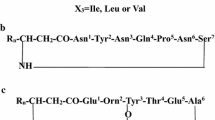Abstract
A soil microorganism identified as Bacillum megaterium was found to produce several antibiotics substances after growth for 20 h at 37°C in a mineral culture medium. Analysis both by electron spray ionization (ESI) and matrix-assisted laser desorption ionization—time of flight (MALDI-TOF) mass spectrometry (MS) identified these substances as lipopeptides. Predominant peaks at m/z 1,041 and m/z 1,065 revealed ions which are compatible with surfactins and lichenysins, respectively. Two other ions m/z 1,057 and m/z 1,464 were further studied by collision-induced dissociation (CID) unveiling an iturin A at the first and fengycins A and B at the second m/z peaks. The CID spectrum of the m/z 1,464 ion also suggests the existence of fengycins A and B variants in which Ile was changed to Val in the position 10 of the peptide moiety. Raw mixtures of all these compounds were also assayed for antibiotic features. The data enlighten the unusual diversity of the lipopeptide mixture produced by a sole Bacillus species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
An intriguing and not completely solved question in the physiology of microorganisms refers to the synthesis and secretion of antibiotics substances. Generally described as products of the secondary metabolism they comprise a wide variety of compounds whose utility inside the cell has never been conclusively reported. However, it is hard to imagine that the intricate reaction sequences of antibiotic biosynthesis, the self protection mechanisms, the externalization processes capable to keep the integrity of the cellular envelope, the commitment of part of the genome, were retained during the evolution, without benefit to the microorganism. Disregarding ideas such as metabolic relics or metabolic aberrations, several functional roles were proposed for these substances, all of them with the purpose to account for the survival abilities of cells located in a broad environmental diversity.
Several other strategies were developed by bacteria to survive in hostile surroundings. One of the most efficient is the endospore formation, universally found in the genus Bacillus. This is thought to be a sophisticated trick for survival in the soil environment, wherein these bacteria predominate. The aerial dispersion of the dormant spores is probably the cause of the Bacillus ubiquity. Thus, the reason why these microorganisms are so prolific in producing an amazing antibiotic variety may be speculated in the sense that these substances could represent an overall colonization effort, which, if not successful, may unleash mechanisms described as “altruistic suicide” [1] or “cannibalism of siblings” [2]. Both mechanisms are genetically regulated and biosurfactant performed, in order to supply emergency feeding for a starving community. Apparently, the strategy consists of delaying spore formation, a highly energy consuming process, which is the last resort to survive, when all the other remedies have failed.
Among antibiotic substances produced by species of the genus Bacillus, there are biosurfactants, such as cyclic lipopeptides. These comprise mainly of surfactins [3], iturins [4, 5], lichenysins [6–8], fengycins [9, 10], pumilacidins [11], and isohalobacillins [12]. These molecules are lipopeptides which have a cyclic portion formed either by the linkage between –NH2 terminal to the hydrophobic portion of a variable length fatty acid or by another amino acid residue of the peptidic moiety. Such compounds were described as acting on the membrane bilayer, thus forming selective pores, which disturb the ionic balance killing the cell, as was observed for iturins [13, 14] or lichenysin [15]. Since this antibiotic effect is exerted at a very fundamental level, the integrity of the cellular membrane, certainly is less dependent on mutations, such as those responsible for the remarkable resistance observed with antibiotics of highly specific action. In principle, these substances should eliminate Gram positive and Gram negative bacteria, and even attack other cell types. Nevertheless, this is a simplistic approach due to the fact that, the barriers which isolate the lipidic bilayer from the surroundings, are very different. This is easily discernable in the case of Gram positive and Gram negative bacteria, in which the nature of these barriers is well established. In Gram negative bacteria, the outer membrane impede certain drugs and antibiotics from penetrating the cell [16, 17]. Efflux systems related to the outer membrane [18, 19] partially account for the reason why these microorganisms are generally more resistant to antibiotics than Gram positive bacteria are. Other distinctive features, as the periplasmic space and the lipopolysaccharide coat which wrap up the Gram negative cells, may also be the cause of this kind of resistance. The antibiotic effect on bacteria, observed in the study of several lipopeptides, including those described in the present work, is probably due to the disruption of the cellular envelope as a consequence of undetermined interactions with several components, which include the lipidic bilayer. The information from the well-studied species Bacillus subtilis, shows a great plasticity in producing antibiotic substances. Among them, cyclic lipopeptides and cyclic peptides such as maltacins [20–23] were identified. The inspection of the amino acidic portion of these substances indicates that all are versions of a central theme, although their activities both “in vivo” and “in vitro” may be different. These substances were generally isolated from separated B. subtilis strains, but the possibility that a unique strain could produce several of them, under a feasible environmental influence, cannot be ruled out. This may lead to synergistic effects since interaction among lipopeptides was observed in vitro [24] and deduced by using mutants of Bacillus amyloliquefaciens [25]. The analysis of artificial lipopeptide blends could unveil properties different from those of the isolated components. For this reason, species which are producers of a wide range of individually well-known compounds, may be well suited for studying their lipopeptide mixtures.
This paper provides further evidence that antibiotic versatility is a widespread feature in bacteria of the genus Bacillus. The data indicate that a single Bacillus megaterium strain is able to spread a battery of lipopeptides, some of them very closely related. Culture supernatants of a B. megaterium strain isolated from the soil, contain iturin, fengicyns A, B as well as modified fengycins in which leucine at the position 10 was changed to valine at both A and B forms. These substances were determined by MALDI-TOF MS techniques. Also peaks indicating the presence of surfactin and lichenysin were detected in these supernatants. The lipopeptide melange produced by a Bacillus strain alone as described in this work, was not detected previously. For this reason, its antibiotic activity against other Bacillus species was also investigated. Further studies on the rich and apparently unique mixture described here, could offer better insights on the actual action of this microorganism on its surroundings, as well as it could reveal unsuspected features of such blend of substances.
Material and Methods
Bacteria Isolation and Identification
Bacteria used in this study were isolated from a soil sample collected at the University campus. Characterization both by morphological and biochemical criteria, was performed in accordance with the Bacillus key characterization procedure described in Bergey’s Manual of Systematic Bacteriology. It gave the following results: gram stain reaction is positive; shape of spores is oval/ellipsoidal; spore placement is central/subterminal; negative swollen sporangium; motility of cells was observed; single cells were observed; anaerobic growth did not occur; nitrate reduction was not observed; Voges-Proskauer (V-P) reaction (seventh day) was negative; pH V-P reaction was 7.5; citrate utilization was observed; propionate utilization was not detected; casein hydrolysis was observed; starch hydrolysis was observed; tyrosine degradation was not observed; and lecithinase degradation was not observed or was weak. According to these data, the bacterium closely resembles B. megaterium. Identification service of the American Type and Culture Collection, Manassas, VA, USA recognized it as a B. megaterium strain (Project Report SC 3488), which throughout this work was named B. megaterium pL6.
Minimal Medium for Bacillus Growth
The following substances were used to prepare 500 ml of a minimal medium in which the growth of several Bacillus species is very efficient: 0.8% MgS04, 10 ml; 5 M NaCl, 17.0 ml; 1 M KCl, 3.4 ml; 85% H3P04, 0.2 ml; 2% asparagine, 12.5 ml; 2% NH4N03, 2.5 ml; 0.05% FeS04, 3 ml; 20% glucose, 25.0 ml. All stock solutions were sterilized individually and added in the specified volumes up to 350 ml of deionized water in the same order shown above. This is important to avoid precipitation. The pH was set to 7.0 with 1 M Tris–HCl, pH 8.0 sterile solution. Volume was complemented up to 500 ml with deionized water.
Culture Conditions and Preparation of the Mixture of Biosurfactants Produced by B. megaterium pL 6
Lyophilized cells of Bacillus megaterium pL6 were added to rich culture medium (1 mg cells to 20 ml of Luria Broth (LB) medium) and incubated for 12 h at 37°C, 200 rpm. This pre-culture was used to inoculate 500 ml of mineral medium in a 2-l Erlenmeyer flask where the new culture was incubated for 20 h at 37°C, 200 rotations per minute (rpm).
Cells were centrifuged at 6,000 rpm (5,800×g) for 10 min in a Hitachi RPR9–2 rotor, the supernatant transferred to a l-l flask and precipitated overnight at pH 2 with 5N HCl at 4°C. Centrifugation at 6,000 rpm (5,800×g) for 15 min in the RPR9–2 rotor recovered a precipitate which was extracted with 50 ml of pure ethanol for 3–4 h.
The solid material resulting from the extraction was discarded after centrifugation at 10,000 rpm (12,000×g) in a Hitachi rotor RPR20–2. The supernatant from this centrifugation was evaporated at 50°C under vacuum, until an oily residue appeared at the bottom of the flask. The residue became soluble by adding 5 ml of deionized water at pH adjusted up to 8.0 with 2 M NaOH. It was transferred to a dialysis bag prepared with a membrane of 10 kDa cut off, previously boiled in a 2% sodium bicarbonate, 1 mM EDTA, pH 8.0 solution and exhaustively dialyzed against deionized water. The bag content was transferred to a 100-ml Erlenmeyer flask where it was lyophilized and stored under vacuum in a desiccator flask at –20°C.
Determination of Phosphate Leakage by Bacillus cereus Cells Exposed to the Biosurfactant Mixture
Phosphate determinations were performed essentially according to Rouser et al. [26]. For this purpose, 5 ml of B. cereus cultures in Luria Broth at the early stationary phase (5 h of growth) were centrifuged in a Hitachi SCR20B refrigerated centrifuge at 8,000 rpm (7,700×g) in the RPR20–2 rotor. Cells were rinsed twice with 0.05 M NaCl and suspended with 2.5 ml of deionized water. The biosurfactant mixture was added to final concentration of 12 mg/l (which is the minimum inhibitory concentration, MIC. See details in Fig. 7). The cell suspension was incubated at 37°C. From this suspension, samples of 100 μl were taken at 30 s, 15 min, 30 min, 60 min, and 180 min and centrifuged in an IEC table centrifuge at 8,000×g in order to obtain clear supernatants that were assayed for inorganic phosphate as follows: the supernatants were transferred to glass test tubes, heated at 100°C for 24 h to completely evaporate water. To each tube, 0.3 ml of concentrated perchloric acid were added. The tubes were sealed with a glass ball and heated in an oven for 1 h at 180°C. After cooling, 1 ml of deionized water and 0.4 ml of a 1.25% solution of ammonium molibdate were added and the contents of each tube carefully shaken in a vortex device. A 0.4 ml aliquot of 5% ascorbic acid solution freshly prepared was placed in all the tubes, which were again sealed with glass balls and boiled for 5 min. After cooling, the optical density at 795 nm of each tube was measured. Controls were done in order to determine the amount of phosphate in samples also from the above suspension, without biosurfactant exposure. A standard calibration curve, i.e., optical density at 795 nm versus amount of phosphate, was performed using a series of glass test tubes containing 10, 30, 50, 70, and 100 μl of 1 mM Na2HPO4 in a final volume of 100 μl.
Surface Tension Measurements
The measurement of surface tension in the culture medium was accomplished by using 5-ml samples which were cleared from cells by centrifugation. The supernatants were transferred to a 4-cm-diameter glass petri dish adapted to a Dynometer tensiometer (BYK Gardiner, Labotrom, Germany) coupled to a Du Nuoy platinum ring. For critical micelle concentration (CMC) measurement the same device was used. The fall of the surface tension was determined after adding fixed volumes of a concentrated stock solution of biosurfactant mixture to 5 ml of freshly distilled and deionized water (initial surface tension 72 mN/m).
Determination of the Minimal Inhibitory Concentration (MIC) Using the Biosurfactant Mixture
A B. cereus culture grown in minimal medium, up to the early stationary phase, was lyophilized and used as a standard source of cells in all measurements. These were accomplished by adding 0.5 mg of the lyophilized cells to tubes with 5 ml of rich culture medium (Luria Broth). These tubes were incubated for 5 h at 37°C under agitation at 200 rpm. Five microliter of the resulting cell suspension were inoculated into 5 ml of fresh LB containing increasing concentrations of the biosurfactant mixture: 2, 3, 5, 8, 10, and 12 mg/l. The tubes were incubated as before for 6 h. At each hour, 0.7 ml samples were taken under sterile conditions in order to assess the optical density of the culture at 650 nm. After each measurement, the samples were replaced into the original flask to keep a constant volume.
Survival Rate of B. cereus Cells Exposed to the Lipopeptide Mixture
Samples were prepared as described under MIC determination. Only the control culture and the one exposed to 12 mg/l (MIC) of the biosurfactant mixture was assayed for the number of surviving colonies after 5 h of culture. Every hour, 10 μl samples were taken both from control and exposed cultures. In order to determine the colony number, these volumes were diluted as follows:
-
a) control—0 h, 1:100; 1 h, 1:100; 2 h, 1:100; 3 h, 1:1000; 4 h, 1:1000; 5 h, 1:10000
-
b) exposed—0 h, 1:100; 1 h, 1:100; 2 h, 1:100; 3 h, 1:100; 4 h, 1:100; 5 h, 1:10
Samples (10 μl) of each dilution were spread onto an 8-cm-diameter petri dish filled out with 15 ml of 1.5% agar in LB. The colony number of each plate was determined after growth at 37°C overnight.
Fourier Transformed-Infra Red (FT-IR) Spectra
FT–IR spectra were recorded in a FT-IR Bomem MB100 spectrometer after inclusion of the lipopeptide mixture in KBr tablets.
Molecular Mass Determination
The molecular mass of the mixture components were determined by MALDI-TOF/MS in a 4700 Proteomics Analyzer with TOF-TOF optics (Applied Biosystems, Framingham, MA, USA). Approximately 1 mg of lyophilized mixture was dissolved in pure ethanol, mixed with a saturated matrix solution of cyano-4-hydroxycinnamic acid (1:3, v/v) and spotted onto a MALDI-TOF sample plate, at room temperature.
Alternately, samples of the same mixture were analyzed in an Ettan MALDI-TOF Pro 2.0 Mass Spectrometer at 8 laser pulses/s, acceleration potential 20 kV, 200 spectra per scan at less than 1.5 × 10–6 bar.
Sequence Analysis
Amino acid sequence of the lipopeptides was developed in a 4700 Proteomics Analyzer with TOF-TOF optics (Applied Biosystems, Framingham, MA, USA). The MS and MS/MS spectra were carried out in reflector mode with external calibration using the 4700 calibration mixture kit (Applied Biosystems). Precursor ion fragmentation occurred using N2 as collision-induced dissociation (CID) gas and the collision cell pressure was kept at 1.8 × 10–6 Torr.
Results
Chemical Composition and Surface Activity of the Lipopeptide Mixture
Cultures of a soil B. megaterium pL 6 strain were grown at 37°C in a mineral medium as described under “Material and Methods”. Raw preparations of lyophilized lipopeptides were obtained from several stages in order to assess the best yield. This was accomplished by determining the variation of surface tension during the culture growth. Usually, the best yield is found after 20 h as is shown in Fig. 1a. Figure 1b shows a typical determination of the critical micelle concentration in lyophilized samples prepared from 20-h cultures. The CMC value of ∼100 mg/l was graphically estimated by taking the intersection of the tangents to the two curve branches.
In a, surface tension changes in culture are plotted as a function of time during a typical Bacillus megaterium pL6 growth. Cells were grown over a 36-h period at 37°C in 500 ml of mineral medium under aeration. Five milliliter samples were collected at the indicated times, assayed for turbidity at 600 nm (black circles), cleared from cells by centrifugation and transferred to 4 cm diameter Petri dishes for surface tension measures (empty circles). In b, surface tension is determined as a function of concentration of the lipopeptide mixture obtained after ethanolic extraction and lyophilization. The critical micelle concentration (CMC) was graphically estimated from the intersection of the two straight lines
Fourier Transformed-Infra Red (FT-IR) Spectrum of the Ethanol Extract
To preliminarily characterize the substances produced by B. megaterium pL 6 cells, the culture cell free supernatant was acidified with 5.5 N HCl, at 4°C. The solution was left overnight at 4°C forming a precipitate which was recovered and extracted with pure ethanol during five or more hours. After completing the solvent evaporation, the oily residue was lyophilized until a yellowish powder was formed. A FT-IR spectrum of this ethanol extract was recorded and is shown in Fig. 2. Several inverted peaks at 3,347; 2,950; 2,926; 1,734; 1,646; 1,541; 1,465; 1,215; 1,073; 1,035; and 593 cm–1 indicate the presence of compounds with the following features: secondary amide, intermolecular H bonds (3,347 cm–1); –CH3 asymmetric stretching (2,950 cm–1); C-H stretch of –CH2- and –CH3, alkyl groups (2926 cm–1); C=0 stretching, ester (δ-lactone) carbonyl band (1,734 cm–1); amide I band frequency (1,646 cm–1 C–O stretching in peptide bond of random coil peptides); amide II band frequency (1,541 cm–1, C–N stretching in peptide bond); –CH3 asymmetric bending (1,465 cm–1); likely C–O ether stretching vibrations (1,073, 1,035 cm–1). Bands 1,215–1,254 cm–1 (for better clarity, are indicated by an inverted arrow in Fig. 2) and bands 1,035–1,073 cm–1 could suggest DNA/RNA presence.
MALDI-TOF Spectra of the Ethanol Extract
In order to investigate the nature of the compounds in the ethanol extract, exploratory MALDI-TOF spectra was accomplished. Figure 3 shows the spectra that were distributed around two main peaks, namely m/z 1,057 and m/z 1,464. The homologous series of m/z 1,065; 1,079; and 1,093 peaks, which differ each other by m/z 14, a putative methylene group, seems not to be related either to the m/z 1,041 peak or to the m/z 1,057 ion. However, the m/z 1,436; 1,450; 1,464; 1,478; 1,492; and m/z 1,506 ions are probably components of a homologous series. In Figs. 4a,b and 5a,b are exhibited MALDI-TOF MS/MS spectra obtained from the two main ions depicted in Fig. 3, i.e., m/z 1,057 and m/z 1,464. Figures 4a and 5a show fragmented ions of low m/z ratios obtained by collision-induced dissociation (CID) of the m/z 1,057 and m/z 1,464 ions respectively. They correspond mainly to immonium ions which originate from individual amino acids. Figures 4b and 5b show the spectra of ions m/z 1,057 and m/z 1,464 respectively, with CID activated. The sequence of amino acids in the peptide moiety is established from these spectra (Fig. 6). Figures 3, 4, and 5 were redrawn from the original by using a graphic computer program in order to exclude the noise peaks and to better clarify the spectra. They are accurate replica of the data provided by the mass spectrometer.
Molecular mass determination by MALDI-TOF mass spectrometry. Data were acquired in the mass range m/z 1,000–2,200 for the ethanol extract. MALDI-TOF conditions: 0.5 μl of the ethanol extract was mixed with 3,5 dimethoxy-4-hydroxycynnamic acid as the organic matrix and spotted on the slide and dried before analysis in an Ettan MALDI-TOF Pro 2.0 mass spectrometer. Data were obtained in the linear mode, 8 laser pulses/s, acceleration potential 20 kV, vacuum less than 1.5 × 10–6 bar and 200 spectra accumulated per scan
MS and MS/MS MALDI-TOF analysis of the m/z 1,057 peak from Fig. 3. In a are displayed peaks corresponding mainly to amino acid immonium ions (H2N+=CHR) over the m/z 60 to 160 range. They were produced by CID and have masses (m/z) which are 27 atomic mass units (a carbonyl group) lighter than the molecular mass of the corresponding amino acid. In b, the same is shown over the m/z 60 to 1,000 range
Open structures were proposed for the m/z 1,464 fengycins using data quoted in [34] and [36]. When detected, y and b ions [33] are marked along the structure scheme. Fengycin “fingerprints” are y8 and y9 ions which identify fengycins A and B. The fengycin variant called [Val10] was suggested by observing y8 (m/z 952), y9 (m/z 1,066) ions for C17 FA fengycin A [Val10] and y8 (m/z 980), y9 (m/z 1,094) for C15 FA fengycin B [Val10]. In C15 FA, the length of the hydrocarbon chain corresponds to 15 carbon unities in the fatty acid moiety (FA) of the respective fengycin
Antibiotic Activity of the Lipopetide Mixture
Figure 7a and b illustrate antibiotic properties of the lipopeptide mixture against B. cereus. Figure 7a depicts the MIC which is 12 mg/l. In Fig. 7b the rate of cell survival at MIC is presented as the number of colonies formed by milliliter, plated onto petri dishes.
Antibiotic activity of the lipopeptide mixture. a Bacillus cereus cultures were exposed to different concentrations over a 5-h period to determine the minimal inhibitory concentration (MIC) which is 12 mg/l. b Rate of cell survival of Bacillus cereus exposed to the lipopeptide mixture at MIC (12 mg/l). The colony number was plotted as a function of time by counting the colonies on the agar plates and correcting for the corresponding dilution
Phosphate leakage was assayed on cells collected in the stationary phase and exposed to the MIC of the lipopeptide mixture up to 200 min. As is shown in Fig. 8, after 30 min. the culture looses the maximum amount of phosphate, given as nanomoles of inorganic phosphate in 100 μl samples. The value remains constant for the subsequent time of the assay. This indicates that the culture does not grow after the exposure to 12 mg/l of the lipopeptide mixture for 30 min.
Leakage of intracellular phosphorylated compounds from cells of Bacillus cereus at an early stationary phase (∼5 h) after exposure to MIC (12 mg/l) of the lipopeptide mixture (black symbols) for increasing time. A control curve (hexagons) was constructed by measuring the phosphorus leakage by the same culture, in absence of the lipopeptide mixture
Discussion
The CMC value of about 100 mg/l estimated in Fig. 1b, is comparable to that of a pure lipopeptide, such as iturin [27], lower than that of common synthetic detergents such as Triton X-100 [28] but higher than that of biological detergents, such as dipalmitoyllecithin [29]. Surface tension values as low as 28–30 mN/m were measured for the B. megaterium pL 6 lipopeptide mixture, as is shown in Fig. 1b.
The overall FT-IR spectrum in Fig. 2 is very similar to some determined previously, as is the case of pumilacidines (cyclic lipopeptides) from Bacillus pumilus described by Naruse et al. [11] and that of maltacine B1 (a cyclic peptide) from B. subtilis described by Hagelin et al. [20]. Therefore, the ethanol extract obtained from B. megaterium pL6 cultures seems to contain mainly substances formed by peptides in random coil, alkyl, ester, and ether groups.
The peaks in Fig. 3 are quite similar to the ones described previously for cyclic lactone lipopeptides such as surfactins, fengycins, and lychenisins, all of them isolated from different Bacillus species.
Together, MALDI-TOF, FT-IR, as well as previous literature data, suggest a reasonable assumption: cyclic lipopeptides, lipids, and glycolipids are likely components of this extract. They all result from the secondary metabolism of the Bacillus cells and may have a role in the overall properties of the bacterial secretion to the environment. Several previous reports describe the first peak series (around m/z 1,057 in Fig. 3) as formed by surfactins and lichenysins, the second (around m/z 1,464 in Fig. 3) by fengycins, whatever Bacillus species were investigated. These substances were thought as being biomarkers and related to the various Bacillus species [30]. According to the m/z peak criterion, it was observed that B. cereus, Bacillus circulans, and B. subtilis are surfactin producers; Bacillus globigii, B. megaterium, B. circulans, and Bacillus sphaericus are sources of both surfactins and fengycins. Apparently, this is a simple assumption since other lipopeptides having biological properties quite different from those displayed by surfactins and fengycins are hidden under the above mentioned peaks as seems to be the case of isohallobacillins from B. subtilis [12], pumilacidins from B. pumilus [11] or lichenysin from B. licheniformis [6]. These substances are versions of surfactins in which one or two amino acids were changed in the peptide moiety. In the center of this diversity are the peptide synthetases, the enzymes fashioned for the non-ribosomal peptide synthesis in bacterial cells. Catalytic site polymorphism or even tolerance to different amino acids by the isomorphous site may, in principle, be responsible for the assembly of a number of peptides of identical residue length. An additional complication to this figure is raised by the results from Hagelin et al. [20–23]. Substances having exactly the same mass as some peaks described in this and in other works (for instance, the peaks at m/z 1,478 and m/z 1,492) are not lipopeptides. Instead, they are cyclodepsipeptides or cyclic lactone homodectic peptides, the difference of m/z 14 is apparently due to the amino acid composition and are produced by a Bacillus subtilis strain. These findings are in accordance with the heart of OSMAC (one strain, many compounds) idea [31]: the same microorganism when grown in different or abnormal conditions could produce a great variety of secondary metabolites. The repertoire of compounds which is now progressively revealed when studying the genus Bacillus, points toward an evolutionary profit. It is an attractive idea that molecular plasticity may be environmentally affected. Several Bacillus species produce culture supernatants that are self-noxious, leading to an intense lytic process which destroys the culture (results not shown). This can be a key mechanism for control purposes which could be putatively induced by a natural surrounding ambient. Meanwhile, the wide spectrum of biological activity which “do not coincide necessarily with the native target in their biotope” [32] may be exploited according to human concerns. As a corollary of these reflections, it can be postulated that a mass spectrum would not be immediately associated either with a particular substance or with particular species when studying lipopetides from the Bacillus genus. MS/MS for a specific ion followed by ion dissociation induced by collision can offer better insights on the nature of those substances.
Treatment at room temperature for 24 h with 1 N NaOH added 18 atomic mass units to several well established peaks in MALDI-TOF MS (results not shown). Substances which gave the spectra presented in Fig. 3 may be cyclic lipopeptides.
The outstanding peaks at m/z 1,057 and m/z 1,464 in the MALDI-TOF spectrum of Fig. 3, were chosen for further analysis by CID tandem MS/MS. An important aspect of these spectra concerns to the low mass region where immonium ions are clearly shown. In a peptide, an internal fragment with a single side chain (R), originated from a type and y type cleavage [33] is called immonium ion (H2N+=CHR). The detected immonium ions are depicted in Figs. 4a and 5a. Their identification is done automatically by the software resident in the mass spectrometer and provides useful information on the amino acid composition of the lipopeptide. Thus, under the above criterion the ion m/z 1,057 contains Pro, Asn, Gln (Lys), and Tyr as is shown in Fig. 4a. A m/z 1,057 peak was described before [34] as a sodiated adduct of a 1,034 Da compound. Since such substance (m/z 1,034) was not detected by MALDI-TOF MS (Fig. 3) and no additional difficulties were found in analyzing the CID spectrum of the m/z 1,057 ion, this was not considered a sodium adduct. It was identified by matching the CID spectrum of Fig. 4b with the data obtained by Pabel et al. [35] from a lipopeptide found in the marine B. subtilis strain A184. A C15FA iturin A (Iturin A having 15 carbon atoms in the fatty acid portion of the molecule) of m/z 1,057.6 cleaved after CID MS/MS between Pro and Gln. The ions P and Y of the CID spectrum [35] have the same mass numbers than those exhibited in Fig. 4b including the base peak m/z 652. The structure described for iturin A [4] in its open form is CH3(CH2)11CHNH2CH2CO-NHAsn-Tyr-Asn-Gln-Pro-Asn-SerCOOH. Therefore, it is proposed that the m/z 1,057 ion identifies an iturin A with C15 FA moiety. According to several data [27] it is possible to bias the peptide composition of lipopeptides by providing the target amino acids as carbon sources in the medium. The high nitrogen content (asparagine, glutamine) may well be an aftermath of the asparagine and ammonium nitrate presence in the medium where the B. megaterium pL6 cells were raised. The m/z 1,464 peak in Fig. 3 was also investigated by CID as is shown in Fig. 5a,b.
The lowest region of the MS/MS spectrum (CID activated) shows immonium ion of several known amino acids as is the case of Pro, Val, Thr, Glu, Gln(Lys), and Tyr. The 115 m/z peak does not correlate with normal amino acids. If it is a true amino acid immonium ion, it would correspond to an ion of m/z 142 or an ion of 128 amu (atomic mass units) plus 14 amu. Thus, the m/z 115 ion could be tentatively identified as the immoniun ion of glutamine or lysine with an additional –CH2-group. No attempts were done in order to determine the substance to which this ion belongs. Other small ions detected in the lower region of the spectrum in Fig. 5a were not used for identification purposes.
Ions m/z 1,080 and m/z 966 (Fig. 5b) were described previously [35] as the fengycin A y9 and y8 ions, respectively. The 114 m/z difference corresponds to asparagine. Another ion pair i.e. m/z 1,108 and m/z 994 (Fig. 5B) are the y9 and y8 ions of fengycine B, respectively. These ion pairs are so distinguishing that were called “fengycin fingerprints” [36]. The spectrum in Fig. 5b show clearly these fingerprints before, as well as, after CID activation. It is concluded that the peak at m/z 1,464 (Fig. 3) is a contribution of at least C16 FA fengycin A and C14 FA fengycin B. This same situation was described before by Wang et al. [36] for fengycins A and B from B. subtilis JA strain. Interestingly, the pair peaks m/z 952, m/z 1,066 and m/z 980, m/z 1,094 which differ by m/z 114, are also present together with the fengycins A and B, before and after CID activation. These ions m/z 1,066 and m/z 952 were also detected before in the spectrum of m/z 1,464 a fengycin A from intact B. globigii spores [34] although no attempts were done to identify them. The above situation suggests the presence of other y9 (m/z 1,094, m/z 1066) and y8 ions (m/z 980, m/z 952). They were interpreted as the fingerprint ions of a C17 FA fengycin A having valine instead isoleucine at the position 10 (m/z 1,066; m/z 952) and a C15 FA fengycin B having valine instead isoleucine at position 10 (m/z 1,094; m/z 980). Supporting this suggestion, ions m/z 1,351 and m/z 1,365 were found. They were interpreted as b9 ions for the normal fengycins A and B having isoleucine (m/z 1,351) at position 10 and for modified fengycins A and B having valine (m/z 1,365) at that position. Other b and y ions detected in the spectrum of Fig. 5b, were positioned over the structures at Fig. 6. They seem to corroborate the suggestions done before. Thus, it is proposed that the prominent ion m/z 1,464 could be actually a contribution of four substances whose structures are depicted in Fig. 6. These compounds were not detected by our reverse phase HPLC examination (results not shown) as was also noticed for fengycins A and B by Akpa et al. [37]. Nevertheless, if the above interpretation is correct, two new variants of fengycins A and B (structures 2 and 4, Fig. 6) were detected in the present work. These data also indicate that the picture of Bacillus synthesizing a well defined set of lipopeptides, which could be used as species biomarkers [38], may be an awry assumption.
The CID analysis done for the ions m/z 1,057 and m/z 1,464 may be extended to other important peaks of the spectrum displayed in Fig. 3. Particularly interesting is the m/z 1,041 ion and the homologous series formed by m/z 1,065; 1,079; 1,093 ions. In principle, they could be associated to surfactins and lichenysins as was done before [34]. These results could indicate that B. megaterium pL6 is a possible producer of iturins, lichenysins, surfactins, and fengycins. This diversity places the soil pL6 strain together with B. subtilis as a highly versatile microorganism regarding lipopeptide production. The distinctive fact is that all of the substances are produced by a unique strain. The suggested four m/z 1,464 fengycins propose a bacterial strategy for the output of a wide range of substances at a minimal metabolic price.
Antibiotic behavior
The biosurfactants described above can be conceived as natural amphiphilic bi-block copolymers because they are formed by two blocks, hydrophobic, and hydrophilic, both covalently joined. The nature and properties of synthetic block copolymers, which are also mixtures of compounds because both the hydrophobic and hydrophilic moieties display variable length, have been the subject of intensive study for the last 30 years [39, 40]. Characteristically, these compounds may be assembled in a broad range of shapes including vesicles, micelles, rods, spheres, tubules, and so on. These geometric properties were dependent on parameters, such as optimal interface, volume occupied by the hydrophobic chains and the maximum length of these chains when the amphiphilic molecules are in aqueous solution [41]. Concepts and knowledge deriving from the study of synthetic bloc copolymers may be useful if applied to the mixture of biosurfactants produced by B. megaterium PL 6. Therefore, our vision of the amphiphilic and antibiotic properties of lipopeptide mixtures as described in this work are based on the above consideration.
Results similar to that shown in Fig. 7 were found by exposing other Bacillus species, such as B. subtilis, Bacillus polymixa, Bacillus laterosporus, B. circulans, B. pumilus, Bacillus brevis, and Bacillus lentus. Other Gram positive bacteria, Staphylococcus aureus and Mycrococcus lysodeikticus are less sensitive. Gram negative bacteria, such as Escherichia coli and Proteus vulgaris showed poor sensitivity but other Gram negative bacteria such as Klebsiella pneumoniae and Salmonella tiphymurium, displayed sensitivity similar to some Bacillus species (Table 1). In short, Bacillus cells seem to be the preferred target of the lipopeptide mixture. Due to its morphology, biochemical characterization, and growth behavior, both B. megaterium ATCC 14581 and B. megaterium pL 6 may be considered typical representatives of these species. However, strain ATCC 14581 does not produce biosurfactants at conditions pL 6 strain does. When put together in the same petri dish at 37°C, the strain pL 6 eliminates the strain ATCC 14581. While B. megaterium pL6 bears complete auto lysis when stored in liquid culture medium for 2 days, this process is not evident in the case of ATCC 14581. Sequence determination of the 16s ribosomal RNA may give insights on the evolutionary distance of these two strains.
Data in Fig. 8 indicate that after exposure to MIC (12 mg/l) the cellular death is very probable due to a lytic process. Events which induced pore formation leading to losses of cellular components can also explain these results. The data shown in Table 2 indicate that the putative hydrolysis of lactone or lactam (in iturins) rings do not remove the detergent power but abolish the antibiotic activity. The observation that MIC value (12 mg/l) is very low when compared with that of the CMC (∼100 mg/l) together with the above data, reinforce the idea that the cell lysis is not due only to a detergent effect on the cellular envelope. Apparently, the rigid conformation resulting from a cyclic peptidic moiety is essential to unleash the lytic process which kill the B. cereus cells. Similar rationalization may explain the lysis of other sensible microorganisms.
Bacillus megaterium is one of the most studied within the genus Bacillus due to its remarkable capacity of producing intact exoenzymes of industrial use [42, 43]. This knowledge will be certainly very useful to modify cells for production of metabolites such as the compounds described in this work.
References
Stein T (2005) Bacillus subtilis antibiotics: structures, synthesis and specific functions. Mol Microbiol 56:845–857
González-Pastor JE, Hobbs EC, Losick R (2003) Cannibalism by sporulating bacteria. Science 301:510–513
Kakinuma A, Sugino H, Isono M, Tamura G, Arima K (1969) Determination of fatty acid in surfactin and elucidation of the total structure of surfactin. Agric Biol Chem 33:973–976
Isogai A, Takayama S, Murakoshi S, Suzuki A (1982) Structures of α-amino acids in antibiotic iturin A. Tetrahedron Lett 23:3065–3068
Peypoux F, Guinand M, Michel G, Delcambe L, Das BC, Lederer E (1978) Structure of iturin A, a peptidolipid antibiotic from Bacillus subtilis. Biochemistry 17:3992–3996
Jenny K, Käppeli O, Fiechter A (1991) Biosurfactants from Bacillus licheniformis: structural analysis and characterization. Appl Microbiol Biotechnol 36:5–13
Yakimov MM, Abraham W-R, Meyer H, Giuliano L, Golyshin PN (1999) Structural characterization of lichenysin A components by fast atom bombardment tandem mass spectrometry. Biochim Biophys Acta 1438:273–280
Grangemard I, Bonmartin J-M, Bernillon J, Das BC, Peypoux F (1999) Lichenysins G, a novel family of lipopeptide biosurfactants from Bacillus licheniformis IM 1307: production, isolation and structural evaluation by NMR and mass spectrometry. J Antibiotics 52:363–373
Vanittanakom N, Loeffler W, Koch U, Jung G (1986) Fengycin, a novel antifungal lipopeptide antibiotic produced by Bacillus subtilis F-29–3. J Antibiot 39:888–901
Schneider J, Taraz K, Budzikiewicz H, Deleu M, Thonart P, Jacques P (1999) The structure of two fengycins from Bacillus subtilis S499. Z Naturforsch 54:859–866
Naruse N, Tenmyo O, Kobaru S, Kamei H, Miyaki T, Konishi M, Oki T (1990) Pumilacidin, a complex of new antiviral antibiotics. J Antibiot 43:267–280
Hasumi K, Takizawa K, Takahashi F, Park JJ, Endo A (1995) Inhibition of acyl-CoA: cholesterol acytransferase by isohalobacillin, a complex o novel cyclic acypeptides produced by Bacillus sp. A1238. J Antibiot 48:1419–1425
Besson F, Peypoux F, Quentin MJ, Michel G (1984) Action of antifungal peptidolipids from Bacillus subtilis on the cell membrane of Saccharomyces cerevisae. J Antibiot 37:172–177
Latoud C, Peypoux F, Michel G (1987) Action of iturin A, an antifungal antibiotic from Bacillus subtilis on the yeast Saccharomyces cerevisae. Modifications of membrane permeability and lipid composition. J Antibiot 40:1588–1595
Mikkola R, Kolari M, Andersson MA, Helin J, Salkinoja-Salonen MS (2000) Toxic lactonic lipopeptide from food poisoning isolates of Bacillus licheniformis. Eur J Biochem 267:4068–4074
Vaara M (1992) Agents that increase the permeability of the outer membrane. Microbiol Rev 56:395–411
Hancock REW (1997) The bacterial outer membrane as drug barrier. Trends Microbiol 5:37–42
Poole K (2000) Efflux mediated resistance to fluoroquinolones in Gram-negative bacteria. Antimicrob Agents Chemother 44:2233–2241
Poole K (2001) Multidrug efflux pumps and antimicrobial resistance in Pseudomonas aeruginosa and related organisms. J Mol Microbiol Biotech 3:255–264
Hagelin G, Oulie I, Raknes A, Udheim K, Clausen OG (2004) Preparative high-performance liquid chromatographic separation and analysis of the Maltacin complex—a family of cyclic peptide antibiotics from Bacillus subtilis. J Chromatogr B 811:243–251
Hagelin G (2005a) Structure investigation of maltacine B1a, B1b, B2a and B2b: cyclic peptide lactones of the maltacine complex from Bacillus subtilis. J Mass Spectrom 40:527–538
Hagelin G (2005b) Structure investigation of maltacine C1a, C1b, C2a and C2b—cyclic peptide lactones of the maltacine complex from Bacillus subtilis. J Mass Spectrom 40:1276–1286
Hagelin G (2005c) Structure investigation of maltacine D1a, D1b and D1c—cyclic peptide lactones of the maltacine complex from Bacillus subtilis. J Mass Spectrom 40:1287–1299
Thimon L, Peypoux F, Maget-Dana R, Roux B, Michel G (1992) Interactions of bioactive lipopeptides, iturin A and surfactin from Bacillus subtilis. Biotechnol Appl Biochem 16:144–151
Koumoutsi A, Chen X-H, Henne A, Liesegang H, Hitzeroth G, Franke P, Vater J, Borriss R (2004) Structural and functional characterization of gene clusters directing nonribosomal synthesis of bioactive cyclic lipopeptides in Bacillus amyloliquefaciens strain FZB42. J Bacteriol 186:1084–1096
Rouser G, Fleischer S, Yamamoto A (1970) Two dimensional thin layer chromatographic separation of polar lipids and determination of phospholipids by phosphorous analysis of spots. Lipids 5:494–496
Maget-Dana R, Peypoux F (1994) Iturins, a special class of pore-forming lipopeptides: biological and physicochemical properties. Toxicology 87:151–174
Lichtemberg D, Robson RJ, Dennis EA (1983) Solubilization of phospholipid detergents. Structural and kinetic aspects. Biochim Biophys Acta 737:285–304
Smith R, Tanford C (1972) The critical micelle concentration of L-α-palmitoylphosphatidylcholine in water and in water/methanol. J Mol Biol 67:75–83
Williams BH, Hathout Y, Fenselau C (2002) Structural characterization of lipopeptide biomarkers isolated from Bacillus globigii. J Mass Spectrom 37:259–264
Bode HB, Bethe B, Höfs R, Zeek A (2002) Big effects from small changes: possible ways to explore nature’s chemical diversity. Chem Bio Chem 3:619–627
Wessjohann LA, Ruijter E, Garcia-Rivera D, Brandt W (2005) What can a chemist learn from nature’s macrocycles? A brief conceptual view. Mol Divers 3:171–186
Biemann K (1990) Nomenclature for peptide fragment ions (positive ions). Methods Enzymol 193:886–887
Madonna AJ, Voorhees KJ, Taranenko NI, Laiko VV, Doroshenko VM (2003) Detection of cyclic lipopeptide biomarkers from Bacillus species using atmospheric pressure matrix-assisted laser desorbtion/ionization mass spectrometry. Anal Chem 75:1628–1637
Pabel CT, Vater J, Wilde C, Franke P, Hofemeister J, Adler, Bringmann G, Hacker J, Hentschel U (2003) Antimicrobial activities and mass-assisted laser desorbtion/ionization mass spectrometry of Bacillus isolates from the marine sponge Aplysina aerophoba. Mar Biotechnol 5:424–434
Wang J, Liu J, Wang X, Yao J, Yu Z (2004) Application of electrospray ionization mass spectrometry in rapid typing of fengycin homologues produced by Bacillus subtilis. Lett Appl Microbiol 39:98–102
Akpa E, Jacques P, Wathelet B, Paquot M, Fuchs R, Budzikiewicz H, Thonart P (2001) Influence of culture conditions on lipopeptide production by Bacillus subtilis. Appl Biochem Biotechnol 91–93:551–561
Frank G, Torsten HS, Barvel K, Peter F, Joachim V (1999) Rapid typing of Bacillus subtilis strains by the secondary metabolites using matrix—assisted laser desorbtion/ionization mass spectrometry of intact cells. Rapid Commun Mass Spectrom 13:943–949
Opsteen JA, Cornelissen J, van Hest JCM (2004) Block copolymer vesicles. Pure Appl Chem 76:1309–1319
Rodriguez-Hernandez J, Babin J, Zappone B, Lecommandoux S (2005) Preparation of shell cross-linked nano-objects from hybrid peptide block copolymers. Biomacromolecules 6:2213–2220
Discher DE, Eisenberg A (2002) Polymer vesicles. Science 297:967–973
Vary PS (1994) Prime time for Bacillus megaterium. Microbiology 140:1001–1013
Vary PS, Biedendieck R, Fuerch T, Meinhardt F, Rohde M, Deckwer W-D, Jahn D (2007) Bacillus megaterium—from simple soil bacterium to industrial protein production host. Appl Microbiol Biotechnol 76:957–967
Acknowledgements
The authors are grateful to Fernanda Manso Prado, Izaura Nobuko Toma, and Sirlei Mendes de Oliveira for technical assistance. Financial support was from Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP, grant No 98/11480-1) and from Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, grant No 473145/2006-5).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Pueyo, M.T., Bloch, C., Carmona-Ribeiro, A.M. et al. Lipopeptides Produced by a Soil Bacillus Megaterium Strain. Microb Ecol 57, 367–378 (2009). https://doi.org/10.1007/s00248-008-9464-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-008-9464-x