Abstract
Unraveling functional genes related to biodegradation of organic compounds has profoundly improved our understanding of biological remediation processes, yet the ecology of such genes is only poorly understood. We used a culture-independent approach to assess the abundance and diversity of bacteria catalyzing the degradation of n-alkanes with a chain length between C5 and C16 at a forest site co-contaminated with mineral oil hydrocarbons and metals for nearly 60 years. The alkB gene coding for a rubredoxin-dependent alkane monooxygenase enzyme involved in the initial activation step of aerobic aliphatic hydrocarbon metabolism was used as biomarker. Within the area of study, four different zones were evaluated: one highly contaminated, two intermediately contaminated, and a noncontaminated zone. Contaminant concentrations, hydrocarbon profiles, and soil microbial respiration and biomass were studied. Abundance of n-alkane-degrading bacteria was quantified via real-time PCR of alkB, whereas genetic diversity was examined using molecular fingerprints (T-RFLP) and clone libraries. Along the contamination plume, hydrocarbon profiles and increased respiration rates suggested on-going natural attenuation at the site. Gene copy numbers of alkB were similar in contaminated and control areas. However, T-RFLP-based fingerprints suggested lower diversity and evenness of the n-alkane-degrading bacterial community in the highly contaminated zone compared to the other areas; both diversity and evenness were negatively correlated with metal and hydrocarbon concentrations. Phylogenetic analysis of alkB denoted a shift of the hydrocarbon-degrading bacterial community from Gram-positive bacteria in the control zone (most similar to Mycobacterium and Nocardia types) to Gram-negative genotypes in the contaminated zones (Acinetobacter and alkB sequences with little similarity to those of known bacteria). Our results underscore a qualitative rather than a quantitative response of hydrocarbon-degrading bacteria to the contamination at the molecular level.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Anthropogenic activities related to the production, transportation, processing, handling, and disposal of oil have resulted in the contamination of numerous sites with aliphatic n-alkanes and other hydrocarbons in the last century [1, 2]. Although much of these contaminations are susceptible to multiple natural biodegradation processes, remediation can be particularly difficult and costly at sites where mixtures of contaminants are present (SERDP and ESTCP [3]). Assessing the genetic potential and functionality of the organisms and processes relevant to the assessment and/or remediation of contaminants in the environment is thus important for evaluating the impact of anthropogenic disturbances on ecosystem performance and understanding mechanisms of natural attenuation.
Rapid advances in microbial ecology over the last years have significantly improved our knowledge of the genes and the enzymes involved in n-alkane metabolism as well as the microbiology of n-alkane degraders [4, 5]. A class of integral membrane nonheme diiron monooxygenases of the alkB type has been proposed as one of the two unrelated classes of enzymes responsible for the aerobic transformation of midchain-length n-alkanes (C5 to C16) and in some cases even longer alkanes [6]. These enzymes catalyze the terminal or subterminal oxidation of n-alkanes to primary or secondary alcohols, which is the initial step in the aerobic degradation of n-alkanes [7]. Based on this knowledge, various oligonucleotide primers and DNA probes targeting alkB genes have been developed over the last years [8–15], which can be applied to the study of specific bacterial groups or general hydrocarbon-degrading bacteria in the environment.
Despite the rapid development of such molecular markers, relatively little is known concerning the ecology of hydrocarbon-degrading populations at abandoned sites which have been contaminated with complex mixtures of hydrocarbons and metals for decades. This is mainly because field studies on complex and mixed contaminations are rare. In addition, most work on alkB homologous genes has been carried out with isolates [8, 9, 12, 14], in microcosms [16], or using group specific primers [17, 18].
Advancing our knowledge on the ecology of hydrocarbon degraders in soil is important to improve reliance on natural attenuation and/or optimize bioremediation processes at contaminated sites. We investigated various zones at a forest site impacted to a different extent (highly, intermediately, and noncontaminated) by a deposit of acid resin, a waste product from the mineral oil industry. Since the resin contains hydrocarbons of chain length amenable to alkB monooxygenases (C10–C16 and possibly larger), we hypothesized that long-term contaminant-driven selective pressure would result in larger abundance and diversity of n-alkane degraders as characterized by alkB along the contamination plume compared to nonaffected areas and, in turn, favor selection of specific groups depending on the extent of the contamination. To test this, at each zone, we investigated hydrocarbon profiles, general biological properties (biomass and respiration), and molecular abundance and diversity of hydrocarbon degraders using the alkB gene as biomarker.
Materials and Methods
Site Description and Contamination History
The site of study is located in an area known as Seelacher Berg near the city of Kronach (Bayern, Germany). It is characterized by a hilly landscape (about 320 m high) of mixed forest dominated by Fagus sylvatica L. and Coryllus avellana L., and to a lesser extent Pinus sylvetris L., Populus tremula L., and Picea abies (L.) Karst. The climate lies between oceanic and continental [19] with mean annual temperature of 8°C and mean annual precipitation of 765 mm. The soil has a sandy loam texture (77% sand, 16% silt, and 7% clay) and is classified as Braunerde [20], which falls within the cambisols family [21].
The site has been contaminated with hydrocarbons and metals for approximately 60 years. The origin of this contamination is a deposit of acid resin, a residue from the mineral oil industry, which was produced during the recycling of used oils by the Benzmann process during the first half of the last century [22]. These resins were very acidic and enriched with hydrocarbons and trace elements, mainly Pb. Due to the lack of environmental legislation at that time, acid resins were dumped into excavated ponds in public areas without further protection. Estimations suggest between 40 and 160 acid resin deposits in Germany alone [23]. A detailed description of the contamination and its effects on soil functionality can be found in Pérez-de-Mora et al. [23, 24]. The most relevant characteristics of the acid resin are pH (1.5 units), total hydrocarbon concentrations (93 g kg−1), As (63 mg kg−1), Cd (24 mg kg−1), Cu (1,835 mg kg−1), Pb (8,100 mg kg−1), S (55,450 mg kg−1), and Zn (873 mg kg−1).
Soil Sampling
Soil was sampled in October 2006. The investigated area was approximately 4,800 m2. A more detailed description of the sampling strategy can be found in Pérez-de-Mora et al. [23]. Four different zones were investigated in the surroundings of the acid resin deposit: (1) a control zone unaffected by the waste and therefore not contaminated (CB) and (2) three zones along the plume of contamination (XB), one highly contaminated (XB1) and two intermediately contaminated (XB2 and XB3). Within each selected zone (control, high, or intermediately contaminated), five different locations were sampled in an area of approximately 20 m2. At each location, a composite sample of 15–20 soil cores (0–15 cm depth) was collected. Since the study focused on long-term contamination effects, the litter layer was excluded to avoid the influence of recently added material to the soil system. No vegetation was directly growing on the surface of sampled zones, but woody species in the area showed no apparent signs of toxicity. Transport to the laboratory was achieved within 3 h after the sampling. For assessment of gravimetric water content, total hydrocarbons, microbial respiration, and microbial N (Nmic) soil subsamples were stored at 4°C until analysis (within 3 weeks after the sampling). For determination of the other chemical properties, soil samples were dried for 4 days at room temperature. Subsamples for DNA analysis were immediately frozen in situ in dry ice and subsequently stored at −80°C until extraction.
General Soil Properties
Soil pH, total hydrocarbons (THC), trace elements (As, Cu, Pb), and S concentrations are shown in Table 1. All of these general properties in addition to microbial respiration were determined as previously described [23, 24]. Microbial biomass was estimated by quantification of microbial N (Nmic) using the chloroform-fumigation extraction procedure [25]. A k EN value of 0.54 was used to convert the N difference between fumigated and nonfumigated samples in biomass N [26]. Nitrogen in the extracts was determined via catalytic high temperature oxidation using a TC/TN DIMATOC Analyzer (Dimatec Analysentechnick GmbH, Germany). All analyses were performed in duplicate.
DNA Extraction from Environmental Samples
Total nucleic acids from soil (0.5 g dw) were extracted using the method of Griffiths et al. [27]. In brief, cells were lysed via mechanical shaking using Precellys-Keramik-Kit Tubes (PeqLab, Germany) with a Precellys 24® Equipment (Bertin Technologies, France). Extracted nucleic acids were resuspended in 50 mL milliQ water (pH = 6.8); concentration of total DNA was measured via a Nanodrop® ND-1000 spectrometer (Nanodrop Technologies, USA) at 260 nm. Extractions were carried out in duplicate. The quality of the DNA extracted was checked by comparing the ratios OD 260/280 and OD 260/230 between samples. DNA extraction quality was also tested via PCR amplification using the universal 16S rRNA gene as target. All samples from control and contaminated locations could be amplified successfully.
Quantification of the alkB Gene via Real-Time PCR
AlkB gene copy numbers in soil samples were quantified via real-time PCR using an ABI Prism 7700 sequence detector (Applied Biosystems, USA) as previously described [28, 29]. Degenerate primers developed by Kloos et al. [10] without any obvious strain-specific discrimination were slightly modified by substituting every fourfold degenerated base with inosine nucleotides to reduce degeneracy and improve amplification: alkB-f (5′AAYACIGCICAYGARCTIGGICAYAA3′) and alkB-r (5′ GCRTGRTGRTCIGARTGICGYTG3′). This primer set yields a PCR product of approximately 550 bp. The PCR cycle program was as follows: initial denaturation at 95°C for 10 min, 5 cycles of 45 s at 95°C, 1 min at 62°C (stepwise reduced to 57°C) and 45 s at 72°C followed by 40 cycles of 45 s at 95°C, 1 min at 57°C, and 45 s at 72°C. Data were collected at 78°C after each cycle was completed. This is to minimize the fluorescence from primer-dimers which might form due to the degeneracy of the primers. For melting curve analysis, a final cycle was performed (15 s at 95°C, 30 s at 60°C, and 15 s at 95°C). The PCR reaction mixture (final volume of 25 μL) included 12.5 μL of Power SYBR® green PCR Master Mix (Applied Biosystems, USA), 1 μL of 50 mM MgCl2, 0.5 μL of 3% bovine serum albumin (BSA), 0.625 μL of a 4-μM solution of each primer, 7.75 μL of milliQ autoclaved water, and 2 μL of the template DNA. The optimal amount of template for maximum quantification was determined in preliminary experiments for each sample. Different dilutions of original extracts ranging from 3 to 25 ng of template were tested for this purpose (data not shown). All samples and standards were amplified in triplicate. As standard, tenfold serial dilutions of plasmid DNA containing the alkB fragment from Pseudomonas putida GPo1 was used. The efficiency of the PCR was 88%. Standard curves were linear (R2 > 0.991) over seven orders of magnitude. No signals were observed in the nontemplate controls (water instead of template; data not shown). Amplification specificity was double-checked by melting curve analysis and agarose-gel electrophoresis (2% gels).
Diversity Analysis of alkB by T-RFLP Fingerprinting
Restriction endonuclease HpaII was selected for T-RFLP fingerprinting following in silico-based restriction analysis of alkB gene sequences from uncultured bacteria and representatives of different phylogenetic groups using the biology workbench 3.2 online application (San Diego Computer Supercenter, USA). For T-RFLP fingerprinting analysis, genomic DNA from soil extracts was first amplified via PCR using the same primer pair and conditions described above for qPCR with the following modifications: (1) 30 cycles (45 s at 95°C, 1 min at 57°C, and 45 s at 72°C) instead of 40, (2) no reading step at 78°C after each cycle, (3) a final extension of 10 min at 72°C, and (4) no final cycle for melting curve analysis. The PCR reaction mixture (final volume of 50 μL) included: 5 μL of 10× Buffer (Invitrogen, Germany), 2 μL of 50 mM MgCl2, 2.5 μL of a 2 mM dNTP mix solution, 1 μL of 3% BSA, 1.25 μL of a 4-μM solution of each primer, 34.5 μL of milliQ autoclaved water, 0.5 μL of a 5-U μL−1 Taq Polymerase solution and 2 μL of template DNA.
Amplified products (550 bp) were run on a 2% agarose gel, cut in the absence of UV light, and purified using the MinGel Extraction Kit (Qiagen, Germany). Approximately 100–150 ng of the purified PCR product was digested with 20 U of HpaII (New England Biolabs, Germany). The reaction mixture included 2 uL of 10× NEBuffer 1, 2 μL of HpaII (10,000 U mL−1), around 8–12 μL of purified amplicons, and milliQ autoclaved water to make up a final volume of 20 uL. DNA digestion was performed for 3 h at 37°C followed by an enzyme inactivation step at 65°C for 20 min. The reaction mix was purified using the MinElute Reaction Purification Kit (Qiagen, Germany). For each sample, a nondigested PCR extract was used as control to ensure specificity of the restriction assay (T-RFLP peaks deriving from no other band than the 550 bp).
For the separation and detection of fragments, 10–15 ng of the labeled and digested DNA or nondigested DNA (controls) was mixed with 0.25 μL GenomeLab™ DNA Size Standard 600 (internal standard; Beckman Coulter GmbH, Germany) and 26.75 mL of SLS loading solution (Beckman Coulter GmbH, Germany). A drop of mineral oil was added to prevent evaporation. Separation and detection of labeled fragments were achieved via capillary gel electrophoresis and molecular fluorescence spectrometry using a CEQ 2000 XL Sequencer (Beckman Coulter GmbH, Germany). Primary electropherogram evaluation was performed using the CEQ™ 8000 Genetic Analysis System Software version 8.0.52 (Beckman Coulter GmbH). Peak recognition was checked and edited manually. Terminal restriction fragment (T-RF) relative abundances (A p) were inferred from peak heights using the following equation:
where n i represents the peak height of one distinct T-RF and N is the sum of all peak heights in a given T-RFLP pattern. Signals with a peak height below 100 relative fluorescence units and with a peak abundance contribution below 2% were considered as background noise and discarded for further analysis [30]. The size of the fragments was estimated by reference to the internal standard using the quartic algorithm. The standard deviation (SD) for specific T-RF expressed as % of peak height varied in general between 5% and 10%. Technical variability was assessed by triplication of individual samples.
Comparison of T-RF profiles between samples was achieved by means of nonmetric multidimensional scaling (NMS, see “Data Analysis”). Richness (R) was calculated as the number of peaks in a sample unit. Diversity (H) was based on Shannon’s diversity index [31] which is calculated as follows:
where pi represents here the frequency of the ith T-RF.
Evenness (J) was computed as proposed by Pielou [32]:
where \( {H_{{{ \max }}}} = \sum\limits_{{i = 1}}^q {1/q} \;{ \log }\;{1}/q = { \log }q \) and q is the number of T-RF.
For calculation of the functional organization index (Fo), Pareto–Lorenz evenness curves [33] were constructed for each location. They resulted from the cumulative normalized number of T-RF or cumulative proportion of OTUs (x-axis) versus their respective normalized intensities (y-axis) [34]. To obtain the index, the y-axis projection of each curve with the 20% x-axis line was scored.
Cloning and Sequencing of alkB Fragments
To assess the phylogenetic diversity of alkB in soil samples, three clone libraries representing each degree of contamination (high, intermediate, and noncontaminated) and each consisting of 44 clones were constructed. Each library was based on a composite soil sample consisting of approximately 1–1.2 kg of soil from 15 pooled soil cores (0–15 cm). PCR amplicons were obtained using the same primer system and PCR program as described in “Quantification of the alkB via Real-Time PCR.” PCR products were purified with the QIAquick PCR purification kit (Qiagen, Germany) and cloned (around 12 ng of DNA) into the pCR2.1 vector using the TA cloning kit (Invitrogen, Karlsruhe, Germany) following the manufacturer’s instructions. After transformation into Escherichia coli TOP10 competent cells (Invitrogen, Germany), positive clones were picked (white-blue screening) and grown in LB + Kanamycin liquid medium for further plasmid purification [35]. Prior to sequencing, validity of purified plasmids was confirmed via PCR as detailed in “Quantification of the alkB via Real-Time PCR.” Sequencing was performed with the BigDye Terminator Kit 3.1 on the ABI 3730 48-capillary sequencer (both Applied Biosystems, USA). Both strands were read from double-stranded plasmid with the opposing vector primers T7 forward and M13 reverse. Nucleotide sequences were translated into protein sequences in silico using the ExPasy proteomic server software [36] and subsequently aligned with the blastp software (NCBI, www.ncbi.nlm.nih.gov). The phylogenetic tree was calculated with the ARB Software [37] using Maximum-Likelihood as well as Parsimony and Neighbor-Joining linkage methods for stability testing (data not shown). Sequences are available under the GenBank accession numbers: HM441092–HM441135 (control zone), HM441136–HM441179 (highly contaminated zone), and HM441180–HM441223 (intermediately contaminated zone). Rarefaction curves for each clone library were also calculated using the free software Analytic Rarefaction 1.3 [38].
Data Analysis
For general soil and microbiological properties (Tables 1 and 2, respectively), and ecological indexes based on molecular fingerprints (Table 3), mean, median, standard deviations, and minimum and maximum values were calculated for comparison between zones. Correlation analysis was performed to determine the relationship between the different microbiological properties and the ecological indexes considered and the various soil and contaminant-related properties. The significance level reported (α = 0.01 and α = 0.05) is based on Pearson’s coefficients using SPSS 16.0 for Windows.
Multivariate analysis of T-RFLP fingerprints was performed using NMS. Among other features, this iterative procedure is well suited to data that are nonnormal and deal effectively with the “zero truncation problem” which affects performance of other ordination techniques. NMS was performed with the PC-ORD™ software [39]. Calculations were made using Sørensen’s distance and a random starting configuration. The number of runs performed was 250 for the real data and 500 for the randomized data. The number of dimensions in the final solution was two. This number was determined by plotting the final stress vs the number of dimensions and confirmed by Monte Carlo test (p = 0.02). The maximum number of iterations for the final solution was 250. The stability of the latter was assessed with a plot of stress vs the iteration number; stress stabilized after 30 iterations. To test for group differences between zones based on the frequencies of the T-RF, a nonparametric multiresponse permutation procedure (MRPP) was also performed with the PC-ORD™ software based on Sørensen’s distance [39]. A detailed description of the method can be found in McCune and Grace [40].
Results
Natural Attenuation of Hydrocarbons
Figure 1 shows the GC/MS chromatograms for total hydrocarbons characteristic for the acid resin deposit (source; Fig. 1a) and the various zones studied (highly, intermediately, and noncontaminated; Fig. 1b–e). A thorough evaluation of biodegradation using biomarkers and changes in hydrocarbon profiles over time was beyond the scope of this study. Within the highly contaminated zone, there was one location with no signs of biodegradation (Fig. 1b) characterized by an almost identical GC chromatographic profile to that of the source (Fig. 1a). This location was the most extreme in terms of contaminants (see max. levels in zone XB1, Table 1). At the other locations within the highly contaminated zone, the disappearance of some of the well-resolved peaks found in the resin suggests some extent of biodegradation (Fig. 1c). This was even more evident at the intermediately contaminated zones, where the majority of the GC-resolved compounds from the resin were no longer observed; instead, the GC-unresolved fraction became more apparent and compounds less amenable to biodegradation accumulated (Fig. 1d) [41]. There were no signs of resin contamination at the control locations (Fig. 1e).
GC–MS chromatograms for total hydrocarbons in a acid resin deposit, b location C in zone XB1 (highly contaminated), c location A in zone XB1 (highly contaminated), d location A in zone XB3 (intermediately contaminated), and e location A in zone CB (control). Peaks at approximately 8 and 79 min correspond to decane and tretacontane, respectively
Abundance of alkB, Soil Microbial Biomass, and Soil Respiration
Microbial biomass was calculated based on microbial N (Nmic). It was observed that Cmic measurements in contaminated zones were biased because of the high C content of the hydrocarbons from the acid resin (data not shown). Despite lower mean and median values of Nmic in the contaminated zones compared to the control, microbial biomass was within the same order of magnitude in all zones (Table 2). The maximum biomass value from the contaminated zones was similar or even higher than that of the control zone (Table 2). Soil respiration both in absolute terms and normalized with Nmic was always higher in the contaminated zones compared to that in the control area (Table 2).
Abundance of the alkB gene in the soil samples was calculated both as the number of copies per gram of soil and per milligram of Nmic (Table 2). Both mean and median values were within the same order of magnitude for all samples. The greater range observed in XB1 reflects the heterogeneity reported for this zone in terms of general soil properties and contaminant concentrations (Table 1). This may be attributed to a greater and patchier distribution of acid resin contaminants compared to the intermediately contaminated zones.
Diversity of alkB
Differences in the community structure of hydrocarbon-degrading microbial populations were assessed via T-RFLP fingerprints of the alkB gene. A total of 39 distinct T-RF were identified and used for community analysis. Results suggest inter- as well as intrazone differences in the community structure (Fig. S1). In general, fingerprinting profiles were dominated by a few T-RFs (between 1 and 5), which accounted for more than 80% of the total number of T-RFs. There was only one T-RF which was present in all samples (T-RF 72) and three other T-RFs that were found in 95% of the locations studied (T-RF 118, T-RF 131, and T-RF 140; Fig. S1). A significant number of T-RFs (a total of 21) were unique to specific samples. In general, the contribution of unique T-RFs within each sample was low (2–3%); only in the case of T-RF-142 in XB1D (10.2%) and T-RF-383 in XB1E (6.2%) a significant contribution of unique T-RFs was found (Figure S1).
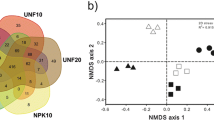
Figure 2 shows the ordination of the 20 samples studied in the multivariate space as a result of the T-RF analysis using nonmetric multidimensional scaling (NMS). The first and second axis accounted for 81% and 14% of the total variance, respectively. Axis 1 of the ordination was strongly correlated with T-RF-72 (τ = −0.926; r 2 = 0.985), and to a lesser extent with T-RF-131 (τ = 0.463; r 2 = 0.422) and T-RF-194 (τ = 0.497; r 2 = 0.409), while axis 2 was mainly correlated with T-RF-238 (τ = 0.316; r 2 = 0.458) and T-RF-240 (τ = −0.476; r 2 = 0.383). In general, ordination analysis showed no clear separation between contaminated and control areas, being samples from the various zones relatively scattered in the multivariate space with the exception of those from the intermediately contaminated zone XB3 (Fig. 2). The MRPP was used to test for differences in the community composition of the different zones and provide statistical support to the ordination approach (data not shown). Results showed that the within-group variation was small for XB3 but relatively large for the other zones. Significant differences based on pairwise comparisons were only found between zone XB3 and the control and XB1 zones (data not shown).
Calculated diversity indexes based on the various T-RF outlined potential ecological trends within the area of study (Table 3). In general, richness, Shannon’s diversity, and Pielou’s evenness were found to be lower in the most contaminated zone (XB1) compared to the other areas, particularly with respect to zone XB3 (Table 3). The functional organization index (Fo) was similar among the different zones, but marginally higher at XB1 and XB2 compared to that in XB3 and the control zone (Table 3).
Correlation Analysis
Bivariate correlation analysis showed significant trends between the different variables studied (Table 4). AlkB gene copy numbers were positively correlated with microbial biomass and microbial respiration, while no trend was found with the total hydrocarbon content and total concentrations of relevant inorganic elements associated with the contamination (Table 4). Microbial biomass, and T-RF-based richness, diversity, and evenness were positively correlated with soil pH and negatively correlated with total concentrations of hydrocarbons, As, Cu, Pb, and S, whereas soil respiration was positively correlated with total concentrations of both organic and inorganic contaminants (Table 4).
Clone Libraries and Cluster Analysis
In order to assess the diversity of alkB at the phylogenetic level, three clone libraries representative of each degree of contamination and each containing 44 clones were constructed. Rarefaction curves indicated that the number of clones sequenced was clearly sufficient to capture most of the diversity present in the highly contaminated zone. Contrarily, rarefaction curves for the control and intermediately contaminated zones did not reach a plateau indicating that alkB bacterial diversity is substantially greater in those areas (Fig. 3), confirming results obtained from the diversity indices calculated based on the T-RFLP data.
Results from the cluster analysis showed very little redundancy between the three clone libraries with only one sequence showing no differences in amino acid composition present in more than one library (sequence represented by clone XB1_2 appearing in the highly and intermediately contaminated zones; Fig. 4). Sequence redundancy differed significantly within each library. In the control zone, there were two sequences accounting for 25% of all clones (sequences represented by clones CB_5, and CB_2 and CB_13; Fig. 4). In contrast, in the highly contaminated zone, four sequences accounted for 79.5% of all clones (XB1_15; XB1_ 5; XB1_2; Fig. 4). In the intermediately contaminated zone, sequence redundancy was moderate with six sequences accounting for 34% of all clones (Fig. 4).
Phylogenetic, bootstrapped amino acid-based PHYLIP tree of clone sequences of alkB obtained from clone libraries representing noncontaminated (C B ), highly contaminated (X B1 ), and intermediately contaminated zone (X B2 ). Asterisk the following clones shared 100% amino acid sequence identity: CB_5 with CB_18, CB_23, CB_24, CB_26, CB_28, CB_35, CB_43, CB_51 and CB_61; XB1_2 with XB1_6, XB1_26, XB1_54, XB1_62, XB1_95, XB1_99, XB1_100, XB1_122 and XB2_67; XB1_4 with XB1_22, XB1_50, XB1_51, XB1_59, XB1_61, XB1_64, XB1_84, XB1_88 and XB1_92; XB1_15 with XB1_28, XB1_32, XB1_34, XB1_49, XB1_53, XB1_86, XB1_103, XB1_104, XB1_107, XB1_108, XB1_109, XB1_113 and XB1_120
Phylogenetic analysis highlighted substantial differences in the community structure of the three zones studied; while the control zone was clearly dominated by alkB sequences most closely related to those of known Gram-positive bacteria, clone libraries from the contaminated zones were entirely composed of alkB sequences most closely affine to those of known Gram-negative bacteria (Fig. 4). Most clones from the control zone were found within cluster IIB_2c2 (82% of all clones from the control area); these were most closely related to alkB sequences from Gram-positive bacteria, mainly Mycobacterium, but also Nocardia and Conexibacter (between 70% and 85% amino acid identity). The remaining 18% was found within other clusters and showed highest similarity to the alkB sequences of Burkholderia xenovorans LB400 (cluster IIB_2c1, Fig. 4), Solimonas sp. S10-1 (cluster IIB_2a, Fig. 4) and the uncultured bacterium ACB11549.1 (cluster IIA, Fig. 4). Clones from the highly contaminated zone were mostly found within one subcluster (64% of all clones; cluster IIB_I, Fig. 4) with highest sequence identity to that of Acinetobacter-like strains (approximately 72% identity) and Marinobacter aquaeolei VT8 (76% identity). The remaining 36% was most affine to the alkB sequence from Alcanivorax dieselolei (75% identity; cluster IIB_2b; Fig. 4). Approximately 60% of all clones from the intermediately contaminated zone was found within cluster IIA (Fig. 4). Sequences from these clones shared little amino acid identity with known alkB sequences (approximately 60% identity with Pseudovibrio sp. JE062), but 85–95% similarity with the alkB sequence from an unknown uncultured bacterium (ACB11549). The rest of clones from this zone showed highest affinity to alkB sequences from Solimonas sp. S10-1 (cluster IIB_2a; Fig. 4) and Acinetobacter sp. ADPI (cluster IIB_1; Fig. 4). A group of three clones from this zone formed a separate cluster (cluster I; Fig. 4) with very little sequence identity to alkB sequences from known organisms (only 55% with Microscilla marina ATCC 23134; Fig. 4) and other unknown uncultured bacteria (65% identity with uncultured bacterium ACB11553; Fig. 4).
Discussion
Abundance of alkB
Given the long-term exposure of the affected area to the contamination source and the fact that the hydrocarbon composition of the acid resin was amenable to biotransformation by alkB-harboring bacteria (chain length between C10–C16 and may be even longer-chain alkanes; [6]; Fig. 1), we expected to find higher alkB copy numbers in the contaminated zone compared to the control locations.
However, our results indicated, that the abundance of alkB in the soil samples was similar in contaminated and control zones. Recently, Powell et al. [42] found similar abundance of alkB gene copy numbers in fuel affected and nonaffected subantarctic soils using the same primers as in this study. Genes related to hydrocarbon degradation seem to be common in the environment, including areas not affected by spills or hydrocarbon contamination [29, 43, 44]. This might be due to the fact that hydrocarbons are naturally produced by plants (e.g., waxes of the cuticle) and microorganisms and, consequently, are ubiquitous in the environment. In addition, many groups of hydrocarbon-degrading bacteria can use other substrates as C sources and therefore do not solely rely on hydrocarbons for survival and proliferation. It should be noted that in this study and in those mentioned above, no transcript analysis was done, and thus, results may not completely reflect activity of hydrocarbon-degrading microorganisms. GC–MS chromatograms of the different zones, however, suggest that there is on-going biodegradation of acid resin hydrocarbons in the affected areas surrounding the acid resin deposit (Fig. 1).
Soil respiration both in absolute terms and normalized with microbial biomass (equivalent to the metabolic coefficient) was higher in the contaminated zones compared to the control sites; positive correlations were found between both normalized and non-normalized respiration rates and alkB copy numbers. Increased respiration rates after oil spills indicate successful hydrocarbon mineralization [17, 45, 46]. However, a high metabolic coefficient also indicates low turnover rates and thus might be also indicative of stress [47, 48]. For instance, the presence of metals or metalloids in the soil might require additional metal-tolerance mechanisms such as energy-dependent efflux of ions by hydrocarbon-degrading microorganisms [49]. The co-contamination with metals and metalloids could therefore limit proliferation of hydrocarbon-degrading bacteria in the contaminated zones. A contribution of both mechanisms to enhanced respiration rates, that is, hydrocarbon metabolism and detoxification, cannot be discarded. The lack of good correlations between microbial biomass measurements (Nmic) and respiration rates and alkB copy numbers might indicate that hydrocarbon-degrading microorganisms as evaluated by alkB are not representative of the total microbial population in soil (Table 4). Recent studies reported that hydrocarbon-degrading microorganisms did not dominate the soil microbial community at hydrocarbon-impacted sites [42, 50], despite no inorganic co-contamination.
T-RFLP-Based Molecular Fingerprints
Molecular fingerprints, in particular T-RFLPs, have been widely used to assess the impact of different stresses on the structure of the soil microbial community [51]. Nonetheless, most studies were based on fingerprints from the 16S rRNA gene [30, 52–54]. As hydrocarbon-degrading populations may not be dominant members of the soil microbial community, even at hydrocarbon-impacted sites [50], we used a T-RFLP approach based on alkB. Our results showed that in most cases, fingerprinting profiles were dominated by a limited number of T-RFs (between 1 and 5 accounting for 80% of the total number of T-RFs). In addition, a substantial number of T-RFs (21 out of 39) were unique to specific samples, although their relative contribution to the total number of T-RFs was usually small (2–3%). Despite the contamination and the different nature of the hydrocarbons present at impacted and control zones, multivariate analysis of the fingerprinting profiles did not show a neat separation of samples into their original groups (areas where they were sampled) (Fig. 2), although there was a certain degree of discrimination between zones, as indicated by some aggregations (e.g., zones XB3 and CB, Fig. 2). Using T-RFLP fingerprints of alkB, Powell et al. [42] were also unable to discriminate between hydrocarbon-contaminated and noncontaminated sites. However, we were able to calculate different biological indexes for the 20 locations studied, which showed interesting and consistent trends. Richness and Shannon’s diversity were generally lower in the most contaminated zone compared to the rest, particularly in the case of the control and the XB3 zones (Table 3). We found negative correlations between both richness and diversity and total hydrocarbon and total inorganic elements; in addition, positive correlations were found between richness and diversity and soil pH (Table 4). This may indicate that co-contaminants and other relevant soil properties may be important for shaping the diversity of the hydrocarbon-degrading community. Shi et al. [55, 56] suggested that metals (Cr and Pb) mainly affected microbial activity, but hydrocarbons were the driving force for changes in the microbial community in their study; nonetheless, the authors did not specifically target hydrocarbon-degrading microorganisms but the whole soil microbial community. Although hydrocarbon-degrading populations have been reported to proliferate in response to hydrocarbon contamination [5, 52], Kuhn et al. [11] found higher diversity of alkB homologous genes in pristine than in contaminated sediments. A reduction in diversity of alkB-bearing organisms might be achieved both through enrichment of particularly successful ecotypes which become dominant and toxicity of less fitted ecotypes [57]. Results from Pielou’s evenness also point toward a more skewed and thus less even alkB community in the highly contaminated zone (XB1) in favor of dominant alkB ecotypes. The functional organization index (Fo), which also reflects the ability of a community to organize in an adequate distribution of dominant and other less frequent members (a value of 0.45 is considered optimal in a scale from 0 to 1; [34]), further supports the abovementioned results.
Phylogenetic Diversity of alkB Homologous Genes in Soil
Results from clone libraries showed important differences between zones suggesting that the structure of the hydrocarbon-degrading community as represented by alkB was affected by the contamination (Fig. 4). Approximately 82% of the clones obtained from the control zone formed a large group (cluster IIB_2c) of sequences most similar to those of actinomycetes. These Gram-positive bacteria possess the ability to utilize short-chain and gaseous alkanes in contrast to the majority of hydrocarbon-degrading bacteria [58]. Dominance of alkB sequences related to actinomycetes has been previously reported in other uncontaminated soils [10, 17, 43], possibly owing to their metabolic versatility which includes n-alkanes and a wide range of organic compounds [7, 59]. In contrast to the control zone, all sequences from the contaminated zones were most closely linked to Gram-negative bacteria. In various studies, predominance of Gram-negative bacteria over Gram-positive bacteria following hydrocarbon contamination was observed [60, 61]. While sequence redundancy was relatively small among clones from the intermediately contaminated zone, four sequences accounted for approximately 79% of all clones from the highly contaminated zone XB1. These results agree with data from the T-RFLP fingerprints, which suggested lower diversity and evenness of alkB-bearing microorganisms in the highly contaminated zone. Nearly half of these clones were grouped in cluster II_A (Fig. 4), showing highest affinity to either Acinetobacter strains or the Gamma-proteobacterium M. aquaeolei VT8, an obligate marine hydrocarbon-degrading bacterium [62]. Notably, a substantial number of clones from both XB1 and XB2 were related to Acinetobacter-like organisms, which are well-known for their ability to degrade medium- and long-chain n-alkanes ranging from C13 to C44 [63, 64]. This might be related to the nature of the hydrocarbons present in the resin (mostly medium-chain) (Fig. 1). The remaining clones (cluster IIB_2a) were similar to another hydrocarbonoclastic marine bacterium A. dieselolei. It is also noteworthy that none of the alkB sequences described here showed a significant sequence identity to alkB from P. putida GPo1, as the occurrence of P. putida-like genes generally increases in contaminated habitats [15, 43, 65]. In addition, the presence of alkB sequences with very little similarity (55–62%), particularly in the intermediately contaminated zone, to those of both known and unknown organisms found in the databases suggests potential novel forms of this gene (Fig. 4).
These results provide new insights into the ecology of hydrocarbon-degrading bacteria under complex mixtures of contaminants at the molecular level and thus contribute to the gradual process of understanding indigenous microbial functionality and natural attenuation mechanisms in contaminated soils. Future studies should address the question whether other alkane-degrading microbes, which perform different pathways of alkane degradation and may harbor other genes different from alkB, might also contribute to the natural attenuation potential of the site, and if such genes do show similar trends to alkB or not. It might be also worth exploring the presence of hydrocarbon-degrading bacteria with metal-tolerant traits through enrichments as suggested by the dominance of certain alkB genotypes in the highly contaminated zone.
Conclusions
-
Abundance of hydrocarbon degraders in soil as quantified by alkB was similar in contaminated and control areas. Potential causes for this might be attributed to the ubiquity of many hydrocarbon degraders and their metabolic versatility to use other C sources.
-
Acid resin contamination resulted in important changes between the structure of the hydrocarbon-degrading microbial community in contaminated (exclusively Gram-negative) and uncontaminated areas (mostly Gram-positives).
-
In terms of community structure, the diversity and evenness of the hydrocarbon-degrading community were greater at intermediately contaminated and control zones compared to the highly contaminated zone, where enrichment of particularly successful ecotypes which become dominant and toxicity (hydrocarbons and metals) of less fitted ecotypes might occur.
-
Results provide evidence suggesting that there is significant natural attenuation potential at the site. Among other numerous features, the success for remediation in this area is limited by continuous contaminant leaching from the source.
References
Dawson JJC, Godsiffe EJ, Thompson IP, Ralebitso-Senior TK, Killham KS, Paton GI (2007) Application of biological indicators to assess recovery of hydrocarbon impacted soils. Soil Biol Biochem 39:164–177
Saterback A, Toy RJ, McMain BJ, Williams MP, Dorn PB (2000) Ecotoxicology and analytical assessment of effects of bioremediation on hydrocarbon-containing soils. Environ Toxicol Chem 19:2643–2652
SERDP ESTCP (2005) Expert panel workshop on research and development needs for the environmental remediation application of molecular biological tools https://www.serdp-estcp.org/News-and-Events/Conferences-Workshops/Past-ER-Workshops
Martin dos Santos VAP, Yakimov M, Timmis KN, Golyshin PN (2008) In: Diaz E (ed) Genomic insights into oil biodegradation in marine systems microbial biodegradation genomics and molecular biology caister. Academic, Norfolk, pp 269–296
Wentzel A, Ellingsen TE, Kotlar HK, Zotchev SB, Throne-Holst M (2007) Bacterial metabolism of long-chain n-alkanes. Appl Microbiol Biotechnol 76:1209–1221
van Beilen JB, Funhoff EG (2007) Alkane hydroxylases involved in microbial alkane degradation. Appl Microbiol Biotechnol 74:13–21
van Beilen JB, Li Z, Duetz WA, Smits THM, Witholt B (2003) Diversity of alkane hydroxylase systems in the environment oil gas. Sci Technol Rev IFP 58:427–440
Baek K-H, Byung-Dae Y, Hee-Mock O, Hee-Sik K (2006) Biodegradation of aliphatic and aromatic hydrocarbons by Nocardia sp H17-1. Geomicrobiol J 23:253–259
Hara A, Baik S, Syutsubo K, Misawa N, Smits THM, van Beilen JB (2004) Cloning and functional analysis of alkB genes in Alcanivorax borkumensis SK2. Environ Microbiol 6:191–197
Kloos K, Munch JC, Schloter M (2006) A new method for the detection of alkane monooxygenase homologous genes (alkB) in soils based on PCR-hybridization. J Microbiol Methods 66:486–496
Kuhn E, Bellicanta GS, Pellizari V (2009) New alk genes detected in Antarctic marine sediments. Environ Microbiol 11:669–673
Mehboob F, Junca H, Schraa G, Stams AJM (2009) Growth of pseudomonas chloritidis mutants AW-1 T on n-alkanes with chlorate as electron acceptor. Appl Microbiol Biotechnol 83:739–747
Powell SM, Ferguson SH, Bowman JP, Snape I (2006) Using real-time PCR to assess changes in the hydrocarbon-degrading microbial community in Antarctic soil during bioremediation. Microb Ecol 52:523–532
Smits THM, Balada SB, Witholt B, van Beilen JB (2002) Functional analysis of alkane hydroxylases from gram-negative and gram-positive bacteria. J Bacteriol 184:1733–1742
Whyte LG, Greer CW, Inniss WE (1996) Assessment of the biodegradation potential of psychrotrophic microorganisms. Can J Microbiol 42:99–106
Hamamura N, Fukui M, Ward DM, Inskeep WP (2008) Assessing soil microbial populations responding to crude-oil amendment at different temperatures using phylogenetic functional gene (alkB) and physiological analyses. Environ Sci Technol 42:7580–7586
Margesin R, Labbe D, Schinner F, Greer CW, Whyte LG (2003) Characterization of hydrocarbon-degrading microbial populations in contaminated and pristine alpine soils. Appl Environ Microbiol 69:3085–3092
Salminen JM, Tuomi PM, Jørgensen KS (2008) Functional gene abundances (nahAc, alkB, xylE) in the assessment of the efficacy of bioremediation. Appl Biochem Biotechnol 151:638–652
Foken T (2007) Das Klima von Bayreuth Status quo und Aufgaben für die Stadtplanung Standort Zeitschrift für Angewandte Geographie 31:150–152
AG Bodenkunde der Geologischen Landesamter un der Bundesanstalt fur Geowissenchaften und Rohstoffe in der Bundesrepublik Deutschland (1982) Bodenkundliche Kartieranleitung (Guidance for Soil Mapping) (in German) 3. Auflage, Hannover, Germany
IUSS Working Group WRB (2006) World reference base for soil resources 2nd edition. World Soil Resources Reports No 103 FAO, Rome, Italy
Kloos K, Schloter M, Meyer O (2006) Microbial activity in an acid resin deposit: biodegradation potential and ecotoxicology in an extremely acidic hydrocarbon contamination. Environ Pollut 144:136–144
Pérez-de-Mora A, Madejón E, Cabrera F, Buegger F, Fuß R, Pritsch K, Schloter M (2008) Long-term impact of acid resin waste deposits on soil quality of forest areas I contaminants and abiotic properties. Sci Total Environ 406:88–98
Pérez-de-Mora A, Madejón E, Cabrera F, Buegger F, Fuß R, Pritsch K, Schloter M (2008) Long-term impact of acid resin waste deposits on soil quality of forest areas II biological indicators. Sci Total Environ 406:99–107
Vance ED, Brookes PC, Jenkinson DS (1987) Microbial biomass measurements in forest soils: determination of Kc values and test of hypothesis to explain the failure of the chloroform fumigation-incubation method in acid soils. Soil Biol Biochem 19:381–387
Joergensen RG, Mueller T (1996) The fumigation–extraction method to estimate soil microbial biomass: calibration of the k(EN) value. Soil Biol Biochem 28:33–37
Griffiths RI, Whiteley A, O’donnell AG, Bailey MJ (2000) Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA and rRNA-based microbial community composition. Appl Environ Microbiol 66:5488–5491
Pérez-de-Mora A, Schulz S, Schloter M (2010) MPN- and real time- based PCR methods for the quantification of alkane-monooxygenase homologous genes (alkB) in environmental samples. In: Cunningham SP (ed) Bioremediation methods in molecular biology 599. Human, New York, pp 59–68
Schulz S, Peréz-de-Mora A, Engel M, Munch JC, Schloter M (2010) A comparative study of most probable number (MPN)-PCR vs real-time-PCR for the measurement of abundance and assessment of diversity of alkB homologous genes in soil. J Microbiol Methods 80:295–298
Osborn AM, Moore ERB, Timmis KN (2000) An evaluation of terminal-restriction fragment length polymorphism (T-RFLP) analysis for the study of microbial community structure and dynamics. Environ Microbiol 2:39–50
Margalef R (1958) Information theory in ecology. Gen Syst 3:36–71
Pielou EC (1966) The measurement of diversity in different types of biological collections. J Theor Biol 13:131–144
Lorenz MO (1905) Methods of measuring the concentration of wealth. Publications of the American statistical association 9:209–219
Marzorati M, Wittebolle L, Boon N, Daffonchio D, Verstraete W (2008) How to get more out of molecular fingerprints: practical tools for microbial ecology. Environ Microbiol 10:1571–1581
Birnboim HC, Doly J (1979) A rapid alkaline extraction procedure for screening recombinant plasmid. DNA Nucleic Acid Res 7:1513–1523
Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A (2003) ExPASy: the proteomics server for in-depth protein knoledge and analysis. Nucleic Acid Res 31:3784–3788
Ludwig W et al (2004) ARB: a software environment for sequence data. Nucleic Acid Res 32:1363–1371
Holland SM (2003) Analytic rarefaction 13 http://strata.uga.edu/software/anRareReadme.html
McCune B, Mefford MJ (2006) PC-ORD Multivariate analysis of ecological data version 5 Gleneden Beach Oregon USA
McCune B, Grace JB (2002) Analysis of ecological communities Gleneden Beach Oregon USA
Wang Z, Stout SA, Fingas M (2006) Forensic fingerprinting of biomarkers for oil spill characterization and source identification. Environ Forensics 7:105–146
Powell SM, Bowman JP, Ferguson SH, Snape I (2010) The importance of soil characteristics to the structure of alkane-degrading bacterial communities on sub-Antarctic Macquarie Island. Soil Biol Biochem 42:2012–2021
Luz AP, Pellizari VH, Whyte LG, Greer CW (2004) A survey of indigenous microbial hydrocarbon degradation genes in soils from Antarctica and Brazil. Can J Microbiol 50:323–333
Vázquez S, Nogales B, Ruberto L, Hernandez E, Christie-Oleza J, Lo Balbo A, Bosch R, Lalucat J, MacCormack W (2009) Bacterial community dynamics during bioremediation of diesel oil-contaminated Antarctic soil. Microb Ecol 57:598–610
Benyahia F, Abdulkarim M, Zekri A, Chaalal O, Hasanain H (2005) Bioremediation of crude oil contaminated soils a black art or an engineering challenge? Proc Saf Environ 83:364–370
Margesin R, Zimmerbauer A, Schinner F (2000) Monitoring of bioremediation by soil biological activities. Chemosphere 40:339–346
Brookes PC (1995) The use of microbial parameters in monitoring soil pollution by heavy metals. Biol Fert Soils 19:269–275
Filip Z (2002) International approach to assessing soil quality by ecologically-related biological parameters. Agr Ecosys Environ 88:169–174
Silver S, Phung LT (1996) Bacterial heavy metla resistance: new surprises. Annu Rev Microbiol 50:753–789
Paisse S, Coulon F, Goni-Urriza M, Peperzak L, McGenity TJ, Duran R (2008) Structure of bacterial communities along a hydrocarbon contamination gradient in a coastal sediment FEMS. Microbiol Ecol 66:295–305
Thies JE (2007) Soil microbial community analysis using terminal restriction fragment length polymorphisms. Soil Sci Soc Am J 71(2):579–591
Coulon F, McKew BA, Osborn AM, McGenity TJ, Timmis KN (2007) Effects of temperature and biostimulation on oildegrading microbial communities in temperate estuarine waters. Environ Microbiol 9:177–186
Röling WFM, Milner MG, Jones DM, Lee K, Daniel F, Swannell RJP, Head IM (2002) Robust hydrocarbon degradation and dynamics of bacterial communities during nutrient-enhanced oil spill bioremediation. Appl Environ Microbiol 68:5537–5548
Winderl C, Anneser B, Griebler C, Meckenstock RU, Lueders L (2008) Depth-resolved quantification of anaerobic toluene degraders and aquifer microbial community patterns in distinct redox zones of a tar oil contaminant plume. Appl Environ Microbiol 74:792–801
Shi W, Becker J, Bischoff M, Turco RF, Konopka AE (2002) Association of microbial community composition and activity with lead, chromium, and hydrocarbon contamination. Appl Environ Microbiol 68:3859–3866
Shi W, Bischoff M, Turco R, Konopka A (2005) Microbial catabolic diversity in soils contaminated with hydrocarbons and heavy metals. Environ Sci Technol 39:1974–1979
Atlas RM (1984) Use of microbial diversity measurements to assess environmental stress. In: Klug MJ, Reddy CA (eds) Current perspectives in microbial ecology. American Society for Microbiology, Washington DC, pp 540–545
Hamamura N, Yeager CM, Arp DJ (2001) Two distinct monooxygenases for alkane oxidation in Nocardiodes sp strain CF8. Appl Environ Microbiol 67:4992–4998
Vomberg A, Klinner U (2000) Distribution of alkB genes within n-alkane-degrading bacteria. J Appl Microbiol 89:339–348
Kaplan CW, Kitts CL (2004) Bacterial succession in a petroleum land treatment unit. Appl Environ Microbiol 70(3):1777–1786
Stephen JR, Chang Y, Gan YD, Peacock A, Pfiffner SM, Barcelona MJ, White DM, Macnaughton SJ (1999) Microbial characterization of a JP-4 fuel-contaminated site using a combined lipid biomarker/polymerase chain reaction–denaturing gradient gel electrophoresis (PCR–DGGE)-based approach. Environ Microbiol 1:231–241
Huu NB, Denner EBM, Ha DTC, Wanner G, Stan-Lotter H (1999) Marinobacter aquaeolei sp nov a halophilic bacterium isolated from a Vietnamese oil-producing well. Int J Syst Bacteriol 49:367–375
Maeng JH, Sakai Y, Tani Y, Kato N (1996) Isolation and characterization of a novel oxygenase that catalyzes the first step of n-alkane oxidation in Acinetobacter sp strain M-1. J Bacteriol 178:3695–3700
Sakai Y, Maeng JH, Tani Y, Kato N (1994) Use of long-chain n-alkanes (C13-C44) by an isolate Acinetobacter sp M-1. Biosci Biotechnol Biochem 58:2128–2130
Heiss-Blanquet S, Benoit Y, Maréchaux C, Monot F (2005) Assessing the role of alkane hydroxylase genotypes in environmental samples by competitive PCR. J Appl Microbiol 99:1392–1403
Acknowledgments
Dr. Pérez-de-Mora thanks the Spanish Ministry of Education and Science (MEC) for the fellowship.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Figure S1
Relative T-RF patterns of the alkB gene in soil samples from the different zones of contamination (small letters a–e indicate field replicates from the corresponding sites). C B control zone, X B1 highly contaminated zone, X B2 and X B3 intermediately contaminated zones (RTF 372 kb)
Rights and permissions
About this article
Cite this article
Pérez-de-Mora, A., Engel, M. & Schloter, M. Abundance and Diversity of n-Alkane-Degrading Bacteria in a Forest Soil Co-Contaminated with Hydrocarbons and Metals: A Molecular Study on alkB Homologous Genes. Microb Ecol 62, 959–972 (2011). https://doi.org/10.1007/s00248-011-9858-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-011-9858-z