, encoding TonB-dependent outer membrane receptor, was up regulated 2.1-fold under zinc treatment. Gram-negative bacteria scavenge Fe3+ ions by employing chelators and express substrate-specific proteins to recognize the ions. Three outer membrane proteins (TonB, ExbB, and ExbD) are required to transport these ferric chelators into the periplasm of the cells and play an important role in the transport of heavy metals such as iron (Skare et al. 1993; Bos et al. 1998; Moeck and Coulton 1998). Under Fe-limiting conditions, the TonB-dependent outer membrane receptor gene was up regulated (Wei et al. 2006a). In contrast, the TonB-dependent outer membrane receptor gene of Caulobacter crescentus was up regulated under excessive concentrations of cadmium, chromium, and uranium (Hu et al. 2005). Therefore, it can be suggested that the TonB-dependent outer membrane receptor is involved in adaptation of the cells in both heavy metal-limited or heavy metal-ample conditions. The transporter genes of Gram-negative bacteria are responsible for the regulation of heavy metal levels between the cytoplasm and the periplasm. Gram-negative bacteria exposed to excessive heavy metals generally express genes encoding transporters for heavy metal resistance (Teitzel et al. 2006). In our research, NE1898, 1899, and 1900, encoding the ABC transporter, were up regulated linking this transporter to heavy metal stress. NE1029, encoding a putative soluble binding metallo-chaperon, was up regulated. The metallo-chaperones sequester metal ions and regulate the metal-dependent gene expression under excess zinc conditions (O’Halloran and Culotta 2000). Therefore, this gene might be up regulated to reduce free zinc ions which can cause toxic effects. The outer membrane proteins of Gram-negative bacteria provide a permeability barrier to protect the cell from toxic agents. Outer membrane protein genes, NE2563 (encoding OmpC) and NE2548 (encoding OmpA), showed statistically significant up regulation (Table 1). OmpA also showed up regulation in 2-D SDS-PAGE of the soluble protein fraction extracted from zinc-exposed cells as shown in Fig. 2, thus confirming the microarray data. The Tol protein in Escherichia coli is related to the stability of the outer membrane by transporting crucial outer membrane components such as LPS, Lpp, or Pal (Lazzaroni et al. 1999). It can be suggested that NE0920 (TolB) was also induced to maintain the function of the outer membrane as a barrier under heavy metal toxicity.
Up regulation of putative oxidative stress genes
Aerobic bacteria such as Bacillus subtilis have enzymes that can remove reactive oxygen species (HO• or HOO••), superoxide dismutase or hydroperoxidases. N. europaea has genes for several comparable enzymes such as katA, katG, and ahpC (Chain et al. 2003) and thioredoxin is a disulfide reductase that removes the protein disulfide bonds produced by reactive oxygen species (Hu et al. 2005). NE1319 encoding thioredoxin showed up regulation when N. europaea were exposed to 3.4 μM ZnCl2 suggesting that 3.4 μM ZnCl2 may lead to oxidative stress in N. europaea. RNA polymerase sigma (σ) factors have been shown to be up regulated to cope with environmental stresses such as oxidative stress (Teitzel et al. 2006). In the current research, NE1071 and NE1096, encoding two extra cytoplasmic function σ-factors, showed greater than 1.9-fold up regulation and the microarray data of NE1096 were confirmed by q-PCR (Table 2). NE1071 (fecI) and NE2143 (rpsD) that also showed up regulation under chloroform stress (Gvakharia et al. 2007) and nitrosative stress (Cho et al. 2006) might be general stress genes. yvqQ (nitrite and sulfite reductase) showed up regulation in our research and it was categorized as an oxidative stress gene in the presence of heavy metals such as cadmium and chromium in previous research (Hu et al. 2005).
Toxins–antitoxins, two-component signal transduction systems and DNA repair genes
Once bacteria are exposed to harmful environment conditions such as nutrient limitation, starvation stress, and chlorinated hydrocarbons stress (Gvakharia et al. 2007), TA loci that are ubiquitous in free-living bacteria regulate the global levels of translation and replication. The mazEF-2 is a representative TA of N. europaea and the PemK belongs to the mazEF family (Gerdes et al. 2005). NE0974 and NE0975, encoding PemK, were up regulated both under zinc stress and under chloroform stress (Gvakharia et al. 2007). Transcripts for Helix-turn-helix protein (GopG) that belongs to mazEF-3 (Oberer et al. 2002) also showed up regulation (>1.5-fold change). TCS, consisting of sensor histidine kinases and phosphorylated cognate target regulators, respond to changes in metal concentrations in the environment to maintain the proper concentration of the metals in the cell (Choudhury and Srivastava 2001). NE0728 and NE0729 encoding TCS were observed to be up regulated in this research. TCS-related genes were also up regulated under uranium and copper stress in previous research (Nies and Brown 1998; Hu et al. 2005). Formamidopyrimidine-DNA glycosylase encoded by NE2552 (mutM), an enzyme for DNA repair, requires zinc for its activity. DNA repair genes were up regulated in response to the toxic conditions caused by TCE oxidation (Yeager et al. 2001). Therefore, the up regulation of this gene in this work may indicate that 3.4 μM ZnCl2 may damage DNA replication in N. europaea. However, the genes may also be up regulated to take up free zinc to fulfill requirements for the functional enzyme.
Validation of microarray trends using q-PCR
The fold changes of twelve genes obtained by q-PCR corresponded with values obtained by Affymetrix microarray analyses, as shown in Tables 1, 2, and 3.
Concluding remarks
This research suggests that although zinc is an essential metal for seven enzymes and two zinc-based domains of N. europaea, 3.4 μM ZnCl2 can cause batch-cultured cells about 60% decrease in nitrite production rate and ammonia-dependent oxygen uptake rate without recovery. In addition, 3.4 μM ZnCl2 changes the transcriptional expression levels of several groups of genes that allow the cells to resist unfavorable environmental conditions. The mercury resistance genes, merTPCAD, showed the highest up regulation (46.2-fold) after 60 min exposure to 3.4 μM ZnCl2, suggesting that the resistance pathway of N. europaea in zinc-ample conditions is comparable to the mercury resistance pathway. In addition, the genes involved in inorganic transport, membrane transport, oxidative stress, TA genes, TCS, and DNA repair were greater than 2.0-fold up regulated. Down-regulated genes were related to RubisCO, transcription, and amino acid transport.
References
Blencowe DK, Morby AP (2003) Zn(II) metabolism in prokaryotes. FEMS Microbiol Rev 27:291–311
Bos C, Lorenzen D, Braun V (1998) Specific in vivo labeling of cell surface-exposed protein loops: a ferrichrome binding site and a ligand-induced conformation change of the Escherichia coli FhuA protein. J Bacteriol 180:605–613
Chain P, et al (2003) Complete genome sequence of the ammonia-oxidizing bacterium and obligate chemolithoautotroph Nitrosomonas europaea. J Bacteriol 185:2759–2773
Cho CM, Yan T, Liu X, Wu L, Zhou J, Stein LY (2006) Transcriptome of a Nitrosomonas europaea mutant with a disrupted nitrite reductase gene (nirK). Appl Environ Microbiol 72:4450–4454
Choudhury R, Srivastava S (2001) Zinc resistance mechanisms in bacteria. Curr Sci 81:768–775
Ely RL, Hyman MR, Arp DJ, Guenther RB, Williamson KJ (1995) A cometabolic kinetics model incorporating enzyme inhibition, inactivation, and recovery 2. Trichloroethylene degradation experiments. Biotechnol Bioeng 46:232–245
Ensign SA, Hyman MR, Arp DJ (1993) In vitro activation of ammonia monooxygenase from Nitrosomonas europaea by copper. J Bacteriol 175:1971–1980
Gerdes K, Christensen SK, Lobner-Olesen A (2005) Prokaryotic toxin–antitoxin stress response loci. Nat Rev Microbiol 3:371–382
Gilotra U, Srivastava S (1997) Plasmid-encoded sequestration of copper by Pseudomonas pickettii strain US321. Curr Microbiol 34:378–381
Gvakharia BO, Permina EA, Gelfand MS, Bottomley PJ, Sayavedra-Soto LA, Arp DJ (2007) Global transcriptional response of Nitrosomonas europaea to chloroform and chloromethane. Appl Environ Microbiol 73:3440–3445
Hu P, Brodie EL, Suzuki Y, McAdams HH, Andersen GL (2005) Whole-genome transcriptional analysis of heavy metal stresses in Caulobacter crescentus. J Bacteriol 187:8437–8449
Hyman MR, Arp DJ (1995) Effects of ammonia on the de novo synthesis of polypeptides in cells of Nitrosomonas europaea denied ammonia as an energy source. J Bacteriol 177:4974–4979
Hyman MR, Russell SA, Ely RL, Williamson KJ, Arp DJ (1995) Inhibition, inactivation, and recovery of ammonia-oxidizing activity in cometabolism of trichloroethylene by Nitrosomonas europaea. Appl Environ Microbiol 61:1480–1487
Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP (2003) Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res 31:e15
Lazzaroni JC, Germon P, Ray MC, Vianney A (1999) The Tol proteins of Escherichia coli and their involvement in the uptake of biomolecules and outer membrane stability. FEMS Microbiol Lett 177:191–197
Moeck GS, Coulton JW (1998) TonB-dependent iron acquisition: mechanisms of siderophore-mediated active transport. Mol Microbiol 28:675–681
Nies DH (1999) Microbial heavy-metal resistance. Appl Microbiol Biotechnol 51:730–750
Nies DH, Brown N (1998) Metal ions in gene regulation. Chapman and Hall, NY
Oberer M, Zangger K, Prytulla S, Keller W (2002) The antitoxin ParD of plasmid RK2 consists of two structurally distinct moieties and belongs to the ribbon-helix-helix family of DNA-binding proteins. Biochem J 41–47
O’Halloran TV, Culotta VC (2000) Metallochaperones, an intracellular shuttle service for metal ions. J Biol Chem 275:25057–25060
Outten CE, O’Halloran TV (2001) Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science 292:2488–2492
Peirson SN, Butler JN, Foster RG (2003) Experimental validation of novel and conventional approaches to quantitative real-time PCR data analysis. Nucleic Acids Res 31:e73
Pomposiello PJ, Bennik MHJ, Demple B (2001) Genome-wide transcriptional profiling of the Escherichia coli response to superoxide stress and sodium salicylate. J Bacteriol 183:3890–3902
Radniecki TS, Ely RL (2007) Zinc chloride inhibition of Nitrosococcus mobilis. Biotechnol Bioeng. doi:10.1002/bit.21672
Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132:365–386
Skare JT, Ahmer BM, Seachord CL, Darveau RP, Postle K (1993) Energy transduction between membranes. TonB, a cytoplasmic membrane protein, can be chemically cross-linked in vivo to the outer membrane receptor FepA. J Biol Chem 268:16302–16308
Stein LY, Arp DJ (1998) Loss of ammonia monooxygenase activity in Nitrosomonas europaea upon exposure to nitrite. Appl Environ Microbiol 64:4098–4102
Stein LY, et al (2007) Whole-genome analysis of the ammonia-oxidizing bacterium, Nitrosomonas eutropha C91: implications for niche adaptation. Environ Microbiol 9(12):2993–3007
Teitzel GM, Geddie A, De Long SK, Kirisits MJ, Whiteley M, Parsek MR (2006) Survival and growth in the presence of elevated copper: transcriptional profiling of copper-stressed Pseudomonas aeruginosa. J Bacteriol 188:7242–7256
Wei X, Sayavedra-Soto LA, Arp DJ (2004) The transcription of the cbb operon in Nitrosomonas europaea. Microbiology 150:1869–1879
Wei X, Vajrala N, Hauser L, Sayavedra-Soto LA, Arp DJ (2006a) Iron nutrition and physiological responses to iron stress in Nitrosomonas europaea. Arch Microbiol 186:107–118
Wei X, et al (2006b) Transcript profiles of Nitrosomonas europaea during growth and upon deprivation of ammonia and carbonate. FEMS Microbiol Lett 257:76–83
Yeager CM, Bottomley PJ, Arp DJ (2001) Requirement of DNA repair mechanisms for survival of Burkholderia cepacia G4 upon degradation of trichloroethylene. Appl Environ Microbiol 67:5384–5391
Acknowledgments
The research in this study was funded through NSF proposal number 0412711. We thank Daniel J. Arp, Luis A. Sayavedra-Soto, and Tyler S. Radniecki for critical reading of the manuscript, and Barbara O. Gvakharia for experimental advice and discussion. We also thank the Center for Genome Research and Biocomputing (CGRB) at Oregon State University for providing facilities for RNA quality and microarray experiments.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Stuart Ferguson.
Rights and permissions
About this article
Cite this article
Park, S., Ely, R.L. Genome-wide transcriptional responses of Nitrosomonas europaea to zinc. Arch Microbiol 189, 541–548 (2008). https://doi.org/10.1007/s00203-007-0341-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-007-0341-7