Abstract
Endophytic streptomycetes have been isolated and characterized from several species of Nothofagus and other plants growing in the southern reaches of Patagonia. No endophytic streptomycete was obtained from any plant species studied in Northern Patagonia. However, from Southern Patagonia, biologically active Streptomyces spp. from several plant species were isolated. Each isolate, as studied by scanning electron microscopy (SEM), has small hyphae, some produce typical barrel-shaped spores in culture and each has some unique hyphal surface structures. Interestingly, although none has any detectable antibacterial killing properties, each has demonstrable killing activity against one or more pathogenic fungi including representative plant pathogenic organisms such as Phytophthora erythroseptica, Pythium ultimum, Sclerotinia sclerotiorum, Mycosphaerella fijiensis, and Rhizoctonia solani. The 16S rDNA sequences of the isolates were distinct from all other genetic accessions of Streptomyces in GenBank. However, isolate C-2 from Chiliotrichum diffusum (Compositae) is identical, in all respects, to isolate C-4 obtained from Misodendrum punctulatum (Loranthaceae). These results confirm that endophytic streptomycetes represent a novel source of biologically active microorganisms.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
As a group, the streptomycetes provide nearly 80% of all of the world's antibiotics [1]. A search reveals that all of these organisms have their origins in the soil. The advent of drug resistance in most bacterial pathogens and the current increase in the number of fungal infections has caused a resurgence of interest in finding other reserves of biologically active compounds from the streptomycetes [12]. One biologically important niche that has been overlooked as a source of novel are streptomycetes within the tissues of higher plants. Microbes that take up residence in the tissues of plants are known as endophytes [2]. Earlier reports on this subject did not include this group of organisms because at the time of publication the number of examples of endophytic streptomycetes was exceedingly limited [2, 6, 10, 13–15]. By definition, these organisms cause no apparent damage to the plants that they inhabit [2, 18, 19]. Presumably, the simplest biological arrangement between these organisms is that the plant provides nutrition for the microbe and the microbe provides some form of protection for the plant [2, 18, 19]. Moreover, because the endophytic streptomycete is associated with a eukaryotic organism, the possibilities of it making products that are not toxic to its host organism seem more likely. Thus, one of the major concerns in drug discovery, relating to the toxicity of a drug candidate, may be averted by dealing with endophytic streptomycetes and their biologically active products.
Although practical needs may drive the search for novel streptomycetes, the mere fact that they seem to have been overlooked as playing some role in the biology of the forest ecosystem is of great concern in itself. Only in the past few years have reports begun to emerge concerning the topic of endophytic streptomycetes and their role in the ecology of the plants and the bioactive products that they produce [3–6, 8]. Many of the newly isolated endophytic streptomycetes possess a unique 16S rDNA sequence, many make totally novel biologically active products, many possess unique hyphal structures, whereas others have interesting and totally novel cultural characteristics [4–6, 8].
An expanded search for other endophytic streptomycetes is underway. One area that may possess such organisms is that part of the world representing what once was the ancient landscape of Gondwanaland [17, 18]. Even today, the fossil record in portions of this early continent, which includes Antarctica, has preserved the fragments and impressions of structures that once represented the flora of this region [20]. Plant families such as Fagaceae, Eucryphiceae, and Podocarpaceae are considered representatives of Gondwanaland plants. Members of these plant families still exist in places such as South America, Australia, Madagascar, and Africa. A rationale has been proposed that modern plant relatives of the ancient landscape plant families are likely to be excellent specimens to begin a search for endophytic microbes [17, 18]. This is based on the premise that ancient plants may have had many more millions of years to establish a plant–microbe relationship than more recently appearing plant families. Thus, the chances of finding such endophytic microbes that have established symbiotic relationships with their respective hosts would be enhanced from plants in these areas of the world.
Today, some areas that were once a part of Gondwanaland have been considered to be “hot spots” of biodiversity [11]. Thus, is it possible that there are previously undiscovered streptomycetes in Gondwanaland plant families and if so, how do they compare with organisms already known from other biological niches? Initially, collection efforts were concentrated in a large representative stretch of Patagonia from the northern areas to the south (South America). Ultimately, genetically unique endophytic streptomycetes having a range of biological activities were isolated from several Patagonian plants and partially characterized. The majority of them were from Nothofagus spp., which are representatives of the Gondwanaland family, Fagaceae. This report describes the details on the isolation and bioactivities of these unique endophytic streptomycetes from Patagonia.
Materials and Methods
Sampling Plants
An extensive collection of various plant materials was set up in November 2001, in an area of Patagonia extending from 41° to 51°S latitude. The rationale for selecting this area is that it represents an extant biological/geological remnant of Gondwanaland. As such, plant families associated with this ancient landmass are related or identical to the ones living there anciently. Presumably, the long time span between then and now would have allowed for microbe–plant interactions to have been established. Soil associated streptomycetes usually have a plethora of biological activities associated with them [1]. Thus, because they have never been isolated and studied, the endophytic streptomycetes of Patagonian plants were the primary focus of this project with the idea that they could be found and they may have promising biological activities.
Usually, the specimens were 15–20 cm long and 0.5 cm in diameter taken from the tips of stems. Some representative tree samples were collected in the area of 41°32′52′′S and 72°35′39′′W, and they were as follows (common name followed by scientific name): Manio, Podocarpus nubigena; Ulmo, Eucryphia cordifolia; Canelo, Drymis winteri; Lahuan, Pilgerodendron uviferum; Taique, Desfontainia spinosa; Pitra, Myrceugenia exsucca; Tepa, Laureliopsis phillipiana; Luma, Amomyrtus luma; Voqui, Lardizabala biternata; Tineo, Weinmannia trichosperma; Alerce, Lahuan lahuen; Palo Muerto, Aextoxicon punctatum. Another sampling of tree and shrub samples was done at the location 50°58′25′′S and 72°52′27′′W, near and around Torres del Paine, and the collected plants were as follows: mistletoe (growing on Nothofagus pumilio), Misodendron punctulatum; Lenga, N. pumilio; Nirre, Nothofagus antarctica; Coigue, Nothofagus betuloides; Romerillo, Chiliotrichum diffusum; Matanegra, Juniellia tridens; Calafate, Berberis buxifolia; Lena dura, Maytenus magellanicum; Notro, Empothrium coccineum. At least four limb samples were obtained from each plant and, in most cases, only one plant was sampled.
Isolating Actinomycetes
Each stem specimen was thoroughly treated with 95% ethanol and flamed until the alcohol was eliminated and the bark removed with a sterilized sharp blade. The inner pieces of the stem, containing the cambium, phloem, and xylem tissues, were plated on different types of solid media to select for actinomycetes [4]. The most notable one was the glycerol/arginine medium (Difco Catalog) at 1/10 strength containing cycloheximide at a concentration of 100 mg/L. As colonies began to grow, they were observed with a binocular microscope and the putative streptomycetes were separated from other microbes by virtue of their morphological differences.
Putative colonies of streptomycetes were optimally grown in nutrient broth (NB) and nutrient agar (NA) (Difco Catalog). Ultimately, to isolate pure cultures, individual cultures were picked with a sterile toothpick and grown in starter cultures of 5 mL NB for 2–3 days before plating. Then, 1:1, 1:10, and 1:1000 concentrations were spread onto plates containing NA. This concentration gradient was adjusted according to the different growth patterns of cultures until single colonies were easily obtained. Many individual streptomycete colonies were similar in color, shape, and size; however, hyphal length and structure, as observed with light and stereobinocular microscopes, allowed them to be segregated into distinct isolates. The process of picking colonies from solid media and growing them in liquid media was repeated until pure cultures were obtained. Ultimately, this allowed for the individual isolates to be examined by scanning electron microscope (SEM) techniques and for their bioactivity as well as their morphological characteristics.
After incubation for 10 days or more at 28°C, individual colonies were removed with a sterile fine-tipped needle and transferred onto NA. Each was photographed after 2–3 weeks of growth. Spore formation of each streptomycete was encouraged via the proper manipulation of growth conditions. The plates were continuously monitored for spore formation by stereo and light microscopy. Small agar pieces, containing hyphae and spores, were stored in 15% glycerol in water (v/v) at −80°C. Cultures were numbered and placed in the permanent microbial culture collection at Montana State University.
Scanning Electron Microscopy
Once microbial isolates were obtained that outwardly resembled actinomycetes (Streptomyces sp.), they were initially characterized by photographing and recording their cultural characteristics and given a designated number. Then, SEM was used to obtain images of selected isolates. The microbe being studied with a high-vacuum SEM was placed into 2% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.2–7.4) with Triton X, a wetting agent, aspirated for 5 min and left overnight [4]. Next, the fixed agar pieces supporting microbial growth were placed into #1 Whatman filter paper packets. The packets were made by folding the filter paper over a piece of cork (1.5 × 1.5 cm). The packets were tied with cotton string, and two removable split shot sinkers (ca. 3.25 g each) were attached next to the packets to hold them under the surface of the dehydrating solutions and liquid carbon dioxide during critical point drying. The next day, they were washed six times each for 15 min in changes of water buffer, followed by a 15-min change in 10% ethanol, a 15-min change in 30% ethanol, a 15-min change in 50% ethanol, and five 15-min changes in 70% ethanol and then left overnight or longer in 70% ethanol. They were then rinsed for 15 min in 95% ethanol and then three 15-min changes in 100% ethanol, followed by three successive 15-min changes in acetone. The dehydration process was carried out slowly to discourage the processes of hyphal shriveling that may occur during rapid dehydration. Ultimately, for SEM the microbial material was critically point dried, gold sputter coated, and images were recorded with an XL30 ESEM FEG in the high-vacuum mode using the Everhart–Thornley detector [4].
Sequence Analysis
A single colony of each Streptomyces sp. to be analyzed was collected from an agar plate and transferred to 5.0 mL of liquid NB medium in a 25-mL flask. The culture was shaken at 150 rpm, 24°C for 5 days. The culture (2 mL) was harvested by centrifugation (13,000 rpm, 1 min in an IEC Micromax RF centrifuge). Next, the pellet was washed twice by resuspending it with 1 mL double-distilled sterile water and centrifuged; finally, the culture was resuspended in 1 mL sterile ddH2O.
Ribosomal 16S DNA amplification was performed via polymerase chain reaction (PCR). The washed culture (2 μL) was added to 18 μL of amplification mixture containing 1 μL (10 nmol) of each primer, 1 μL (10 mM dNTPs mix), 2 μL of 10× reaction buffer (JMR 420; Fermentas Inc., USA), and 1 μL of 15 mM MgCl2, 0.2 μL 5 unit/μL Taq polymerase (super-therm; Fermentas Inc.), and 12.8 μL ddH2O (UltraPure PCR grade; Fisher Biotec, Australia). After the initial denaturation of 5 min at 96°C, 35 cycles of amplification were performed: 45 s at 95°C denaturation, 45 s at 50°C annealing and 1 min at 72°C polymerization. These cycles were followed by a final 5-min elongation cycle at 72°C. The PCR reaction was performed in a Biometra personal thermocycler (Biometra, Germany). The PCR products were purified by UltraClean™ PCR purification Kit (MoBio laboratory Inc., California, USA) and directly sequenced by BigDye Terminator Cycle Sequencing Kit from ABI (Hay Laboratories Ltd., Rehovot, Israel). The primers used in the PCR reaction were BSF8/20, BSR1114/16, and BSR1525/21 (Table 1). The sequence data from each of the streptomycetes in this study was compared to those at GenBank by using the BLAST software (blastn) on the National Center Biotechnology Information (NCBI) web site (http://www.ncbi.nlm.nih.gov/).
Bioassays of the Streptomycetes
Each streptomycete isolate was grown as a ca. 2-1cm colony for 10–14 days on petri plates containing NA. Then small (3 mm3) plugs of agar containing freshly grown cultures of Pythium ultimum, Sclerotinia sclerotiorum, Rhizoctonia solani, Phytophthora erythroseptica, Fusarium solani, Mycosphaerella fijiensis, and Aspergillus fumigatus were placed about 1.0 cm from the edge of the streptomycete colony. Yeasts and bacteria, on the other hand, were streaked about 1–1.5 cm from the edge of the colony being tested and these included Bacillus subtilus, Escherichia coli, Saccharomyces cereviseae, and Candida albicans. The plant infecting fungi were chosen on the basis that they represent the complete spectrum of the various families of the fungal class—Fungi Imperfecti. Furthermore, the assay bacteria have both a Gram-positive and a Gram-negative organism represented. At least two yeastlike fungi were also used as bioassay test microbes. Growth of the test organisms was evaluated after 24, 48, and 72 h, and recorded as growth, inhibition, and no growth as compared to a control plate containing no streptomycete colony. Finally, a sample of each test organism was taken from the treatment plate and placed on a plate of potato dextrose agar (PDA) to determine if the test organism had been killed after having been exposed to it for 72 h. The tests were repeated at least thrice. The plant associated test organisms were obtained from Dr. Don Mathre, Department of Plant Sciences, MSU, and the bacteria were from the Department of Microbiology at MSU. The culture of P. erythroseptica was supplied by Dr. John Menge, UC Riverside, and M. fijiensis from the CBS in Holland.
Results
Patagonian Endophytic Streptomycetes
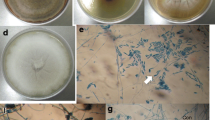
Fungal and bacterial endophytes were recovered from each Patagonian plant specimen, and details on some of these observations have been made in other reports [16–18]. Most interestingly, no streptomycete isolate appeared from any of the plants that were sampled in the northern reaches of Patagonia, i.e., at 41°S latitude. However, six streptomycete isolates were obtained from different plant species at the southern end of Patagonia at 51°S latitude. Each isolate appeared only once from the plant that was sampled and from no other plant in the collection. They were labeled as C-1 through C-6 (Table 2). Each possessed hyphae of a small diameter, i.e., 0.5–0.8 μm, and the SEM data yielded some information about spore formation or potential spore formation as being through hyphal fragmentation (Fig. 1a–f). Sequence analyses of 16S rDNA indicated that each isolate was unique although some were related, but not identical, to other streptomycete sequences reported in GenBank (Table 2). Each isolate possessed some biological activity against the test fungal organisms and none was lethal to either the Gram-positive or the Gram-negative bacteria (Table 3). Most importantly, each organism tested had one or more activities against the filamentous fungi or yeast isolates, but none was active against F. solani (Table 3). Each of the endophytic Streptomyces spp. recovered from southern Patagonia is specifically reported below along with its sequence relationship to other streptomycetes reported in the GenBank repository.
Scanning electron micrographs of the Patagonian streptomycetes. (a, Upper left) C-1 shows early spore formation; (b, upper right) C-2 shows early spore formation and young hyphal cells; (c, center left) C-3 shows distinct segmentation of hyphal cells into spores; (d, center right) C-4 shows young hyphae; (e, lower left) C-5 shows ropy strands of hyphae; (lower right) C-6 shows ropy net-like hyphae. Compare C-2 with the images in C-4.
Streptomyces sp. Isolate C-1.
This organism was recovered from one of the southern beech species—N. pumilio. It produced whitish, erumpent, sectored, fuzzy colonies on NA. Sequence analysis of its 16S rDNA showed 99% identity to Streptomyces seoulensis (1464/1477 bp) and 98% identity to S. galilaeus (1458/1477 bp). The SEMs of the hypha of this organism showed a unique relatively rough, almost particulated hyphal surface with some indication of spore formation arising from segmenting hyphae (Fig. 1a). This organism was the least active among the six isolated from Patagonian tree species because it was only lethal to S. sclerotiorum (Table 3).
Streptomyces sp. Isolate C-2.
This streptomycete was isolated from Chilotrichum diffisum, a shrubby composite plant growing in open woodlands of Patagonia. On NA, it produced grayish colonies having a smooth whitish perimeter with a pitted central portion in each colony. It possessed 99% sequence identity to S. seoulensis (1467/1477) and 98% identity to S. galilaeus (1461/1477). This sequence is very similar to C-1 and differs in only three nucleotide positions. Interestingly, the sequence is perfectly identical to isolate C-4. The SEM shows a relatively smooth hyphal surface with some tendency toward segmentation leading to sporulation (Fig. 1b). Also, the images of the hyphae appear identical to those of C-4 (Fig. 1d). This organism caused lethality to P. ultimum, S. sclerotiorum, R. solani, S. cereviseae, and P. erythroseptica, basically having the same bioactivity as C-4 (Table 3).
Streptomyces sp. Isolate C-3.
This microbe was obtained as an endophyte from another southern beech, N. betuloides. On NA it produced typical fuzzy whitish colonies, each with nondescript fuzzy overgrowths scattered on the culture surface. The 16S rDNA sequence has 98% identity to Streptomyces sp. NRRL 5799 (998/1013) (AJ 391814) and a slightly lower similarity to S. antiquitatis with 968/987 bp identity. The organism has very definite segmentation of its hyphae into spore forms. The SEMs reveal that the hyphal surface is almost echinulated (Fig. 1c). The biological activity of C-3 shows lethality to P. ultimum, S. sclerotiorum, P. erythroseptica, S. cereviseae, M. fijiensis, and R. solani (Table 3).
Streptomyces sp. Isolate C-4.
This organism was recovered from Misodendrum punctulatum (mistletoe) that was growing in the crown of N. pumilio. The colony description is identical to that of C-2 (above). Sequence analysis of its 16S rDNA shows 99% identity to S. seoulensis (1464/1474 bp) and 98% identity to S. galilaeus (1458/1474 bp). The SEMs of this organism indicate that it has a relatively smooth hyphal surface with some indication of spore formation arising from segmenting hyphae (Fig. 1d). This organism was active against some of the test organisms in causing lethality to P. ultimum, S. sclerotiorum, R. solani, and P. erythroseptica, and it had a bioactivity profile equal to that of C-2 (above) (Table 3). This organism was not isolated from N. pumilio, the host of M. punctulatum that had been gathered and studied at several other locations in Patagonia. Although it has high 16S rDNA similarity to C-1 and identity to C-2 (above), it shows hyphal structural characteristics that are identical to C-2 (Fig. 1).
Streptomyces sp. Isolate C-5.
This organism was also isolated from N. betuloides. Colonies on NA possessed smooth margins with raised whitish fuzzy growth toward the center of the colony. Its 16S rDNA sequence is 99% similar to Streptomyces NRRL 5799 with 1017/1022 bp being identical. It is 98% similar to Actinomycetales bacterium HPA 72 with 1011/1026 bp having identity. This isolate is 98% similar to C-3 with 1021/1035 bp being identical. The SEMs of this organism show clumping of the hyphae into ropelike strands (Fig. 1e). The hyphal surfaces appear smooth and there is only a hint of segmentation of the hyphae into spores. The organism only caused death of P. ultimum and M. fijiensis in the bioassay tests (Table 3).
Streptomyces sp. Isolate C-6.
This endophytic streptomycete was also isolated from N. betuloides. On NA the colonies of this streptomycete possessed white margins having fuzzy brownish growth toward the colony center. The 16S rDNA analysis showed 99% identity to Streptomyces NRRL 5799 with 1449/1457 bp identity. In addition, it was 98% similar to S. intermedius (1453/1472 bp). Furthermore, it has 98% identity to C-3 and 99% to C-5. The hyphae of the organism seem to clump into ropelike strands having an overall uneven appearance (Fig. 1f). The hyphae show hints of segmenting into spores. The biological activity was the most extensive of all of the Patagonian streptomycetes with killing of the following test organisms being observed: P. ultimum, S. sclerotiorum, P. erythroseptica, and R. solani. A. fumigatus, S. cereviseae, and C. albicans were each almost totally inhibited by C-6 but not killed (Table 3).
Overall Phylogenetic Analysis of the Endophytes
Sequences obtained from this study and GenBank were aligned by using the DNA alignment program ClustalW (http://www.ebi.ac.uk/clustalw/). Phylogenetic analyses were performed with TreeViewX V.5.0 software developed by Roderic D.M. (http://darwin.zoology.gla.ac.uk/~rpage/treeviewx/). S. intermedius (AB184277), S. seoulensis (Z71365), S. galilaeus (AB122719), S. scabiei (AB184828), S. kanamyceticus (AB217602), Streptomyces sp. NRRL (AJ391814), and Streptomyces sp. (NRRL 5799) was each used in order to compare and locate the C1–C6 sequences to known streptomyces sequences in the GeneBank (Fig. 2). The Streptomyces sp. NRRL 5799 sequence was the closest to C3 and C5. However, the C6 sequence was closer to C3 and C5 than to C1, C2, and C4. Both isolates C4 and C2 have a very high similarity and we consider them to be the same organism. It is obvious from the phylogram that the streptomyces collected in this study can be divided into two major groups. The first group is (C3, C5, and C6) with closer similarity to S. intermedius, a soil bacterium. The second group is (C1, C2, and C4) with closer similarity to S. seoulensis, an actinomycete strain isolated from a Korean soil [7].
Discussion
Six streptomycete isolates were obtained as endophytes from various plants found in southern Patagonia. They were confirmed as authentic streptomycetes by virtue of their hyphal characteristics, spore forms, and—most importantly—their partial 16S rDNA sequence similarities to other sequences in GenBank (Fig. 2). Genetically, structurally, and on the basis of biological activities, isolates C-2 and C-4 were considered as identical even though they had been isolated from different plants in different locations (Tables 2 and 3) (Fig. 1a–f). In addition, phylogenic analysis showed that C-3 and C-5 are almost identical and close to C-6. On the other hand, C-1 is closest to C-2 and C-4, while more distant to C-3, C-5, and C-6 (Fig. 2).
It is not known why it was possible to only obtain endophytic streptomycetes from southern Patagonia and not from the north. It may be the case that the southern areas, which have vast continuous forests of Nothofagus spp., better allow for a more suitable condition to maintain a microbe–plant relationship as would be true in most plant pathogen–host plant interactions. Thus, in a situation of a continuous forest with primarily one species, the spores of the endophyte will have ready access to nearby plants to invade and develop within the host. Ultimately, this will allow a continuation of the microbe–plant relationship. In the northern areas, for instance, around Porto Monte, Chile, the Nothofagus spp. are only sparely scattered in the mixed forests. This may preclude the establishment or a continuation of the microbe/plant interaction. On the other hand, the results could simply be related to all matters related to technique and plant sampling methods. The observation is worthy of being pursued given the important biological activities associated with these streptomycetous endophytes (Table 3).
The differential antimicrobial activity of these Patagonian streptomycetes is of some interest. Although none kills common Gram-negative and Gram-positive bacteria and none kills F. solani, each has some antifungal activity toward major plant pathogens (Table 3). This observation is almost contrary to what is commonly observed with soilborne streptomycetes being antibacterial and not necessarily overwhelmingly antifungal [1]. This observation is also a major deviation from the general antimicrobial activities that have been noted with other endophytic streptomycetes studied in this laboratory especially those from snakevine [4–6]. Most unusual and yet striking, in this series of organisms, is the antifungal specificity exhibited to certain target organisms. For instance, C-1 only kills S. sclerotiorum and has no killing effect on any other microorganism (Table 3). To the other extreme, C-6 either inhibits or kills all test fungal organisms except M. fijiensis (Table 3). In between are all of the other streptomycetes having killing or inhibition effects on varying numbers of the test fungi. This antifungal activity has both academic as well as practical interest because it provides tools to potentially differentiate between pathogenic fungi for selection purposes and for disease control.
It is also noteworthy that these isolates came exclusively from Nothofagus (a Gondwanaland plant genus), a mistletoe on Nothofagus, a composite C. diffisum. Attempts to isolate streptomycetes from other tree and shrub species in Northern Patagonia did not meet with success. At this point, this study is too limited to make general conclusions about the relative distribution of endophytic streptomycetes in Patagonia, but at least there is some indication that the initial hypothesis deserves further and more intense study. Furthermore, the isolation and characterization of the biologically substances produced by these endophytic streptomycetes will provide a greater understanding of the diverse nature of the outcome in the bioassay tests (Table 3).
One of the most biologically interesting discoveries is that both C-5 and C-3 can make one or more products that kill M. fijiensis (Table 3). This fungus is the cause of Black Sigatoka disease in bananas and it appears that the streptomycetous endophyte, its culture broth, or its purified antimycotic(s) should be considered as a means of treating this disease via a biocontrol method. The same argument applies for the other bioactivities observed in these endophytes (Table 3). It appears that some of these organisms, on the basis of their biological spectrum, may be making different antimycotics. For instance, compare the activity of C-1 to each of the other organisms. The major exception would seem to be the identical biological activities of C-2 and C-4 because they are probably the same microbe, and thus probably producing the same antimicrobial compounds (Table 3, Fig 2). Another interesting antifungal streptomyete is C-6, whose biological activity includes the inhibition of A. fumigatus and C. albicans. Both of these organisms are considered lethal human parasites.
The exact biological role of these endophytic streptomycetes in their respective hosts can only be a matter of speculation at this point. It seems that some measure of the endophytic nature of each of these Patagonian streptomycetes can be surmised on the basis of their antifungal activity. Without exception, each makes products that diffuse into the agar base causing death to one or more fungal pathogens (Table 3). Basically, the plant pathogens used in this bioassay test series represent pathogen genera that most likely exist in southern Patagonia. For instance, Pythium, Sclerotinia, and Phytophthora are virtually ubiquitous pathogens and constantly challenge the existence of the plant species all over our planet. Perhaps, these endophytic Streptomyces spp. help forestall disease and death that may threaten the well being of these Patagonian plant species.
References
Arai, T (1976) Actinomycetes: The Boundary Microorganisms. Toppan, Singapore
Bacon, C, White, JF (2000) Microbial Endophytes. Marcel Dekker, New York
Bieber, B, Nuske, J, Ritzau, M, Grafe, U (1998) Alnumycin a new naphthaquinone antibiotic produced by an endophytic Streptomyces sp. J Antibiot 51: 381–382
Castillo, U, Giles, S, Browne, L, Strobel, GA, Hess, WM, Hanks, J, Reay, D (2005) Scanning electron microscopy of some endophytic streptomycetes in snakevine—Kennedia nigriscans. J Scanning Microsc 27: 305–311
Castillo, U, Harper, JK, Strobel, GA, Sears, J, Alesi, K, Ford, E, Lin, J, Hunter, M, Maranta, M, Ge, H, Yaver, D, Jensen, JB, Porter, H, Robison, R, Millar, D, Hess, WM, Condron, M, Teplow, D (2003) Kakadumycins, novel antibiotics from Streptomyces sp. NRRL 30566, an endophyte of Grevillea pteridifolia. FEMS Lett 224: 183–190
Castillo, U, Strobel, GA, Mullenberg, K, Condron, MM, Teplow, D, Folgiano, V, Gallo, M, Ferracane, R, Mannina, L, Viel, S, Codde, M, Robison, R, Porter, H, Jensen, J (2006) Munumbicins E-4 and E-5: novel broad spectrum antibiotics from Streptomyces NRRL 3052. FEMS Lett 255: 296–300
Chun, J, Youn, HD, Yim, YI, Lee, H, Kim, MY, Hah, YC, Kang, SO (1997) Streptomyces seoulensis sp. nov. Int J Syst Bacteriol 47: 492–498
Ezra, D, Castillo, U, Strobel, GA, Hess, WM, Porter, H, Jensen, J, Condron, M, Teplow, D, Sears, J, Maranta, M, Hunter, M, Weber, B, Yaver, D (2004) Coronamycins, peptide antibiotics produced by a verticillated Streptomyces sp. (MSU-2110) endophytic on Monstera sp. Microbiology 150: 785–793
Hain, T, Ward-Rainey, R, Kroppenstedt, E, Stackebrandt, E, Rainey, FA (1997) Discrimination of Streptomyces albidoflavus strains based on the size and number of 16S–23S Ribosomal DNA intergenic spacers. Int J Syst Bacteriol 47: 202–206
Kunoh, H (2002) Endophytic actinomycetes: attractive biocontrol agents. J Gen Plant Pathol 68: 249–252
Mittermeier, RA, Myers, N, Gil, PR, Mittermeier, CG (1999) Hotspots: Earth's Biologically Richest and Most Endangered Ecoregions. CEMEX Conservation International, Washington DC
NIAID Global Health Research Plan for HIV/AIDS, Malaria and Tuberculosis (2001) U.S. Department of Health and Human Services, MD, Bethesda
Redlin SC, Carris LM (Ed.) (1996) Endophytic Fungi in Grasses and Woody Plants. APS Press, St. Paul
Sardi, P, Saracchi, M, Quaroni, S, Petrolini, B, Borgonovi, GE, Merli, S (1992) The isolation of endophytic streptomyces strains from surface-sterilized roots. Appl Environ Microbiol 58: 2691–2693
Shimizu, M, Nakagawa, Y, Sato, Y, Furumai, T, Igarashi, Y, Onaka, H, Yoshida, R, Kunoh, H (2000) Studies on endophytic actinomycetes(1) Streptomyces sp. isolated from rhododendron and its antifungal activity. J Gen Plant Pathol 66: 360–366
Stinson, M, Ezra, D, Hess, WM, Sears, J, Strobel, GA (2003) An endophytic Gliocladium sp. of Eucryphia cordifolia producing selective volatile antimicrobial compounds. Plant Sci 165: 913–922
Strobel, GA (2002) Rainforest endophytes and bioactive products. Crit Rev Biotechnol 22: 315–333
Strobel, GA, Daisy, B (2003) Bioprospecting for microbial endophytes and their natural products. Microbiol Mol Biol Rev 67: 491–502
Tan, RX, Zou, WX (2000) Endophytes: a rich source of functional metabolites. Nat Prod Rep 18: 448–459
Veblen, TT, Hill, RS, Read, J (Ed.) (1996) The Ecology and Biogeography of Nothofagus Forests. Yale Univ. Press, New Haven, CT
Wilmotte, A, Van der Auwera, G, De Wachter, R (1993) Structure of the 16S ribosomal RNA of the thermophilic cyanobacterium Chlorogloeopsis HTF (Mastigocladus laminosus HTF) strain PCC7518, and phylogenetic analysis. FEBS Lett 317: 96–100
Acknowledgments
The authors appreciate the support of a Vaadia-BARD grant No FI-321-2001 to David Ezra. Financial support was also supplied by the NSF and the Montana Department of Commerce-Board of Research and Commercialization Technology. Help from Ms. Gladys Garay and Mr. Oscar Guineo of Torres del Paine, as well as Gary James and Jano Igor of the Alerce Lodge in Northern Patagonia, in collecting and identifying plant specimens, is greatly appreciated. The Montana Agricultural Experiment Station also provided financial support.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Castillo, U.F., Browne, L., Strobel, G. et al. Biologically Active Endophytic Streptomycetes from Nothofagus spp. and Other Plants in Patagonia. Microb Ecol 53, 12–19 (2007). https://doi.org/10.1007/s00248-006-9129-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-006-9129-6