Abstract
The effect of nutrient and inocula amendment in a bioremediation field trial using a nutrient-poor Antarctic soil chronically contaminated with hydrocarbons was tested. The analysis of the effects that the treatments caused in bacterial numbers and hydrocarbon removal was combined with the elucidation of the changes occurring on the bacterial community, by 16S rDNA-based terminal restriction fragment length polymorphism (T-RFLP) typing, and the detection of some of the genes involved in the catabolism of hydrocarbons. All treatments caused a significant increase in the number of bacteria able to grow on hydrocarbons and a significant decrease in the soil hydrocarbon content, as compared to the control. However, there were no significant differences between treatments. Comparison of the soil T-RFLP profiles indicated that there were changes in the structure and composition of bacterial communities during the bioremediation trial, although the communities in treated plots were highly similar irrespective of the treatment applied, and they had a similar temporal dynamics. These results showed that nutrient addition was the main factor contributing to the outcome of the bioremediation experiment. This was supported by the lack of evidence of the establishment of inoculated consortia in soils, since their characteristic electrophoretic peaks were only detectable in soil profiles at the beginning of the experiment. Genetic potential for naphthalene degradation, evidenced by detection of nahAc gene, was observed in all soil plots including the control. In treated plots, an increase in the detection of catechol degradation genes (nahH and catA) and in a key gene of denitrification (nosZ) was observed as well. These results indicate that treatments favored the degradation of aromatic hydrocarbons and probably stimulated denitrification, at least transiently. This mesocosm study shows that recovery of chronically contaminated Antarctic soils can be successfully accelerated using biostimulation with nutrients, and that this causes a change in the indigenous bacterial communities and in the genetic potential for hydrocarbon degradation.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Petroleum hydrocarbon contamination is a serious environmental problem all over the world including the Antarctic continent. Antarctica suffers from chronic contamination, introduced during the times where there was no legislation about waste disposal, and from the impact of current human activities (such as scientific stations, fisheries, and tourism). These activities require the transport and storage of diesel oil and other oil-derived fuels to be used in the generation of electric power and heat, determining a high risk of pollution. Accordingly, several contamination problems have been reported from many Antarctic stations [3]. At the Jubany Argentinean Scientific Station, the constant transport and use of diesel and other fuels (JP1, naphtha) has resulted in chronic hydrocarbon contamination of several areas [22]. This occurs especially at the vehicle transit paths, pipes for fuel transport, boat shoring sites, and in areas of fuel tank replenishment and storage. Also, these activities imply the risk of sudden acute contaminations in case of accidental spills.
The Protocol on Environmental Protection to the Antarctic Treaty [4] states that the generators of wastes and the users of polluted sites must assume the cleanup of the fore mentioned contaminations. In that sense, all the countries managing stations or carrying out any kind of activities in Antarctica should develop strategies and have the means to remove contaminants from the sites they manage and use [46].
Bioremediation has emerged as one of the most important tools to eliminate or reduce the contamination caused by diverse compounds of anthropogenic origin that are spilled into the environment, either accidentally or deliberately [12]. It is known that the extent of biodegradation can be limited by the low availability of nitrogen and phosphorous and the concomitant unbalance in the C:N:P ratio caused by the high carbon levels provided by the spilled hydrocarbons [1]. These conditions reduce the capacity of microbes to form biomass from the carbon source represented by the pollutant, and often occur in Antarctic soils where the natural levels of nitrogen and phosphorous are frequently low [40, 47]. In consequence, to make a bioremediation process successful, appropriate quantities of the inorganic P and N should be applied by fertilizing with a source of these nutrients (biostimulation) in order to enhance the growth of hydrocarbon-degrading bacteria [18, 33]. On the other hand, the success of the addition of pollutant-degrading bacteria (bioaugmentation) is ambiguous, and thus it should be tested for each system, as environmental conditions, soil characteristics, or the effect of predation and competition cannot be inferred in advance. Therefore, inoculation of polluted soils with previously isolated hydrocarbon-degrading bacteria is considered as an alternative when the autochthonous microbiota of contaminated areas under treatment is scarce or inadequate to metabolize the pollutants [51]. However, it should be taken into account that, regarding bioremediation in Antarctica, only indigenous bacteria can be used for bioaugmentation as the Antarctic Treaty states that either allochthonous or genetically modified organisms cannot be introduced in the Antarctic continent.
The presence of hydrocarbons in soils selectively promotes the growth of those microorganisms capable of utilizing them [36]. Thus, in chronically contaminated soils, the microbiota changes towards the dominance of hydrocarbon degraders, with respect to the natural microbiota of pristine soils [42]. In that sense, the changes that might occur in the composition of those adapted microbial communities as a result of a bioremediation process involving different treatments are something to be analyzed. This should occur before the implementation of the process at full scale to minimize the risk of a big shift in community structure and diversity. Additional studies are also needed to understand the ecological significance of these shifts and how long it might take for the initial microbial community to recover after the soil is bioremediated to its maximum extent. In this way, it is useful to study the diversity of a soil microbial community at a particular site, to evaluate the effects that anthropogenic contaminants or exogenously added autochthonous or allochthonous bacteria might have on the structure of that community.
Few studies report the effect of oil spills on soil microbial communities in Antarctica, describing mostly the changes observed by culture-dependent methods in bacterial counts [2, 9] or identifying the predominant bacterial groups [36]. At present, no studies using culture-independent methods to evaluate the effect that an on site bioremediation process carried out in land plots has on the composition and structure of microbial communities in Antarctic soils have been reported. Besides, there are no reports to date on the effect of the addition of bacterial consortia to bioremediation plots in Antarctica at a mesocosm scale. In a land-plot mesocosm approach, drainage of water and nutrients and free-living predators and competitors affect the systems more than in land-isolated microcosms, even if they were left on site. In this work, an on site bioremediation assay was performed in mesocosms prepared in plots with soil chronically polluted with diesel oil from Jubany Station, Antarctica. Our aim was to analyze the effect of biostimulation and bioaugmentation with two autochthonous bacterial consortia (one isolated from the same site and another from a different location in Antarctica) on the hydrocarbon-biodegradation activity of the bacteria present in a chronically polluted Antarctic soil. We focused on the hydrocarbon removal throughout the assay and the changes occurred in bacterial diversity and structure of communities.
Methods
Study Area and Soil Collection
Studies were carried out during the Antarctic summer (January–March 2003) at the Argentinean Antarctic Scientific Station Jubany [62°14′S, 58°40′W, located in Potter Peninsula, King George Island (Isla 25 de Mayo), South Shetland Islands]. Jubany Station has some limited areas under pollution risk due to the manipulation of jet fuel and diesel oil used for transportation and management of the station [22]. Soil was collected with a shovel from the upper 50 cm of the soil layer in an area close to the station facilities. Next, the soil was thoroughly mixed and weighed to distribute the same amount in each mesocosm. This soil was chronically contaminated with diesel fuel spilled during a pipe leak produced 2 years before this experiment was undertaken. The weather conditions during the experiment had been predominantly cloudy days with abundant rains and snowfalls. The fluctuations in pressure, temperature, relative humidity, and wind speed are shown in Figure S1 in the Electronic Supplementary Material.
Mesocosm Design and Sampling
The experiment was performed in ten 1-m2 land plots, each containing 50 kg of soil (5 cm deep), placed in an isolated area separated one from the other by a distance of around 2 m and marked by using stainless steel poles. The plots were delineated by wood enclosures firmly settled into the ground. After that, the soil inside the enclosures was removed to give the desired height and the plots were filled with the contaminated soil. Plots were arranged according to a randomized block design of three treatments distributed in three blocks plus one control. All plots were exposed to the same environmental conditions. The response of natural soil bacterial communities to nutrient amendment and the additional effect of bioaugmentation with two different bacterial consortia were tested. Untreated control plot (CC) was used as control for the autochthonous community. Plots designated as AB, M10, and J13 were biostimulated with 27.6 g N and 2.9 g P per plot, to give an initial C:N:P ratio of approximately 100:10:1 (considering the carbon content of the hydrocarbons present in the soil), by adding NH4NO3 and KH2PO4/Na2HPO4 as nitrogen and phosphorous sources. Plots M10 and J13 were also bioaugmented with the bacterial consortia M10 and J13, respectively. Consortium M10 was obtained from chronically contaminated soil from the surroundings of diesel fuel storage tanks at Argentinean Antarctic Station Marambio (64°14′S, 56°37′W). Enrichment was made after repetitive culturing in basal medium containing a mixture of phenanthrene, anthracene, fluorene, and dibenzothiophene as carbon sources [39]. J13 consortium was obtained by the same means from chronically contaminated soil taken from the pools below the diesel fuel tanks in Jubany Station. Both consortia are able to degrade the main aliphatic and aromatic hydrocarbons present in the fuel contaminating the studied soil [24]. Inocula for soil bioaugmentation were prepared from 8-day cultures on saline basal medium [13] with diesel oil (2% v/v) as carbon source and incubated at 15°C and 250 rpm. Bioaugmented plots were inoculated with cell suspensions to reach a cell density of approximately colony-forming units per gram of soil dry weight 5 × 106 CFU g−1 dw.
Every 8 days and for a period of 48 days (T0 to T6), plots were aerated by mixing the soil with a shovel. Also, 90 ml of a nutrient solution containing 11.3 g N and 1.2 g P were sprayed onto biostimulated plots to compensate the washing of nutrients due to rain and snowfall. Before the addition of nutrients, two replicate samples per plot were collected. Each replicate contained 250 g of soil and was composed of five subsamples taken randomly that were pooled, mixed, and placed in an aluminum pot previously treated overnight at 450°C. Each subsample consisted of a cylindrical portion of soil which extended from surface to 5 cm deep. A portion of 5 g of each replicate soil sample was used separately for pH, moisture, and determination of culturable bacterial counts. Portions of 10 g were pooled and stored in sterile glass vials at −20°C for bacterial community analysis by molecular methods. The rest of the samples were kept at −20°C for total hydrocarbon concentration measurement, which were done on T0, T3, and T6 samples. The first sample, T0, was taken 1 day (24 h) after the preparation of the mesocosms.
Soil Analysis
Water content in soil samples was determined gravimetrically after desiccation at 105°C for 48 h. The pH was measured on the supernatant of a suspension of 1 g of soil in 10 ml of diluent (8.5 g l−1 NaCl in water), after vortexing for 5 min and letting the soil particles settle. Total hydrocarbon concentration (THC) in soil samples were measured following the EPA 418.1 method [50]. Briefly, approximately 1 g of soil was accurately weighed and placed into a 20-ml glass vial with a metal-covered hermetic cap. Then, 10 ml of HPLC-grade CCl4 and a spatula tip of anhydrous Na2SO4 were added to each flask. The flasks were placed into an ultrasonic bath and treated overnight. After that, the samples were transferred to a quartz cell and analyzed directly in a Buck Model HC 404 hydrocarbon analyzer IR spectrometer.
Enumeration of culturable bacteria was performed by the plate count method. One gram of soil was suspended in 10 ml of the same diluent as for pH measurement and vortexed for 3 min. Once the soil particles settled down, 10-fold serial dilutions of the supernatant were made and 100 μl of each dilution were spread (in duplicate) on Petri dishes containing the appropriate media. Plates were incubated at 15°C and the number of CFU g−1 dw was evaluated after 15 days and 1 month. Enumeration of culturable heterotrophic aerobic bacteria (HAB) was performed on casein–peptone–starch agar, as was suggested by Wynn-Williams [53] for Antarctic soils analysis. For hydrocarbon-degrading bacteria (HDB), saline basal medium [13] supplemented with 1% diesel oil as sole carbon source was used.
DNA Extraction from Soil
Total community DNA was extracted using a modified version of the protocol described by Nogales et al. [28]. Approximately 2 g of soil were placed in 15-ml sterile plastic tubes containing 2.4 ml of extraction buffer (0.1 M Tris–HCl pH 8.0, 0.1 M EDTA, 0.1 M phosphate buffer pH 8.0), 0.4 mg proteinase K, and 7.2 mg lysozyme and incubated at 37°C for 10 min. After that, sodium dodecyl sulfate (SDS) was added (2% w/v final concentration) and samples were incubated at 37°C for 15 min. Then, NaCl was added (1.5 M final concentration) and tubes were mixed by inversion before the addition of a preheated CTAB solution (0.8% w/v, final concentration). Samples were heated at 65°C for 15 min in a water bath and then subjected to three cycles of freezing on liquid nitrogen and thawing at 65°C. Finally, samples were centrifuged at 6,000 ×g for 10 min to separate soil particles and the supernatant was transferred to a sterile 15-ml plastic tube and kept on ice. A second extraction was performed by adding 800 μl of extraction buffer and SDS to the soil pellet, mixing by inversion and incubation at 65°C for 10 min. Following incubation, samples were centrifuged as described before and both supernatants were pooled and extracted twice with an equal volume of phenol:chloroform:isoamyl alcohol (25:24:1). The aqueous phase was transferred to a sterile tube and 0.1 vol of 3 M sodium acetate (pH 4.8) and 0.7 vol of isopropanol were added. Extracts were incubated for 10 min at room temperature and centrifuged for 30 min at 13,000 ×g. DNA pellets were washed with 70% ethanol, air-dried, and resuspended in 500 μl of sterile milli-Q water. Further purification of the soil DNA extracts was done with the Wizard® SV Genomic DNA Purification System (Promega) following the manufacturer’s instructions. Genomic DNA quality and quantity was verified by agarose gel electrophoresis and staining with ethidium bromide.
PCR Amplification and T-RFLP Profiling of Bacterial Community
The structure of bacterial communities in soils from the different treatments, as well as that of the inoculated consortia, was analyzed by terminal restriction fragment length polymorphism (T-RFLP) of amplified 16S rRNA genes (16S rDNA). Soil samples were analyzed from the control plot (CC) and three plots from each treatment (AB, M10, and J13) at three different times along the experiment, T0, T3, and T6 (see Figure S1 of the Electronic Supplementary Material). Bacterial 16S rDNAs were amplified from total community genomic DNA by polymerase chain reaction (PCR) using primers 27f [19] and 1389r [29], labelled at the 5′-end with the phosphoramidite dyes 6-FAM and HEX, respectively (Applied Biosystems and Invitrogen). PCR amplifications were done as described by Osborn et al. [29] in a Mastercycler personal thermal cycler (Eppendorf). PCR products from two replicate reactions were pooled and purified using PureLink™ PCR Purification Kit (Invitrogen) to remove salts, unincorporated nucleotides, and labelled primers. Aliquots of purified PCR products (10 μl) were digested separately with 20 U of restriction endonucleases AluI and CfoI (Roche Applied Science) in a total volume of 15 μl for 3 h at 37°C [29]. Restriction fragments were mixed with a ROX-labelled size standard GeneScan 500 (Applied Biosystems), denatured by heating at 96°C for 5 min in formamide, and loaded in an ABI 310 Genetic Analyzer (Applied Biosystems) for electrophoretic separation. From each digested DNA, two replicate T-RFLP profiles were obtained. Electrophoretic patterns were analyzed with the GeneScan software v.3.1 (Applied Biosystems). Profiles were checked for possible artifacts and incorrect peak determination. Fluorescence data were standardized as described by Fahy et al. [14] to compensate for different amounts of DNA loaded on the genetic analyzer. Replicate profiles were aligned to create consensus profiles using the web-based program T-Align [44]. Sample consensus profiles were then compared in order to generate a comparison profile among all the samples, listing all the terminal restriction fragments (T-RFs) observed and the relative abundance (fluorescence) in the different samples analyzed [44], which was used for further analyses.
Comparison of T-RFLP Profiles and Statistical Analyses
For comparisons of 16S rDNA T-RFLP profiles from bacterial communities in different soil samples, we used the proportional area of T-RFs as indicator of the proportional abundance of the bacterial populations originating the terminal fragments. The processed data from the two separate restriction digestions (AluI and CfoI) and from the two end fragments of 16S rRNA genes (5′-T-RFs and 3′-T-RFs) were combined to obtain a matrix with a unique description for each soil sample. The relative abundance data in the matrix were log transformed and used to compute the distance values between profiles based on the Bray–Curtis coefficient. Distance values were used to calculate dendrograms by hierarchical clustering analysis using the unweighted pair-group moving average method and to ordinate profiles by non-metric multidimensional scaling (NMDS) analysis using the software PAST—Paleontological Statistics v.1.29 [17]. The statistical comparison of the T-RFLP profiles using analysis of similarity (ANOSIM) test (one-way) was also performed with the PAST software. The results of bacterial counts and hydrocarbon concentration data from the different mesocosms were analyzed by one-way analysis of variance and Tukey–Kramer multiple comparison test, using the InfoStat software, version 2004 (Grupo InfoStat, FCA, Universidad Nacional de Córdoba, Argentina).
Detection of Hydrocarbon Catabolic Genes by Dot-Blot Hybridization
DNA extracts from soil samples were blotted onto nylon membranes (Hybaid) using a MilliBlot™-D vacuum manifold (Millipore) according to the manufacturer’s instructions. Hybridizations with probes for different catabolic genes were done following standard procedures as described by Sambrook and Russell [41] at a low stringency temperature (37°C). Gene-specific probes for Pseudomonas-like naphthalene dioxygenase (nahAc), catechol 2,3-dioxygenase (nahH), and catechol 1,2-dioxygenase (catA) were prepared from genomic DNA of strain Pseudomonas stutzeri AN10 [35]. Probes for alkane monooxygenase gene (alkB) were prepared from genomic DNA of Pseudomonas putida GPo1 [43]. An additional probe for nitrous oxide reductase gene, nosZ, was used (this probe was obtained from P. stutzeri AN10). Finally, a probe for 16S rRNA gene was used as control. This probe was obtained using a mixture of genomic DNAs as template in the PCR reaction. These genomic DNAs were obtained from a collection of phylogenetically distinct isolates, most of them pseudomonads. Probes were generated by PCR amplification following protocols described previously [8, 15, 16, 45]. Probe labelling and chemoluminescent detection were carried out with the ECL Direct Nucleic Acid Labelling and Detection System (GE HealthCare) according to the manufacturer’s instructions. Signal intensities in dots obtained for the different gene probes were quantified using the GeneTools analysis program (SynGene) and were given as relative values to the signal obtained for the 16S rRNA probe.
Results
Soil Characteristics and Bacterial Counts
The soil used in the bioremediation trial in mesocosms had an initial total hydrocarbon concentration of 5.95 mg g−1, a HAB of 9.6 × 106 CFU g−1 dw, and a HDB of 8.2 × 104 CFU g−1 dw. These results showed that there were indigenous bacteria in the soil capable of growing on hydrocarbons. In bioaugmented plots, 4.9 × 106 and 6.8 × 106 CFU g−1 dw per plot of consortia M10 and J13, respectively, were added, resulting in estimated bacterial concentrations in the soil of approximately 1.5 × 107 CFU g−1 dw. Soil pH ranged between 6.0 and 7.5 during the time of the experiment, remaining lower in the treated plots than in the control (CC) probably due to the addition of nutrients. The water content was highly variable (between 1.5% and 48%) because of fluctuations in weather conditions (wind, rain, snow and blizzards) (see Figures S1 and S2 of the Electronic Supplementary Material). Therefore, to compensate the effect of the differences in soil moisture, bacterial counts and THC values were expressed per gram of dry soil.
At the beginning of the experiment (T0, 24 h after preparation of soil plots), HAB in the control (CC) and the biostimulated soil plots (AB) were 7.3 × 107 and 1.0 × 108 CFU g−1 dw, respectively, while for the bioaugmented plots, values of 7.4 × 107 CFU g−1 dw (M10) and 3.5 × 108 CFU g−1 dw (J13) were measured. Nevertheless, all values showed no significant differences (p > 0.05). HAB in biostimulated and bioaugmented plots increased during the experiment up to 2.3 × 109 CFU g−1 dw (AB), 1.4 × 109 CFU g−1 dw (M10), and 8.9 × 108 CFU g−1 dw (J13) at d 48 (T6), without significant differences between them. On the contrary, HAB counts in the CC plot did not change significantly during the experiment (1.1 × 108 CFU g−1 dw at d 48). Increases in HDB (Fig. 1) were pronounced, showing that quantitative changes in hydrocarbon-degrading populations were in part responsible for the increase in the bacterial counts in treated plots. This was reflected by the increase in the number of degrading bacteria relative to total heterotrophic aerobic bacterial counts (HDB/HAB ratio). This ratio was higher between T3 and T5, reaching values from 17.5 and 26.8 in treated plots. Conversely, in CC, this ratio fluctuated between 0.2 and 2.7 during the trial (from T0 to T6), these values being similar to those registered in the treated plots at T0. It is important to mention that no significant differences were observed either in HAB or HDB counts when plots under biostimulation were compared with those where biostimulation was combined with bioaugmentation. From this point of view, for the purpose of bacterial counting interpretations, the three treatments analyzed (AB, M10, and J13) behaved in the same way. HDB counts in control samples (CC), although also increasing during the duration of the experiment, were always about two orders of magnitude lower than those in the treated plots. During the 24-h period elapsing between the preparation of land plots and the first sampling (T0), a 7-fold increase in culturable bacteria and a 1.5-fold increase in hydrocarbon-degrading bacteria were observed in CC. This was presumably due to the soil manipulation carried out. This increase was still more pronounced in the biostimulated plots (around 10-fold in HAB and around 9-fold in HDB). THC detected in the soil sampled from the plots ranged from 5.95 mg g−1 (T0) to 2.05 mg g−1 (AB at T6). As can be seen in Fig. 1, no significant decrease in THC (p > 0.05) was observed by d 24 (T3) in the control plot, while all treated plots registered a significant decrease of 14–18% in THC (p < 0.01). At the end of the experiment (T6, d 48), the percentage of hydrocarbon removal in the control plot was only 10% while the treated plots ranged between 52% (M10) and 65% (AB) (p < 0.001), with no significant differences between the three treatments.
Bacterial Community Structure and Dynamics in Soil Plots
The structure of bacterial communities and their temporal variations were studied by T-RFLP of amplified 16S rRNA genes. Despite the natural biological variability between samples and the bias that might be introduced mainly by DNA extraction and PCR amplification, the T-RFLP profiles of soil replicates showed a very high reproducibility, since at each sampling time, electropherograms from the three replicate plots of each treatment were highly similar.
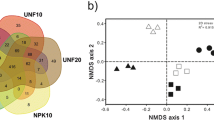
A comparison of the T-RFLP profiles obtained for control soils and those from the different treatments was done taking into account the presence and relative fluorescence of the different T-RFs observed. Cluster analysis of data sets obtained from 5′-end and 3′-end T-RFs of 16S rRNA genes (for enzymes AluI and CfoI), separately and all together, showed strong similarities in dendrogram topologies with respect to the grouping of profiles according to soil treatment and time course of the trial (data not shown). The same grouping of the T-RFLP profiles was observed in bi-dimensional NMDS plots (Fig. 2), where the differences in microbial community composition can be estimated from the distances between samples in the plot. Profiles grouped strongly according to sampling time and there were no remarkable differences between the different treatments (AB, M10, and J13), but there were differences with the control. Thus, the profiles for all samples at T0 and those for treated plots at T3 and at T6 formed three separate groups with high levels of intra-group similarity (>80%, Bray–Curtis coefficient). The analysis of similarity (one-way ANOSIM) used to compare groups of samples (Table 1) provided statistical significance to the grouping shown in Fig. 2. This analysis confirmed that the T-RFLP profiles of the treatments were not significantly different at the initial, intermediate, and final time of the bioremediation trial (group A in Table 1), and that there were significant temporal changes in bacterial populations from one sampling time to the next one (group B in Table 1). Profiles from CC plot were not directly comparable in ANOSIM to those of the treatments because there were no replicate plots for the control. Therefore, we made a comparison between the three profiles from CC plots (T0, T3, and T6) with average profiles from all treatments at the three time points (group D in Table 1). Although the differences were not significant, the R values for this comparison were indicative of a certain degree of discrimination between the samples, as seen in Fig. 2.
NMDS showing the comparison between T-RFLP profiles from soil samples from the different treatments and the community control, at the initial, intermediate, and final times of the field assay. All samples were analyzed in duplicate and the four T-RFs (5′- and 3′-T-RFs from AluI and CfoI digestions) were used to compute the Bray–Curtis similarity indices. Circles, control plot (CC); squares, biostimulated plots (AB); diamond, M10-bioaugmented plots (M10); triangles, J13-bioaugmented plots (J13). Lines encircling samples indicate profile similarity values higher than 80%
At each time of the experiment, changes in the abundance of some T-RFs occurred while some fragments appeared and others disappeared. This reflected the differences in profiles observed in soils from the different treatments and the dynamics of bacterial populations. The relative abundances of the more relevant 5′-end T-RFs obtained with the enzyme AluI are shown (as an example) in Fig. 3. To simplify the figure, only the most abundant peaks and those that changed in the different conditions (even if they were not particularly abundant) are shown. At T0, 1 day after the preparation of plots, the profiles were characterized by the presence of five abundant T-RFs and many less abundant ones, especially in CC. Some differences were observed at this time between the profiles of bacterial communities in the treated plots and those in the control. Mainly, the relative abundance of the predominant T-RF in CC (204.8) diminished and those of T-RFs 67.7 and 246.2 increased considerably, suggesting that rapid changes occurred within the first day as a consequence of aeration and addition of nutrients and consortia to soil. At T3, the relative abundance of the major T-RF in CC (204.8) diminished even more in the treated plots while increasing in abundance in CC. The T-RF 246.2 appeared in CC at this time. Minor T-RFs disappeared in the treated plots while some fragments increased slightly in abundance (T-RFs 134.4, 195.9, and 233.6) or considerably (T-RFs 228.5 and 230.9). At T6, only minor changes occurred in the treated and control plots with respect to T3.
Relative abundance of main 5′-T-RFs (AluI) obtained from T-RFLP profiles from soil samples from the different treatments and the community control, at the initial (a), intermediate (b), and final (c) times of the field assay, showing the temporal variation of bacterial communities in treated and control plots
In order to analyze if the inoculated consortia were established as members of the bacterial community in bioaugmented soil plots, T-RFLP profiles of the inoculated consortia were obtained and the presence of T-RFs characteristic for those consortia was analyzed in profiles from bioaugmented plots. The T-RFLP profiles of the inocula suggested that the bacterial composition of consortia M10 and J13 was highly similar, even when they had a different origin and their degradation efficiency seemed to be different (Vázquez et al., unpublished results). There was hardly any correspondence between the profiles of consortia and those of the soils, except for four T-RFs, highly abundant the consortia, which were observed in all soil profiles, including those of uninoculated plots (CC and AB). The fragments corresponded to 5′- and 3′-end fragments from AluI (67.7, 127.5) and CfoI digestions (204.1, 298.5). The identity of the bacterial population producing these four T-RFs has been inferred as Pseudomonas sp. after analyzing the composition of consortia by culture-independent methods (data not shown). As can be seen in Fig. 3, the relative abundance of T-RF 67.7 was considerably higher in treated plots at T0 than in control plots, and was higher in bioaugmented plots, particularly in J13 plots, consistent with the addition of consortia. Within 24 days (T3), the abundance of T-RF 67.7 decreased in treated plots to about the levels observed in control plots, remaining like that at T6.
Detection of Catabolic Genes
The genetic potential for hydrocarbon catabolism was analyzed in DNA extracts from the consortia and the different soils by using a dot-blot hybridization approach. Detection of genes involved in the degradation of linear (alkane monooxygenase gene, alkB) and aromatic hydrocarbons (naphthalene dioxygenase, nahAc; catechol 2,3-dioxygenase, nahH, and catechol 1,2-dioxygenase, catA) was tested at times T3 and T6, when hydrocarbon degradation was observed (see Fig. 1). We included in the analysis a key gene of the denitrification process, nosZ (coding for nitrous oxide reductase), as a control for a gene not involved in hydrocarbon degradation, and for two additional reasons: a nitrate salt was used for fertilization, and denitrification has been linked to hydrocarbon degradation in fertilized Antarctic soils [30]. Since the probes used for detection were originated from Pseudomonas species, we expected detection of catabolic genes similar to those found in bacteria from this genus. High hybridization signal was obtained for genes nahAc and nahH in samples from consortium M10 but only for nahAc in consortium J13, indicating that there was Pseudomonas-like genetic potential for naphthalene degradation in the consortia used for inoculation. The results of the detection of catabolic genes in the different soil samples are shown in Fig. 4. In all soils, nahAc gene was detected at high level, and there were no significant differences in the results obtained from control or treated soils. However, in treated soils at T3, there was an increase in hybridization signal for catechol dioxygenase genes nahH and catA (Fig. 4a). At the end of the experiment (T6), there were no relevant differences in the results obtained for catabolic genes in control and only biostimulated plots (AB) (Fig. 4b). In contrast, the detection levels for nahH were still high in both types of bioaugmented plots (M10 and J13). The hybridization signal for the denitrification gene, nosZ, was higher in all treated plots (especially AB and J13) at T3 when compared to control plots.
Detection of catabolic genes involved in the degradation of linear (alkane monooxygenase gene, alkB) and aromatic hydrocarbons (naphthalene dioxygenase, nahAc; catechol 2,3-dioxygenase, nahH, and catechol 1,2-dioxygenase, catA) in total DNA extracted from soil samples of the treated (AB, M10, and J13) and control (CC) plots taken at T3, after 24 days of treatment (a), and at T6, at the end of the bioremediation field assay (b)
Discussion
In in situ bioremediation of Antarctic soils, degrading bacteria have to face an intrinsic problem, i.e., the low temperatures and consequent low metabolic activities. In addition, the winds, rains, snowfalls, and temperature regimes change considerably over the year and even from one day to the other. Also, the freezing and thawing cycles of the soil influence the availability of liquid water, which is essential to support the growth and metabolism of bacteria. All the abovementioned characteristics determine a highly variable environment (Figure S1 of the Electronic Supplementary Material). All these factors represent additional stress for the soil microbiota, beyond that caused by the presence of contaminants, and might have a negative effect on the success of bioremediation treatments. However, as it was previously reported [2, 3, 10, 11], the results of this study indicate that in Antarctic chronically polluted soils, such as the one used here, there is a well-adapted microbiota able to overcome the harsh and fluctuating conditions, and still capable of decreasing the hydrocarbon concentration in soils. As we found previously in microcosms with soil from Jubany Station [36, 38], bioaugmentation with cultured hydrocarbon-degrading bacteria is a strategy that usually makes no difference compared to biostimulation-only strategy in bioremediation of chronically contaminated soils. However, it is mainly successful when applied on acutely contaminated soils, where there is not a microbiota adapted to the pollutants. Nevertheless, some authors reported that inoculation may improve the rate of elimination of hydrocarbons from polar soils [26], or at least reduce the lag phase [52]. As the success of adding inocula seems to be uncertain, we tested the addition of two different consortia previously enriched from two Antarctic soils from the same and a distinct site. This strategy of analyzing the possible site specificity of degrading autochthonous bacteria from Antarctica applied to on site field experiments was, to our knowledge, not previously reported.
The rise in bacterial counts observed between the day when the plots were prepared and the next day (first sampling, T0) could be due to aeration of soil in the case of the control, while the rise observed in the rest of the plots could have been produced also by the balance of the C:N:P ratio caused by the nutrient amendment. These results show that aeration and especially biostimulation contributed to the rapid growth of indigenous hydrocarbon-degrading bacteria present in the contaminated soil, as the rise observed in 24 h was of about one to two orders of magnitude (in control and treated plots, respectively). In the non-biostimulated control, culturable heterotrophic bacterial counts increased to a value similar to the biostimulated systems but the degrading bacterial counts increased slightly in comparison, indicating that without the addition of nutrients some bacteria, not necessarily hydrocarbon degraders, could grow using the bioavailable substrates present in soil when other environmental factors are improved (i.e., oxygen supply by aeration).
During this trial, the rise in culturable heterotrophic and degrading bacterial numbers in all biostimulated plots displayed almost the same behavior, with no major differences between the inoculated and non-inoculated treatments. This was previously observed in microcosms with chronically diesel-contaminated soil from Marambio Station [37] and from maritime Antarctica [46]. Reports by researchers working in the Arctic with chronically hydrocarbon-contaminated soils [52] and with weathered diesel-contaminated soil [48] are also in agreement. This rise in CFU numbers in treated soils seemed to be due to the selective growth of the degrading populations of the indigenous microbiota, without evidence of growth of the added inocula, as far as it can be inferred from bacterial count data and T-RFLP analysis (see below).
Part of the observed removal of hydrocarbons could be due to abiotic loss of the lighter fraction of the diesel oil, as the soil was aerated and mixed every sampling day and periods of strong winds occurred frequently during the trial. In any case, the important decrease in hydrocarbon content in treated soils, in comparison with the control, indicates that most of the hydrocarbons disappeared as a result of the treatments. In treated plots, at T3 (after 3 weeks), the growth of hydrocarbon degraders had been stimulated, but the elimination of TPH was low. From T3 to T6, the increase of hydrocarbon degraders proceeded at a lower rate but the metabolization of hydrocarbons by the active microbiota was enhanced, resulting in a final THC reduction of 52–65%. A similar level of hydrocarbon removal was also obtained by Margesin et al. [23] in biostimulated, acutely contaminated soil from an arable site in Austria and by Mishra et al. [25] in oily-sludge, chronically contaminated soils from a refinery after bioaugmentation. In this last case, a better hydrocarbon removal was observed when adding bacteria together with nutrients. This contrasts our results, which did not reflect differences in hydrocarbon reduction between treatments.
The analysis of bacterial community composition by T-RFLP of 16S rDNA allowed assessing the response of whole communities (including culturable and non-culturable bacteria) to the different treatments. Among the different techniques used at present to analyze microbial communities, T-RFLP has proved to be a valuable and sensitive method for rapidly and reproducibly comparing the relationships and changes in bacterial communities from environmental samples as well as their temporal variations [27]. In this work, the T-RFLP profiles of soil replicates showed a very high reproducibility. This result, not usual for studies dealing with complex environmental samples such as soils [34, 49], indicated the homogeneity of the soil plots and that small distances in the field did not determine different bacterial community structures. The soil profiles obtained at T0 agreed with the results of bacterial counts in showing a rapid response of bacterial communities to the treatments applied as compared with the control. When testing bioremediation of acutely hydrocarbon-contaminated sediments, Röling et al. [34] found the same rapid changes in one of their plots, which had significant differences in community composition 1 day after fertilization with liquid inorganic nutrients. This observation reveals the ability of the natural adapted microbiota to quickly respond to nutrient balance and aeration when there is a ready-to-use carbon source present in soil.
The treatments done in the bioremediation trial reported here caused an important change in soil bacterial community composition (T3 and T6). T-RFLP profiles obtained from biostimulated and bioaugmented plots were not significantly different, and therefore it seemed that the addition of nutrients was the main driver of the observed changes in bacterial communities in treated plots. These results, together with those of the bacterial counts and hydrocarbon degradation (which were not significantly higher in bioaugmented plots than in only biostimulated ones), raise the question of the survival of added consortia. The establishment and survival of inocula is as important as any other factor for the success of bioaugmentation [25]. After comparison of the T-RFLP profiles from the soils and the consortia, we concluded that none of the two consortia used survived at detectable levels in the soil. The only T-RFs from consortia observed in soils were also found in all soil profiles, including non-inoculated plots (although it was slightly more abundant in biostimulated and particularly in bioaugmented plots). Besides, the relative abundance of these T-RFs, identified by inference as Pseudomonas, decreased during the trial to levels found in the control, after what seemed a quick response to nutrient addition at T0. It should be kept in mind that one of the consortia, J13, was enriched from the same polluted soil that was used in the trial, and that this soil had a history of pollution of about 2 years before the experiment started. Therefore, we hypothesized that the indigenous bacterial community was already adapted to the presence of hydrocarbons, and hence enriched in bacteria capable of degrading them (as was also evidenced by bacterial counts). In the presence of nutrients that provided an adequate balance between carbon, nitrogen, and phosphorous, the indigenous microbiota could grow and compete with the added inocula, likely restraining its growth. Competition for resources, as well as predation, has been recognized as factors involved in the inability of inocula to survive during bioaugmentation treatments [5, 6]. In this sense, Mishra et al. [25] reported successful addition of inocula to a soil but with low degrading bacterial counts (103 CFU g−1 dw or less), where the effect of competition was reduced.
The hypothesis of the presence of a bacterial community well-adapted to hydrocarbon pollution in the soil used for the trial was also consistent with the results of the detection of hydrocarbon degradation genes by hybridization. Genes involved in the degradation of aliphatic and aromatic hydrocarbons were detected even in control, untreated samples. This indicates that there was genetic potential for hydrocarbon degradation in this soil, particularly for the upper pathway of naphthalene catabolism. These results agree with those reported by Luz and collaborators, who detected the presence of alkane monooxygenase and aromatic dioxygenases in diesel fuel-polluted soils taken from the proximity of storage tanks at a Brazilian Antarctic station [21]. Bioremediation treatments seemed to have an effect in increasing the genetic potential for catechol degradation in soils, which would result in a more efficient channeling of intermediates, favoring the degradation of polyaromatic hydrocarbons. Since the probes used for gene detection were mainly targeting Pseudomonas-like degradation genes, we could presume the presence of bacteria from this genus in the soils used for the trial. This is in agreement with the results of Luz and collaborators [21]. Besides, there are previous reports in which Pseudomonas spp. have been widely cited as hydrocarbon-degrading bacteria present in the natural Antarctic microbiota [7, 46] and also as main components of contaminated soils in Antarctica and other cold regions [31, 40]. This genus was also reported as predominant in chronically contaminated soils from the Alpes by Margesin et al. [23]. The specificity of the probe used for alkane monooxygenase avoids the detection of Rhodococcus-like genes, which have been shown to be relevant in Antarctic polluted soils as well [21]. Fertilization may have also stimulated denitrification in treated soils at some times, such as at T3 (as evidenced by detection of gene nosZ). Although soil aeration by mixing was done at each sampling time, denitrification could have occurred in anaerobic pockets in soil, especially in moments when water content was higher (i.e., 2 weeks before T3 sampling, see Figure S2 of the Electronic Supplementary Material). One possibility is that fertilization may have stimulated the growth of strains of P. stutzeri, known hydrocarbon-degrading and denitrifying bacteria in polar soils [1]. In fact, an increase in the detection of catA gene was also observed at T3 and this gene, together with nosZ, is considered to be characteristic of this species [20]. An effect of fertilization on denitrification may have contributed to some of the rapid changes in bacterial populations observed in treated soils as compared to control and it is also important in the interpretation of the results on the efficiency of the different treatments for hydrocarbon bioremediation since stimulation of fuel degradation in denitrification conditions has been reported [30, 32].
In summary, our results show that hydrocarbon bioremediation in treated mesocosms was successful and it resulted in changes in diversity and structure of bacterial communities, compared with the control, but not between the different treatments: biostimulation alone or biostimulation combined with bioaugmentation. Apart from a likely failure of the added consortia to establish in the soil, our results point to the presence of a well-adapted autochthonous bacterial community in this chronically polluted soil, with genetic and metabolic potential for hydrocarbon degradation, able to respond rapidly to nutrient addition. Further research should be done to optimize the success of the inoculation by immobilizing the consortia as well as to test the effect of the addition commercial fertilizers, mainly the slow-nutrient releasers, to avoid rapid washing of the soil. The evidences obtained in relation to a likely stimulation of denitrification opens a gate to the study of anaerobic processes in nitrate-amended bioremediation trials of hydrocarbon-contaminated soils from the Antarctic Peninsula.
References
Aislabie JM, Saul DJ, Foght JM (2006) Bioremediation of hydrocarbon-contaminated polar soils. Extremophiles 10:171–179
Aislabie J, Fraser R, Duncan S, Farrell RL (2001) Effect of oil spills on microbial heterotrophs in Antarctic soils. Polar Biol 24:308–313
Aislabie JM, Balks MR, Foght JM, Waterhouse EJ (2004) Hydrocarbon spills on Antarctic soils: effects and management. Environ Sci Technol 38:1265–1274
Antarctic Treaty Consultative Parties (1991) Protocol on environmental protection to the Antarctic treaty. CM 1960. Her Majesty’s Stationery Office, London
Bouchez T, Patureau D, Dabert P, Juretschko S, Doré J, Delgenès P, Moletta R, Wagner M (2000) Ecological study of a bioaugmentation failure. Environ Microbiol 2:179–190
Bouchez T, Patureau D, Dabert P, Wagner M, Delgenès JP, Moletta R (2000) Successful and unsuccessful bioaugmentation experiments monitored by fluorescent in situ hybridization. Water Sci Technol 41:61–68
Cavanagh JE, Nichols PD, Franzmann PD, McMeekin TA (1998) Hydrocarbon degradation by Antarctic coastal bacteria. Antarct Sci 10:386–397
Cladera AM, Bennasar A, Barceló M, Lalucat J, García-Valdés E (2004) Comparative genetic diversity of Pseudomonas stutzeri genomovars, clonal structure, and phylogeny of the species. J Bacteriol 186:5239–5248
Delille D (2000) Response of Antarctic soil bacterial assemblages to contamination by diesel fuel and crude oil. Microb Ecol 40:159–168
Delille D, Pelletier E (2002) Natural attenuation of diesel-oil contamination in a subantarctic soil (Crozet Island). Polar Biol 25:682–687
Delille D, Coulon F, Pelletier E (2004) Biostimulation of natural microbial assemblages in oil-amended vegetated and desert sub-Antarctic soils. Microb Ecol 47:407–415
Dua M, Singh A, Sthunathan N, Johri AK (2002) Biotechnology and bioremediation: successes and limitations. Appl Microbiol Biotechnol 59:143–152
Espeche ME, Mac Cormack WP, Fraile ER (1994) Factors affecting growth of an n-hexadecane degrader Acinetobacter sp. isolated from a highly polluted urban river. Int Biodeter Biodegr 33:187–196
Fahy A, Lethbridge G, Earle R, Ball AS, Timmis KN, McGenity TJ (2005) Effects of long-term benzene pollution on bacterial diversity and community structure in groundwater. Environ Microbiol 7:1192–1199
Ferrero M, Llobet-Brossa E, Lalucat J, García-Valdés E, Rosselló-Mora R, Bosch R (2002) Coexistence of two distinct copies of naphthalene degradation genes in Pseudomonas strains isolated from the western Mediterranean region. Appl Environ Microbiol 68:957–962
García-Valdés E, Castillo MM, Bennasar A, Guasp C, Cladera AM, Bosch R, Engesser KH, Lalucat J (2003) Polyphasic characterization of Pseudomonas stutzeri CLN100, which simultaneously degrades chloro- and methylaromatics: a new genomovar within the species. Syst Appl Microbiol 26:390–403
Hammer Ø, Harper DAT, Ryan PD (2001) PAST: palaeontological statistics software package for education and data analysis. Palaeo Electronica 4:9
Kerry E (1993) Bioremediation of experimental petroleum spills on mineral soils in the Vestfold Hills, Antarctica. Polar Biol 13:163–170
Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. Wiley, Chichester, pp 115–175
Lalucat J, Bennasar A, Bosch R, García-Valdés E, Palleroni NJ (2006) Biology of Pseudomonas stutzeri. Microbiol Mol Biol Rev 70:510–547
Luz AP, Pellizari VH, Whyte LG, Greer CW (2004) A survey of indigenous microbial hydrocarbon degradation genes in soils from Antarctica and Brazil. Can J Microbiol 50:323–333
Mac Cormack WP, Fraile ER (1997) Characterization of a hydrocarbon degrading psychrotrophic Antarctic bacterium. Antarct Sci 9:150–155
Margesin R, Hämmerle M, Tscherko D (2007) Microbial activity and community composition during bioremediation of diesel-oil contaminated soil: effects of hydrocarbon concentration, fertilizers, and incubation time. Microb Ecol 53:259–269
Mestre MC, Vazquez SC, Ruberto LAM, Mac Cormack WP (2007) Identificación de los componentes cultivables del consorcio bacteriano degradador de hidrocarburos M10. Proceedings of VI Jornadas Nacionales y III Latinoamericanas de Comunicaciones sobre Investigaciones Antárticas (Epub: http://www.dna.gov.ar/CIENCIA/SANTAR07/CD/PDF/CVRE407.PDF)
Mishra S, Jyot J, Kuhad RC, Lal B (2001) Evaluation of inoculum addition to stimulate in situ bioremediation of oily-sludge-contaminated soil. Appl Environ Microbiol 67:1675–1681
Mohn WW, Radziminski CZ, Fortin MC, Reimer KJ (2001) On site bioremediation of hydrocarbon-contaminated Arctic tundra soils in inoculated biopiles. Appl Microbiol Biotechnol 57:242–247
Nocker A, Burr M, Camper AK (2007) Genotypic microbial community profiling: a critical technical review. Microb Ecol 54:276–289
Nogales B, Moore E, Abraham W, Timmis K (1999) Identification of the metabolically active members of a bacterial community in a polychlorinated biphenyl-polluted moorland soil. Environ Microbiol 1:199–212
Osborn AM, Moore ERB, Timmis KN (2000) An evaluation of terminal-restriction fragment length polymorphism (T-RFLP) analysis for the study of microbial community structure and dynamics. Environ Microbiol 2:39–50
Powell SM, Ferguson SH, Snape I, Siciliano SD (2006) Fertilization stimulates anaerobic fuel degradation of Antarctic soils by denitrifying microorganisms. Environ Sci Technol 40:2011–2017
Powell SM, Ma WK, Siciliano SD (2006) Isolation of denitrifying bacteria from hydrocarbon-contaminated Antarctic soil. Polar Biol 30:69–74
Rakhimova ER, Osipova AL, Zaripova SK (2004) Purification of soil from oil pollutants with the use of denitrifying hydrocarbon-oxidizing microorganisms. Appl Biochem Microbiol 40:563–567
Röling WFM, Milner MG, Jones DM, Lee K, Daniel F, Swannell RJP, Head IM (2002) Robust hydrocarbon degradation and dynamics of bacterial communities during nutrient- enhanced oil spill bioremediation. Appl Environ Microbiol 68:5537–5548
Röling WFM, Milner MG, Jones DM, Fratepietro F, Swannell R, Daniel F, Swannell RJP, Head IM (2004) Bacterial community dynamics and hydrocarbon degradation during a field-scale evaluation of bioremediation on a mudflat beach contaminated with buried oil. Appl Environ Microbiol 70:2603–2613
Rosselló R, García-Valdés E, Lalucat J, Ursing J (1991) Genotypic and phenotypic diversity of Pseudomonas stutzeri. Syst Appl Microbiol 14:150–157
Ruberto LAM, Vázquez SC, Mac Cormack WP (2003) Effectiveness of the natural bacterial flora, biostimulation and bioaugmentation on the bioremediation of hydrocarbon contaminated Antarctic soil. Int Biodeter Biodegr 52:115–125
Ruberto LAM, Vázquez SC, Lobalbo A, Mac Cormack WP (2004) Biorremediación de suelos contaminados con hidrocarburos utilizando bacterias Antárticas sicrotolerantes. Actas del V° Simposio Argentino y 1° Latinoamericano sobre Investigaciones Antárticas. On-line publication in http://www.dna.gov.ar/CIENCIA/SANTAR04/CD/PDF/206BH.PDF, 5 pp
Ruberto LAM, Vázquez SC, Lobalbo A, Mac Cormack WP (2005) Psychrotolerant hydrocarbon-degrading Rhodococcus strains isolated from polluted Antarctic soils. Ant Sci 17:47–56
Ruberto L, Vázquez S, Curtosi A, Mestre MC, Pelletier E, Mac Cormack W (2006) Phenanthrene biodegradation in soils using an Antarctic bacterial consortium. Biorem J 10:191–201
Ruberto LAM, Vázquez SC, Mac Cormack WP (2008) Bacteriology of extremely cold soils exposed to hydrocarbon pollution. In: Dion P, Chandra Shekhar N (eds) Microbiology of extreme soils, soil biology, vol. 13. Springer, Berlin, pp 247–274
Sambrook J, Russell DW (2001) Molecular cloning, a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Saul DJ, Aislabie JM, Brown CE, Harris L, Foght JM (2005) Hydrocarbon contamination changes the bacterial diversity of soil from around Scott Base, Antarctica. FEMS Microbiol Ecol 53:141–155
Schwartz RD, McCoy CJ (1973) Pseudomonas oleovorans hydroxylation–epoxidation system: additional strain improvements. Appl Microbiol 26:217–218
Smith CJ, Danilowicz BS, Clear AK, Costello FJ, Wilson B, Meijer WG (2005) T-Align, a web-based tool for comparison of multiple terminal restriction fragment length polymorphism profiles. FEMS Microbiol Ecol 54:375–380
Smits TH, Röthlisberger M, Witholt B, van Beilen JB (1999) Molecular screening for alkane hydroxylase genes in gram-negative and gram-positive strains. Environ Microbiol 1:307–317
Stallwood B, Shears J, Williams PA, Hughes KA (2005) Low temperature bioremediation of oil-contaminated soil using biostimulation and bioaugmentation with Pseudomonas sp. from maritime Antarctica. J Appl Microbiol 99:794–802
Tarnocai C, Campbell IB (2002) Soils of the polar regions. In: Lal R (ed) Encyclopaedia of soil science. Marcel Dekker, New York, pp 1018–1021
Thomassin-Lacroix EJM, Eriksson M, Reimer KJ, Mohn WW (2002) Biostimulation and bioaugmentation for on-site treatment of weathered diesel fuel in Arctic soil. Appl Microbiol Biotechnol 59:551–556
Tom-Petersen A, Leser TD, Marsh TL, Nybroe O (2003) Effects of copper amendment on the bacterial community in agricultural soil analyzed by the T-RFLP technique. FEMS Microbiol Ecol 46:53–62
USEPA (1983). Petroleum hydrocarbons, total recoverable. In: Methods for chemical analysis of water and waste. EPA-600-4-79-020, Cincinnati, Ohio, 418·1·1–418·1·3
Walter MV (1997) Bioaugmentation. In: Hurst CJ, Knudsen GR, McInerney MJ, Stetzenbach LD, Walter MV (eds) Manual of environmental microbiology. ASM, Washington DC, pp 753–757
Whyte LG, Bourbonniere L, Bellerose C, Greer CW (1999) Bioremediation assessment of hydrocarbon-contaminated soils from the high Arctic. Biorem J 3:69–79
Wynn-Williams DD (1992) Counting bacterial colony-forming units in soil. In: Wynn-Williams DD (ed) BIOTAS manual of methods for Antarctic terrestrial and freshwater research. Scientific Committee on Antarctic Research, Cambridge, p 3
Acknowledgements
This research was supported by grants from the Argentinean Antarctic Institute (IAA no. 42), the National Agency for Scientific and Technical Researches (PICTO 11555), and the University of Buenos Aires (UBACyT U007) as well as by a short-term travel fellowship from the Argentinean Research Council (CONICET). Research at the University of the Balearic Islands (UIB) was supported by grants VEM2003-20565 and CTM2005-01783 from the Spanish Ministry of Education and Science (MEC). B.N. was supported by a contract from the program “Ramón y Cajal” from MEC, and J.C-O by a fellowship of the Balearics Autonomous Government. The experience and assistance of logistic and technical personnel at Tte. Jubany Antarctic Station is also highly appreciated, as well as the support of the Scientific-Technical Services of the UIB during the operation of the genetic analyzer for fragment analysis.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link of electronic supplementary material.
ESM Fig. 1
Weather conditions at Jubany Station, King George Island (Isla 25 de Mayo), South Shetland Islands, Antarctica, during the field experiment. Arrows indicate the sampling times (GIF 28 kb)
ESM Fig. 2
Relative moisture content (%) of soil during the field experiment. Black bars: community control plot (CC); white bars: biostimulated plots (AB); light gray bars: biostimulated and bioaugmented plots with M10 consortia (M10); and dark gray bars: biostimulated and bioaugmented plots with J13 consortia (J13). In the treated plots, the first bar corresponds to plot 1, the second one to plot 2, and the third one to plot 3 (GIF 20 kb)
Rights and permissions
About this article
Cite this article
Vázquez, S., Nogales, B., Ruberto, L. et al. Bacterial Community Dynamics during Bioremediation of Diesel Oil-Contaminated Antarctic Soil. Microb Ecol 57, 598–610 (2009). https://doi.org/10.1007/s00248-008-9420-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-008-9420-9